Summary
hBRE1/RNF20 is the major E3 ubiquitin ligase for histone H2B. RNF20 depletion causes a global reduction of monoubiquitylated H2B (H2Bub) levels and augments the expression of growth promoting, pro-oncogenic genes. Those genes reside preferentially in compact chromatin, and are inefficiently transcribed under basal conditions. We now report that RNF20, presumably via H2Bub, represses selectively those genes by interfering with chromatin recruitment of TFIIS, a factor capable of relieving stalled RNA polymerase II. RNF20 inhibits the interaction between TFIIS and the PAF1 complex and hinders transcriptional elongation. TFIIS ablation selectively abolishes the upregulation of those genes upon RNF20 depletion, and attenuates the cellular response to EGF. Consistent with its positive role in transcription of pro-oncogenic genes, TFIIS expression is elevated in various human tumors. Our findings provide a molecular mechanism for selective gene repression by RNF20, and position TFIIS as a key target of RNF20's tumor suppressor activity.
Introduction
The eukaryotic DNA is packaged into a chromatin structure consisting of repeating nucleosomes, comprising 146 base pairs of DNA wrapped around an octamer of core histone proteins. The histone tails are subjected to several post-translational modifications (PTM), which are crucial in regulation of transcription, DNA repair and other chromatin-associated processes (Berger, 2007; Jenuwein and Allis, 2001; Weake and Workman, 2008). One such PTM is the monoubiquitylation of histone H2B on lysine 123 in yeast, or lysine 120 in mammals, carried out by the yeast E3 ligase BRE1, or the mammalian RNF20(hBRE1)/RNF40 complex (Hwang et al., 2003; Kim et al., 2005; Zhu et al., 2005). The ubiquitin-conjugating (E2) enzymes involved in that process were reported to be RAD6 in yeast, and its mammalian orthologs RAD6A and RAD6B (Kim et al., 2009; Robzyk et al., 2000).
Transcription by RNA polymerase II is a highly regulated process. This applies equally not only to the extensively studied transcription initiation step, but also to the elongation step. During elongation, RNA polymerase II (Pol II) may often become paused or arrested, in a manner that can be rate limiting for efficient transcription (Rahl et al., 2010; Selth et al., 2010; Sigurdsson et al., 2010). Many elongation factors are required to stimulate Pol II's progression; some are general and apply to many genes, while others are selective to specific subsets of genes (Arndt and Kane, 2003; Reines et al., 1999; Selth et al., 2010).
TFIIS is a transcription elongation factor that is highly conserved in the eukaryotic and archaeal kingdoms. TFIIS helps Pol II pass through transcriptional blocks on template DNA. It can reactivate arrested elongation complexes by stimulating endonucleolytic cleavage by Pol II of the nascent RNA (Fish and Kane, 2002). This activity of TFIIS is required for efficient read-through at arrested sites (Ghavi-Helm et al., 2008; Labhart and Morgan, 1998; Reinberg and Roeder, 1987). It is also suggested that TFIIS has a role in overcoming the arrest induced by nucleosomes, thereby promoting chromatin transcription in vitro (Guermah et al., 2006; Kireeva et al., 2005). Genome-wide analysis revealed that TFIIS plays a role also in RNA polymerase III transcription (Ghavi-Helm et al., 2008).
In the yeast S.cerevisiae, TFIIS is encoded by DST1, and is not essential for viability under normal conditions. DST1 deletion mutant strains grow like wild type, but are sensitive to drugs that compromise elongation efficiency (Nakanishi et al., 1992; Nakanishi et al., 1995; Ubukata et al., 2003). Recently, it was shown that a yeast TFIIS mutant that lacks transcript cleavage stimulatory activity and inhibits Pol II unstimulated cleavage is lethal, indicating that escape from transcriptional arrest via transcript cleavage is essential for efficient transcript elongation and cell viability (Sigurdsson et al., 2010). In mammals, two different forms of TFIIS were originally identified: a ubiquitously expressed form encoded by the TCEA1 gene, and a testis-specific form encoded by TCEA2 (Xu et al., 1994). A third form, encoded by TCEA3, was later identified in vertebrates (Labhart and Morgan, 1998). Knockout of mouse TCEA1 leads to embryonic lethality (Ito et al., 2006).
Several studies in yeast and mammals addressed a role for RNF20 and H2B ubiquitylation in regulation of transcription in general, and of transcriptional elongation in particular. In yeast, BRE1, RAD6 and consequently monoubiquitylated H2B (H2Bub) were shown to associate with elongating Pol II (Xiao et al., 2005). H2Bub was found to be necessary for efficient reassembly of nucleosomes and restoration of the chromatin structure during the elongation process (Osley et al., 2006). In mammals, H2Bub was found to associate preferentially with the transcribed region of highly expressed genes (Minsky et al., 2008), and in vitro transcription elongation assays established a role for H2Bub in facilitating elongation by Pol II (Pavri et al., 2006). Moreover, H2B ubiquitylation was recently shown to disrupt chromatin compaction in an in vitro system of chemically defined nucleosome arrays (Fierz et al., 2011). Other studies, however, have suggested a negative role for H2Bub in regulating transcriptional elongation. For example, yeast H2Bub was shown to serve as a barrier to transcriptional elongation by blocking the recruitment of the kinase Ctk1, which phosphorylates Pol II on serine 2 of its carboxy terminal domain (CTD) repeat (Wyce A, 2007). Recent work implicated H2Bub in enhancing nucleosome stability, proposing that this is a mechanism by which ubH2B plays a role in gene repression (Chandrasekharan et al., 2009). Studies in mammals showed that RNF20 suppresses the induction of interferon regulatory factor 1 (IRF1) in response to STAT1 signaling (Buro et al., 2010). Furthermore, H2Bub was implicated in gene repression through USP22, an H2Bub-specific deubiquitylating enzyme and a subunit of the human SAGA complex, which is necessary for activation of SAGA-dependent genes and contributes to heterochromatin silencing (Zhang et al., 2008; Zhao et al., 2008).
Previously, we showed that RNF20 selectively regulates a subset of genes in human cells and acts as a putative tumor suppressor. Genes whose expression was positively regulated by RNF20 (RNF20-dependent genes) include the p53 tumor suppressor as well as numerous histone H2A and H2B genes. In contrast, RNF20 suppresses the expression of several proto-oncogenes and growth-related genes, including many genes that are induced by epidermal growth factor (EGF). RNF20-suppressed genes are characterized by a rather low rate of transcription, despite high occupancy by Pol II. Notably, these genes are preferentially associated with compact chromatin fibers, which presumably makes them more difficult to transcribe and may result in frequent pausing of Pol II at multiple points along the transcribed gene (Shema et al., 2008). Given the reported role of TFIIS in relieving Pol II pausing and facilitating transcriptional elongation through “difficult” regions, we asked whether it might be involved in the positive regulation of RNF20-suppressed genes. We now describe a novel mechanism by which RNF20, presumably via H2B ubiquitylation, represses transcription elongation through inhibition of TFIIS recruitment to chromatin. Depletion of TFIIS abolishes RNF20-mediated suppression of growth-related genes, and attenuates the cellular response to EGF. Our findings thus position TFIIS as a key target in RNF20-mediated gene repression, and propose a molecular mechanism that mediates the tumor suppressor activity of RNF20.
Results
Transcriptional regulation by RNF20 requires its E3 ligase activity
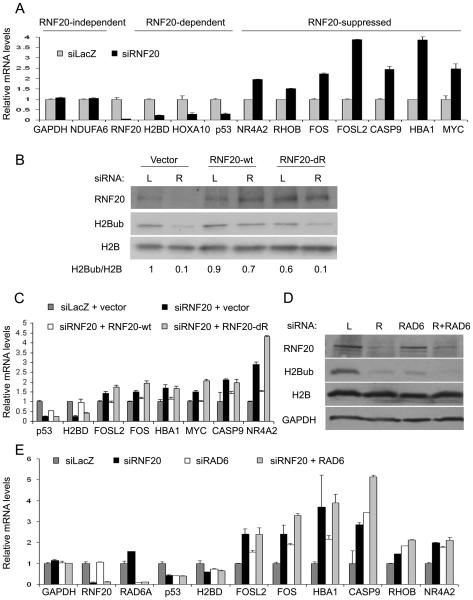
We have previously shown that RNF20 selectively regulates the expression of a subset of genes in human cells (Shema et al., 2008). These genes can be divided into two distinct subgroups: (1) genes that are dependent on RNF20 for their optimal expression, and (2) genes that are suppressed by RNF20. Genes representative of each subgroup are shown in Figure 1A: The RNF20-dependent genes H2BD, HOXA10 and TP53 (p53) are strongly downregulated upon depletion of RNF20 by siRNA, while the RNF20-suppressed genes NR4A2, C-FOS, FOSL2, C-MYC and others are upregulated. As shown earlier, this is due to altered transcription rates (Shema et al, 2008). Of note, the bulk transcriptome of the cells, represented here by GAPDH and NDUFA6, is not significantly affected by transient RNF20 depletion, suggesting that H2B ubiquitylation is not rate limiting for global transcription (Shema et al, 2008). These effects were validated with 3 different RNF20 siRNA oligonucleotides (Supplemental Fig. S1A,B), ruling out off-target effects.
Figure 1. The E3 ligase activity of RNF20 is necessary for its regulation of gene expression.
(A) qRT-PCR analysis of RNF20-independent, RNF20-dependent and RNF20-suppressed genes, 48 hours after transient transfection of HeLa cells with RNF20 siRNA (siRNF20) or LacZ siRNA (siLacZ). Also shown is RNF20 itself, as a measure of knockdown efficiency. Gene values were divided by the control siLacZ samples, which were set as 1. Prior to the division, values were normalized to GAPDH mRNA in the same sample, except for the bars indicating the raw values of GAPDH itself. Bars indicate averages of values from duplicate qPCR reactions; error bars represent standard deviation. Similar data was obtained in 10 independent experiments. See Supplemental Fig. S1A,B for validation with 3 different RNF20 siRNA oligonucleotides.
(B) HeLa cells were transiently transfected with siRNA oligonucleotides directed to the 5′ UTR of endogenous RNF20 (R) or with siLacZ (L). 24 hours later, cells were transfected with expression plasmids encoding either wild-type RNF20 (RNF20-wt) or a delta RING mutant thereof (RNF20-dR), both lacking the 5′ UTR of RNF20 mRNA and therefore resistant to the siRNA; empty vector was transfected as control. Cells were harvested 48 hours later for Western blot analysis with the indicated antibodies. The relative abundance of H2Bub, corrected for total H2B in the same sample, is shown at the bottom; the H2Bub/H2B ratio in the control sample (first lane) was taken as 1.0.
(C) qRT-PCR analysis of RNF20-dependent genes (H2BD and p53) and RNF20-suppressed genes (FOSL2, FOS, HBA1, MYC, CASP9 and NR4A2), from the experiment described in (B). Values and error bars were calculated as in (A). Similar data was obtained in 3 independent experiments.
(D) Western blot analysis of RNF20 and H2Bub 48 hours after transient transfection of HeLa cells with siLacZ (L), siRNF20 (R), siRAD6A (RAD6) or a combination of siRNF20 and siRAD6A (R+RAD6). GAPDH and H2B serve as loading controls.
(E) qRT-PCR analysis of the experiment described in (D) for the RNF20-independent gene GAPDH, the RNF20-dependent genes H2BD and p53, and RNF20-suppressed genes FOSL2, FOS, HBA1, CASP9, RHOB and NR4A2. Also shown are RNF20 and RAD6A, as a measure of knockdown efficiencies. Values and error bars were calculated as in (A). Similar data was obtained in 3 independent experiments.
See Supplemental Fig. S1C, D for analysis of RNF40 depletion.
Next, we sought to determine whether transcriptional regulation of those genes by RNF20 requires its E3 ubiquitin ligase activity. To that end, we knocked down endogenous RNF20 using siRNA directed against its 5′ UTR and reintroduced exogenous wild type (wt) RNF20 or a mutant RNF20 lacking the RING domain (ΔRING; dR). Knockdown of endogenous RNF20 in combination with an empty vector led to a marked decrease in H2Bub levels (Figure 1B), which was largely rescued by reintroduction of wt RNF20 but not the catalytically inactive ΔRING (dR) mutant. Reintroduction of wt RNF20 reversed the transcriptional effects of RNF20 depletion on both gene subgroups (Fig. 1C). However, this did not occur with the ΔRING mutant, suggesting that RNF20 regulates gene expression in a manner dependent on its E3 ligase activity, possibly through monoubiquitylation of histone H2B.
RAD6A is the main E2 enzyme recruited by RNF20, and is responsible for RNF20-mediated H2B ubiquitylation (Kim et al., 2009). Indeed, RAD6A knockdown led to a strong reduction in H2Bub levels, comparable to that seen upon RNF20 knockdown (Fig. 1D; knockdown validation in Fig. 1E). Notably, RAD6A depletion reproduced faithfully the transcriptional effects of RNF20 knockdown (Fig. 1E), further supporting the conclusion that RNF20-mediated transcriptional regulation depends on its ubiquitin ligase activity. Moreover, knockdown of RNF40, the heterodimeric partner of RNF20 that together with it forms the active E3 ligase, effectively inhibited H2B monoubiquitylation and also reproduced the selective effects of RNF20 knockdown on gene expression (Supplemental Fig. S1C,D). However, the interpretation of this result is limited by the fact that RNF40 depletion also caused a marked reduction in RNF20 protein (Supplemental Fig. S1C), suggesting that formation of an RNF20-RNF40 dimer is necessary for stabilization of each partner in the dimer, in line with earlier observations (Kim et al., 2009).
RNF20 and TFIIS counter the transcriptional effects of each other on the expression of a specific subset of genes
RNF20-suppressed genes reside preferentially in closed chromatin and are poorly transcribed despite bearing histone marks usually associated with active transcription and despite their high association with RNA-polymerase II, supporting the possibility that their transcription is being persistently hindered (Shema et al., 2008). Given the known function of TFIIS in relieving transcriptional blocks, we wished to determine whether TFIIS can selectively modulate the transcription of this subgroup of genes. In addition to TCEA1, the most prominent TFIIS member in HeLa cells, these cells express also moderate amounts of TCEA2 mRNA, but negligible amounts of TCEA3 mRNA (data not shown). We therefore performed a combined knockdown of RNF20 and TFIIS (TCEA1 plus TCEA2 genes) followed by qRT-PCR analysis of mRNA levels. Knockdown validation is shown in Figure 2A and Supplemental Fig. S2A. Of note, whereas RNF20 depletion exerted the expected effects on the RNF20-suppressed, RNF20-independent and RNF20-dependent genes (Fig 2, panels B,C and D, respectively; compare siLacZ to siRNF20), TFIIS depletion by itself did not affect the basal expression levels of any of those genes (compare siLacZ to siTCEA1+2), implying that basal transcription in this experimental system is not dependent on TFIIS. However, TFIIS depletion abrogated almost completely the positive effect of RNF20 knockdown on the RNF20-suppressed genes (Fig. 2B, compare siRNF20 to siTCEA1+2+siRNF20), while making no difference to the other subgroups (Fig. 2C,D). Analysis of heterogeneous nuclear RNA with intron-derived primers, which provides a better representation of relative transcription rates (Kuroda et al., 2005; Phelps et al., 2006), revealed an essentially similar picture (Supplemental Fig. S2B), strongly arguing that TFIIS is required for efficient transcription of the RNF20-repressed genes upon RNF20 depletion. Knockdown of either TCEA1 or TCEA2 alone had a partial effect (Supplemental Fig. S2C,D), indicating that both contribute to the transcriptional response in these cells, as opposed to knockdown of TCEA3 which had no effect (Supplemental Fig. S2E,F). Hence, TFIIS is selectively required for the transcriptional induction of the RNF20-suppressed genes following RNF20 knockdown.
Figure 2. TFIIS depletion abolishes the increased transcription of RNF20-suppressed genes following RNF20 knockdown.
(A) qRT-PCR analysis of RNF20, TCEA1 and TCEA2 48 hours after transient transfection of HeLa cells with the indicated siRNA oligonucleotides. Values and error bars were calculated as in Fig. 1A. Similar data was obtained in 6 independent experiments. See also Supplemental Fig. S2A for Western blot analysis of the same experiment.
(B-D) qRT-PCR analysis of RNF20-suppressed genes (B), RNF20-independent genes (C) and RNF20-dependent genes (D) 48 hours after transfection as in (A). Values and error bars were calculated as in Fig. 1A. Gene values were divided by the control siLacZ samples, which were set as 1. For (B) and (D), prior to the division, values were normalized to GAPDH mRNA. (C) depicts the raw values of GAPDH and NDUFA6 mRNA, used for normalization of the other values. Similar data was obtained in 6 independent experiments. See also Supplemental Fig. S2B for analysis of heterogeneous nuclear RNA of RNF20-suppressed genes, and Supplemental Fig. S2C-F for the effects of individual knockdown of either TCEA1, TCEA2 or TCEA3.
The involvement of TFIIS in the regulation of RNF20-suppressed genes was further assessed by overexpression experiments. Augmentation of TFIIS levels by overexpression of either HA-tagged or flag-tagged TCEA1 did not affect RNF20 or H2Bub levels (Fig. 3A). Notably, excess TFIIS elicited a modest upregulation of all tested RNF20-suppressed mRNAs (Figure 3B), while not affecting RNF20-independent genes (NDUFA6) or RNF20-dependent genes (p53 and H2BD; Figure 3C). Thus, RNF20 and TFIIS selectively exert opposing effects on the transcriptional regulation of RNF20-suppressed genes.
Figure 3. TFIIS overexpression increases the transcription of RNF20-suppressed genes.
(A) Western blot analysis of RNF20, TFIIS and H2Bub 48 hours after transient transfection of HeLa cells with control empty vector, HA-tagged TCEA1 or flag-tagged TCEA1 (TFIIS-HA and TFIIS-flag, respectively). Tubulin served as a loading control.
(B) qRT-PCR analysis of RNF20-suppressed genes, from the experiment described in (A). Also shown is RNF20 mRNA. Values and error bars were calculated as in Fig. 1A. Similar data was obtained in 3 independent experiments.
(C) qRT-PCR analysis of the RNF20-dependent genes p53 and H2BD and RNF20-independent gene NDUFA6, from the experiment described in (A). Values and error bars were calculated as in Fig. 1A. Similar data was obtained in 3 independent experiments.
RNF20 depletion increases TFIIS occupancy on chromatin
To further elucidate the crosstalk between TFIIS and RNF20 in the transcriptional regulation of RNF20-suppressed genes, we examined the effect of RNF20 knockdown on TFIIS. We found that while RNF20 depletion had no appreciable effect on overall TFIIS levels (Figure 4A, compare lanes 1 and 2; see also Supplemental Fig. S2A), it increased strongly the association of TFIIS with chromatin (Figure 4B, compare lanes 3 and 4).
Figure 4. RNF20 knockdown increases the occupancy of TFIIS on chromatin.
(A) Western blot analysis of RNF20, TFIIS, H2Bub and H2B 48 hours after transient transfection of HeLa cells with the indicated siRNA oligonucleotides. Equal amounts of total protein were loaded in each lane.
(B) HeLa cells were transiently transfected with the indicated siRNA oligonucleotides. 48 hours later, the chromatin bound fraction was isolated as described under Experimental procedures, and subjected to Western blot analysis with the indicated antibodies. Whole cell extract aliquots (Total) were analyzed in parallel.
(C) Chromatin immunoprecipitation (ChIP) analysis of relative TFIIS abundance on RNF20-suppressed genes (NR4A2, RhoB, FOS, FOSL2, CASP9, HBA1), the RNF20-independent gene GAPDH and the RNF20-dependent gene H2BD, in siLacZ and siRNF20-transfected HeLa cells. Immunoprecipitated DNA and input DNA were quantified by qPCR with primers specific for the 5′ transcribed regions of the indicated genes. Results are presented as percentage of input. Bars indicate averages of data from duplicate qPCR reactions; error bars represent standard deviation. Similar data was obtained in 3 independent experiments.
Consistent with the increased global association of TFIIS with chromatin in RNF20-depleted cells, chromatin immunoprecipitation (ChIP) analysis confirmed the enhanced recruitment of TFIIS to the transcribed regions of numerous genes following RNF20 knockdown (Figure 4C). This increase was global and was observed not only for the RNF20-suppressed genes but actually for all genes tested. This implies that RNF20, presumably through H2B ubiquitylation, can interfere with the recruitment of TFIIS to chromatin. While this interference is general, only the RNF20-suppressed genes appear to rely on TFIIS for their efficient transcription (Fig. 2 C-E); hence, they selectively benefit from the relief of this interference and the resultant increased recruitment of TFIIS. In sum, TFIIS likely provides a distinct transcription-facilitating function that is essential for the RNF20-suppressed genes but is dispensable for the other subgroups of genes.
Downregulation of RNF20 enables more efficient elongation of RNF20-suppressed genes and increases the interaction between TFIIS and the PAF1 complex
Since TFIIS is an elongation factor, we next sought to examine the rate of elongation of RNF20-suppressed genes following RNF20 depletion alone or in combination with TFIIS depletion. Knockdown of RNF20 alone augmented the rate of transcriptional elongation of RNF20-suppressed genes, as deduced from the increase in the ratio between RNA polymerase II occupancy near the 3′ end relative to its occupancy near the 5′ end of a gene (Figure 5A). This suggests a negative role for RNF20/H2Bub in regulation of transcriptional elongation. This effect was not observed for RNF20-independent genes such as GAPDH and NDUFA6 implying that it is specific for RNF20-repressed genes. Importantly, in line with the transcriptional effects shown in Figure 2, the increase in elongation rate was abolished when RNF20 knockdown was combined with TFIIS knockdown (Fig. 5A, siTCEA1+2+RNF20). Hence, the increased transcriptional elongation rate of the RNF20-suppressed genes after RNF20 depletion is dependent on the activity of TFIIS.
Figure 5. Downregulation of RNF20 facilitates the elongation of RNF20-suppressed genes and increases the interaction between TFIIS and the PAF1 complex.
(A) Ratio between RNA polymerase II occupancy near the 3′ end versus near the 5′ end of RNF20-suppressed genes (NR4A2, RhoB, FOS, FOSL2, CASP9, MYC) and RNF20-independent genes (GAPDH and NDUFA6). HeLa cells were transiently transfected with siLacZ, siRNF20, siTCEA1+2 or siTCEA1+2+siRNF20. 48 hours later, cells were subjected to ChIP analysis with antibodies against Pol II, followed by qPCR analysis with primers corresponding to the 5′ and 3′ ends of each of the transcribed regions. Bars indicate averages of data from duplicate qPCR reactions; error bars represent standard deviation. Similar data was obtained in 3 independent experiments.
(B, C) HeLa cells were transiently transfected with RNF20 (R) or LacZ (L) siRNA oligonucleotides in (B), or with RAD6A (Rad) siRNA oligonucleotides in (C). 24 hours later, cells were transfected with flag-tagged TCEA1 (TFIIS-flag) or empty vector as control. After an additional 48 hours, cells were harvested for immunoprecipitation (IP) with anti-flag antibodies; 3% of the total cell extract served as input. The IP and input samples were subjected to Western blot analysis with the antibodies indicated on the left.
The graph in B represents quantification of the hPAF1 signal in the indicated IP samples, derived from three independent experiments (including the one shown in B). The hPAF1 signal in the siLacZ sample for each experiment was set as 1, and the fold increase in the corresponding siRNF20 sample was calculated accordingly. Quantification was with ImageJ. Error bars represent standard deviation.
RNF20 binds RNA polymerase II through the hPAF1 subunit of the PAF1 complex (Kim et al., 2009). TFIIS binds directly to both RNA polymerase II and the hPAF1 complex (via the hPAF1 and hLEO1 subunits); the direct interaction between TFIIS and hPAF1C results in their cooperative binding to Pol II and a strong synergistic effect on transcriptional elongation (Kim et al., 2010). To assess whether RNF20 activity may impede transcriptional elongation by attenuating the TFIIS-hPAF1 interaction, we performed co-immunoprecipitation analysis. As seen in Figure 5B, the interaction between TFIIS and hPAF1 became more apparent upon RNF20 depletion (left panel; quantification of three independent experiments shown in the right panel). Moreover, depletion of RAD6A, which exerts similar effects on H2Bub levels and gene expression (Figure 1 D,E), likewise augmented the TFIIS-hPAF1 interaction (Figure 5C). Together, these results are strongly consistent with the conclusion that the catalytic activity of the RNF20-RAD6A complex, presumably through H2B ubiquitylation, impedes the hPAF1-TFIIS interaction.
TFIIS and RNF20 counter each other in the transcriptional response to EGF
RNF20 preferentially suppresses the expression of many EGF-inducible genes (Shema et al., 2008). To investigate a possible involvement of TFIIS in that context, we examined whether TFIIS is recruited to such EGF target genes upon induction by EGF. As shown in Figure 6A, this was indeed found to be the case; moreover, the kinetics of recruitment paralleled those of transcription (see Fig. 6B). In contrast, no increase in TFIIS binding was observed for GAPDH and p21, which are not transcriptionally induced by EGF (Figure 6A). Furthermore, depletion of TFIIS (TCEA1 and TCEA2) mildly but reproducibly attenuated the transcriptional response of those genes to EGF, as assessed by analysis of steady state mRNA (Figure 6B), as well as of heterogeneous nuclear RNA indicative of relative transcription rates (Figure 6C). Depletion of TCEA1 alone excreted similar effects (Supplemental Fig. S3A,B). Hence, recruitment of TFIIS to a subset of EGF target genes is necessary for their optimal transcriptional induction, implicating TFIIS as a positive regulator of this response. Some EGF target genes are not suppressed by RNF20 (Shema et al., 2008). Interestingly, examination of representative genes from that group revealed that they are refractory to TFIIS ablation (Supplemental Fig. S3C), further supporting the specific dependence of RNF20-suppressed genes on TFIIS activity.
Figure 6. TFIIS is recruited to EGF targets, and counters RNF20 in the transcriptional response to EGF.
(A) ChIP analysis of relative TFIIS abundance in HeLa cells stimulated with EGF (20 ng/ml) for 0, 0.5 hours, 1 hour or 2 hours. Prior to EGF addition, cells were serum starved for 16 hours. Immunoprecipitated DNA and input DNA were quantified by qPCR with primers for the EGF target genes RHOB, FOSL2, NR4A2, FOS and TM4SF1, and also for GAPDH and p21, which are not responsive to EGF. Bars indicate averages of data from duplicate qPCR reactions; error bars represent standard deviation. Similar data was obtained in 3 independent experiments.
(B,C) qRT-PCR analysis of total mRNA (B) or heterogeneous nuclear precursor RNA (C) levels of the indicated EGF target genes at various times after exposure to EGF of HeLa cells transiently transfected with TCEA1+TCEA2 siRNA (light blue) or LacZ siRNA (black). All values are normalized to GAPDH mRNA. Precursor RNA levels were measured with primer pairs derived from intronic sequences of the indicated genes. Values indicate averages of data from duplicate qPCR reactions; error bars represent standard deviation. Similar data was obtained in 4 independent experiments. See also Supplemental Fig. S3 for parallel analysis after TCEA1 knockdown alone.
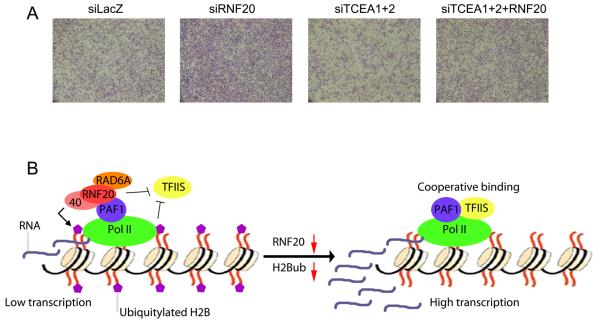
MCF10A cells are stimulated to migrate by addition of EGF, and depletion of RNF20 results in a robust increase in cell migration, consistent with its effects on EGF-induced gene expression (Shema et al., 2008). We therefore next asked whether the induction of cell migration by RNF20 depletion is also dependent on TFIIS. As expected, knockdown of RNF20 led to a marked increase in cell migration in response to EGF (Figure 7A). Knockdown of TFIIS alone (siTCEA1+2) caused a slight decrease in EGF-induced cell migration. Remarkably, concomitant depletion of TFIIS (siTCEA1+2+RNF20) abolished almost completely the stimulatory effect of RNF20 knockdown. Overall, these findings imply that the opposing effects of RNF20 and TFIIS on the expression of the RNF20-repressed genes have also biological consequences.
Figure 7. Enhancement of EGF-induced cell migration by RNF20 depletion is dependent on TFIIS.
(A) MCF10A cells were transiently transfected with siRNA oligonucleotides for LacZ (siLacZ), RNF20 (siRNF20), TFIIS (siTCEA1+2) or a combination of TFIIS and RNF20 (siTCEA1+2+RNF20). 48 hours later, 70,000 cells were plated in the upper compartment of a 24-well Transwell tray (Corning), and allowed to migrate for 16hr towards EGF (added to the bottom compartment). Migrating cells were visualized by staining the membrane with methyl violet, followed by photography under a binocular. See also Supplemental Fig. S4 for analysis of TCEA1 mRNA expression in human tumors.
(B) Proposed model for the regulation of a subset of genes by RNF20 and H2Bub. RNF20-suppressed genes are associated with high levels of H2Bub (purple), and transcribed with low efficiency owing to inefficient elongation. Under those conditions, binding of TFIIS is inhibited, either by H2Bub or/and via additional activities of the RNF20/RNF40/RAD6A complex. Upon RNF20 depletion, H2Bub levels decline, enabling recruitment of TFIIS to the chromatin and cooperative binding of TFIIS to the PAF1 complex and to Pol II. This leads to an increase in transcription rates. The chromatin cartoon was adapted from Marmorstein (Marmorstein, 2001).
Discussion
RNF20, in complex with RNF40, is the main E3 ligase that targets histone H2B for monoubiquitylation. Despite the general association of H2Bub with transcription, only a subset of genes seem to be regulated by that modification (Shema et al., 2008). Here, we show that the E3 activity of RNF20 is essential for that regulation. This conclusion is based on the failure of a RING-deficient RNF20 mutant to rescue the effects of RNF20 knockdown, as well as the fact that depletion of RAD6A, which we find to be the principal E2 enzyme in that process, exerts the same transcriptional effects as depletion of RNF20. This is in agreement with the finding that both human RAD6 proteins (hRAD6A and hRAD6B), but not hUbcH6 which can also serve as an E2 enzyme for H2Bub (Kim et al., 2005; Zhu et al., 2005), fully complement the function of yeast RAD6 in H2B ubiquitylation (Kim et al., 2009). RAD6A comprises the majority of the endogenous RAD6 protein pool (Koken et al., 1996); thus, it is not surprising that its depletion leads to a marked decrease in H2Bub levels.
Significantly, our findings strongly implicate the elongation factor TFIIS in the mechanism of RNF20-mediated gene repression. In particular, we show that upon RNF20 depletion, RNF20-suppressed genes are upregulated only in the presence of TFIIS. Furthermore, overexpression of TFIIS alone leads to selective upregulation of RNF20-suppressed genes, while not affecting other genes. Interestingly, whereas TFIIS is recruited more effectively to all tested genes upon RNF20 depletion (Fig. 4C), only a minority of those genes (the RNF20-suppressed subgroup) are functionally dependent on the increased recruitment of TFIIS. Together, our results imply a novel mechanism for the regulation of a subset of genes by RNF20 and H2B ubiquitylation (Figure 7B). RNF20-suppressed genes reside within relatively closed chromatin, characterized also by high H2Bub content (Shema et al., 2008). According to the proposed model, the catalytic activity of the RNF20/RNF40/RAD6A, presumably through H2B ubiquitylation, interferes with TFIIS binding to the PAF1 complex, resulting in inefficient transcriptional elongation of genes that require TFIIS for this process. Upon RNF20 depletion, H2Bub levels are drastically reduced and TFIIS can therefore bind the PAF1 complex. The interaction between TFIIS and the PAF1 complex results in their cooperative binding to Pol II and a synergistic effect on transcriptional elongation (Kim et al., 2010), augmenting the accumulation of transcripts. For the majority of the genes that are not regulated by RNF20, however, the increased binding of TFIIS to Pol II upon RNF20 depletion has no effect on transcription, suggesting that these genes do not require TFIIS for optimal transcription elongation. The selective requirement of TFIIS by the RNF20-suppressed genes may be due to the fact that they reside within relatively closed chromatin, which is presumably harder to transcribe. This conjecture is supported by the observation that the RNF20-suppressed genes are associated with high constitutive Pol II levels, despite being expressed at relatively low levels (Shema et al., 2008). TFIIS may therefore be specifically needed to reactivate the arrested elongation complexes on these genes. Indeed, we found that depletion of RNF20 specifically increased the elongation rate of these genes, in a TFIIS-dependent manner. Presumably, genes that are not dependent on RNF20 and reside in a more open chromatin context do not require TFIIS binding for optimal transcription elongation.
H2B is the main known target for the catalytic activity of the RAD6A-RNF20-RNF40 complex. It is therefore plausible that the transcriptional effects explored in this study are mediated by H2B ubiquitination. However, at present we can not formally rule out the possibility that some of those transcriptional effects are due to ubiquitylation of other proteins by that complex, or even that the complex exerts some unknown mode of transcriptional interference that requires the RING domain of RNF20 but does not rely on protein ubiquitylation. In yeast, such question can be addressed directly through the construction of strains carrying a point mutation in the ubiquitylation site of the single H2B protein (Henry et al., 2003; Tanny et al., 2007). Indeed, by applying this strategy to the fission yeast S.pombe, Tanny et al. found that the gene expression profile of cells carrying such point mutation exhibited substantial overlap with that of strains deleted for BRL1 and BRL2, the S.pombe BRE1 orthologs. This suggests that BRL1 and BRL2 regulate transcription mostly through ubiquitylation of H2B. Nevertheless, the transcriptional profiles were not completely identical between the H2B mutant strain and the BRL1+BRL2-deficient strain. Hence, it was proposed that these E3 ligases likely have additional ubiquitylation targets that also contribute to gene regulation (Tanny et al., 2007). Unfortunately, this strategy cannot be implemented effectively in mammalian cells, whose genomes carry numerous H2B genes with nucleotide sequence differences that severely hinder the use of siRNA to knock all of them down (Marzluff et al., 2002).
Our findings are reminiscent of earlier observations with regard to the negative elongation factor (NELF), which has been shown to inhibit TFIIS binding to Pol II, presumably by a competition model (Palangat et al., 2005); raising TFIIS concentration gradually overcame this inhibition. It is noteworthy that a recent study, performed in embryonic stem cells, found over 90% of the genes to be regulated at the elongation level by promoter-proximal pausing, mediated by the negative elongation factors NELF and DSIF (Rahl et al., 2010). Stalling of Pol II, bound to NELF, mediates cytokine gene expression (Adelman and Rogatsky, 2010). Interestingly, the induction of the cytokine IL-8 in response to EGF signaling is suppressed by RNF20 (Shema et al., 2008). Here we show that TFIIS is required for optimal induction of IL-8, demonstrating the opposing effects of RNF20 and TFIIS in its transcriptional regulation.
Interestingly, c-Myc, shown here to be regulated by RNF20 and TFIIS, was reported to play a key role in releasing paused Pol II, by recruiting the positive elongation factor P-TEFb (Rahl et al., 2010). This may therefore contribute to a positive feedback mechanism whereby, upon downregulation of RNF20 or signal-dependent attenuation of its catalytic activity, TFIIS-dependent c-Myc induction further enhances transcriptional elongation of paused genes; similar effects may be exerted by increased activity of H2Bub-specific deubiquitinating enzymes (DUBs) (Zhang et al., 2008). It will be of great interest to gain further insight into the intricate interplay between positive and negative elongation factors, as well as the signals and cellular contexts that modulate this interplay to selectively regulate specific gene expression programs.
Finally, our findings may also pertain to human cancer. The misregulation of a variety of histone modifications can actively contribute to cancer (Chi et al., 2010). RNF20 has attributes of a putative tumor-suppressor, chiefly through selective suppression of pro-oncogenic genes that promote proliferation (e.g. c-Myc, c-Fos) along with transcriptional activation of tumor suppressors such as p53 (Espinosa, 2008; Shema et al., 2008). TFIIS, on the other hand, was proposed to have a positive link to cancer: its depletion in cancer cell lines was shown to inhibit cell proliferation and induce apoptosis (Hubbard et al., 2008). In addition, homozygous null mutants for TFIIS show a dramatic increase in apoptotic cells in the liver (Ito et al., 2006). This is partly due to the fact that TFIIS directly upregulates the transcription of the Bcl-XL gene, which encodes a potent anti-apoptotic protein implicated in the survival of cancer cells (Ito et al., 2006; Nagata et al., 2009). Remarkably, examination of the Oncomine database (www.oncomine.org) reveals significant upregulation of TCEA1 expression in a variety of human tumors (Supplemental Figure S4). It is also noteworthy that the TCEA2 gene resides within a region of chromosome 20 that is frequently amplified in cervical cancer, and TCEA2 mRNA is often overexpressed in these tumors, giving rise to the suggestion that it may play a role in cervical carcinogenesis (Scotto et al., 2008). Indeed HeLa cells, derived from a cervical carcinoma, express substantial amounts of TCEA2 mRNA, and knockdown of TCEA2 alone is already sufficient to yield measurable transcriptional effects in these cells (Supplemental Fig. S2D). Thus, excessive TFIIS activity may promote cancer, at least in some contexts.
In sum, our results strongly suggest that part of the tumor suppressor activities of RNF20 may be mediated via inhibition of TFIIS binding, with consequent downregulation of cancer-promoting genes whose transcriptional elongation relies on TFIIS. This calls for further analysis of the interplay between TFIIS and RNF20 in human cancer.
Experimental procedures
Cell culture and transfections
Human cervical carcinoma HeLa cells were grown in DMEM with 10% bovine serum supplemented with antibiotics, and MCF10A human mammary epithelium-derived cells were grown in DMEMF12 medium with 5% heat-inactivated horse serum, supplemented with antibiotics and 10 μg/ml insulin, 0.1 μg/ml cholera toxin, 0.5 μg/ml hydrocortisone and 10 ng/ml EGF. SMARTpool oligonucleotides for siRNA transfection were purchased from Dharmacon. Transfections were carried out with Dharmafect 1 (HeLa) or Dharmafect 4 (MCF10A) reagent according to the manufacturer's protocol. The siRNA sequence targeting the 5′ UTR of RNF20 mRNA was CTGCACGGGCCTTGGAAAA. Transfection of expression plasmids was carried out using the jetPEI DNA transfection reagent (101-10, Polyplus Transfection), according to the manufacturer's protocol.
Antibodies
The following commercial antibodies were used: anti-H2B (07-371, Millipore); Anti-Pol II (ab5408, Abcam), Anti-RNF20 (ab32639, Abcam), Anti-Paf1 (Western-blot: A300-172A, IP: A300-173A, Bethyl Laboratories), Anti-GAPDH (MAB374, Millipore), and Anti-flag (F3165, Sigma). Anti-H2Bub is described in Minsky et al. (Minsky et al., 2008). Anti-TFIIS is described in Kim et al. (Kim et al., 2010).
RNA purification and quantitative Real-Time PCR
For Real-time reverse transcription (RT-PCR) analysis, total RNA was extracted with the NucleoSpin kit (Macherey Nagel, Germany). One microgram of each RNA sample was reverse transcribed with Moloney murine leukemia virus reverse transcriptase (Promega, Madison, WI) and random hexamer primers (Applied Biosystems). Real-time PCR was performed in a StepOne real-time PCR instrument (Applied Biosystems, Foster City, CA) with SYBR Green PCR Supermix (Invitrogen). The primers used are listed in Supplemental table 1.
Chromatin immunoprecipitation (ChIP)
ChIP analysis was performed as described (Minsky and Oren, 2004). The primers used are listed in Supplemental table 1.
Fractionation of chromatin-bound proteins
HeLa cells were harvested and washed twice with ice-cold PBS, then incubated on ice for 5 minutes with 100mM NaCl, 300mM sucrose, 3mM MgCl2, 10mM Pipes pH 6.8, 1mM EGTA, 0.2% Triton X-100 and protease inhibitor mix (Sigma). Cells were centrifuged, and the supernatant was collected as the “unbound” fraction. The remaining “bound” fraction was washed with PBS and then collected in 100mM NaCl, 300mM sucrose, 3mM MgCl2, 10mM PIPES pH 6.8, 1mM EGTA and 0.5U/μl DNaseI, supplemented with protease inhibitor mix (Sigma).
Co-immunoprecipitation (co-IP) analysis
HeLa cells were harvested, washed with ice-cold PBS and lysed on ice in NET lysis buffer (50mM Tris-HCl, pH7.4, 150mM NaCl, 1mM EDTA 0.1% NP40) supplemented with protease inhibitor mix (Sigma). Cells were sonicated in a Bioruptor sonicator (Diagenode) for a total of 5 minutes, including intervals of 30 seconds on and 1 minute off, and then centrifuged at 13000RPM for 10 minutes at 4°C. Total protein content was determined with the BCA protein assay kit (PIERCE, #23227). Anti-flag antibodies (A2220, Sigma), covalently attached to beads, were washed once with ice-cold PBS and twice with NET buffer. Equal amounts of protein from each sample were incubated with the antibody beads at 4°C overnight. Next, beads were washed three times with NET buffer, and elution was carried out using flag peptide (F3290, Sigma) in PBS. Samples were resolved by SDS-PAGE.
Cell migration assay
Cell migration analysis was as described (Katz et al., 2007).
Supplementary Material
Acknowledgments
We thank Gilad Fuchs, Itay Tirosh, Yael Aylon, Lior Golomb and Efrat Lidor-Nili for great help and stimulating discussions. This work was supported in part by grant R37 CA40099 from the National Cancer Institute, the Dr. Miriam and Sheldon Adelson Medical Research Foundation, The Lower Saxony-Israeli Association, and the Yad Abraham Center for Cancer Diagnosis and Therapy. MO is incumbent of the Andre Lwoff chair in Molecular Biology. Efrat Shema is supported by the Adams Fellowship Program of the Israel Academy of Sciences and Humanities.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adelman K, Rogatsky I. RNA polymerase II stalling mediates cytokine gene expression. Cell Cycle. 2010;9:630–631. doi: 10.4161/cc.9.4.10841. [DOI] [PubMed] [Google Scholar]
- Arndt KM, Kane CM. Running with RNA polymerase: eukaryotic transcript elongation. Trends Genet. 2003;19:543–550. doi: 10.1016/j.tig.2003.08.008. [DOI] [PubMed] [Google Scholar]
- Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- Buro LJ, Chipumuro E, Henriksen MA. Menin and RNF20 recruitment is associated with dynamic histone modifications that regulate signal transducer and activator of transcription 1 (STAT1)-activated transcription of the interferon regulatory factor 1 gene (IRF1) Epigenetics Chromatin. 2010;3:16. doi: 10.1186/1756-8935-3-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekharan MB, Huang F, Sun ZW. Ubiquitination of histone H2B regulates chromatin dynamics by enhancing nucleosome stability. Proc Natl Acad Sci U S A. 2009;106:16686–16691. doi: 10.1073/pnas.0907862106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi P, Allis CD, Wang GG. Covalent histone modifications--miswritten, misinterpreted and mis-erased in human cancers. Nat Rev Cancer. 2010;10:457–469. doi: 10.1038/nrc2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinosa JM. Histone H2B ubiquitination: the cancer connection. Genes Dev. 2008;22:2743–2749. doi: 10.1101/gad.1732108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierz B, Chatterjee C, McGinty RK, Bar-Dagan M, Raleigh DP, Muir TW. Histone H2B ubiquitylation disrupts local and higher-order chromatin compaction. Nat Chem Biol. 2011 doi: 10.1038/nchembio.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fish RN, Kane CM. Promoting elongation with transcript cleavage stimulatory factors. Biochim Biophys Acta. 2002;1577:287–307. doi: 10.1016/s0167-4781(02)00459-1. [DOI] [PubMed] [Google Scholar]
- Ghavi-Helm Y, Michaut M, Acker J, Aude JC, Thuriaux P, Werner M, Soutourina J. Genome-wide location analysis reveals a role of TFIIS in RNA polymerase III transcription. Genes Dev. 2008;22:1934–1947. doi: 10.1101/gad.471908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guermah M, Palhan VB, Tackett AJ, Chait BT, Roeder RG. Synergistic functions of SII and p300 in productive activator-dependent transcription of chromatin templates. Cell. 2006;125:275–286. doi: 10.1016/j.cell.2006.01.055. [DOI] [PubMed] [Google Scholar]
- Henry KW, Wyce A, Lo WS, Duggan LJ, Emre NC, Kao CF, Pillus L, Shilatifard A, Osley MA, Berger SL. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev. 2003;17:2648–2663. doi: 10.1101/gad.1144003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard K, Catalano J, Puri RK, Gnatt A. Knockdown of TFIIS by RNA silencing inhibits cancer cell proliferation and induces apoptosis. BMC Cancer. 2008;8:133. doi: 10.1186/1471-2407-8-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang WW, Venkatasubrahmanyam S, Ianculescu AG, Tong A, Boone C, Madhani HD. A conserved RING finger protein required for histone H2B monoubiquitination and cell size control. Mol Cell. 2003;11:261–266. doi: 10.1016/s1097-2765(02)00826-2. [DOI] [PubMed] [Google Scholar]
- Ito T, Arimitsu N, Takeuchi M, Kawamura N, Nagata M, Saso K, Akimitsu N, Hamamoto H, Natori S, Miyajima A, Sekimizu K. Transcription elongation factor S-II is required for definitive hematopoiesis. Mol Cell Biol. 2006;26:3194–3203. doi: 10.1128/MCB.26.8.3194-3203.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Katz M, Amit I, Citri A, Shay T, Carvalho S, Lavi S, Milanezi F, Lyass L, Amariglio N, Jacob-Hirsch J, et al. A reciprocal tensin-3-cten switch mediates EGF-driven mammary cell migration. Nat Cell Biol. 2007;9:961–969. doi: 10.1038/ncb1622. [DOI] [PubMed] [Google Scholar]
- Kim J, Guermah M, McGinty RK, Lee JS, Tang Z, Milne TA, Shilatifard A, Muir TW, Roeder RG. RAD6-Mediated transcription-coupled H2B ubiquitylation directly stimulates H3K4 methylation in human cells. Cell. 2009;137:459–471. doi: 10.1016/j.cell.2009.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Guermah M, Roeder RG. The human PAF1 complex acts in chromatin transcription elongation both independently and cooperatively with SII/TFIIS. Cell. 2010;140:491–503. doi: 10.1016/j.cell.2009.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Hake SB, Roeder RG. The human homolog of yeast BRE1 functions as a transcriptional coactivator through direct activator interactions. Mol Cell. 2005;20:759–770. doi: 10.1016/j.molcel.2005.11.012. [DOI] [PubMed] [Google Scholar]
- Kireeva ML, Hancock B, Cremona GH, Walter W, Studitsky VM, Kashlev M. Nature of the nucleosomal barrier to RNA polymerase II. Mol Cell. 2005;18:97–108. doi: 10.1016/j.molcel.2005.02.027. [DOI] [PubMed] [Google Scholar]
- Koken MH, Hoogerbrugge JW, Jasper-Dekker I, de Wit J, Willemsen R, Roest HP, Grootegoed JA, Hoeijmakers JH. Expression of the ubiquitin-conjugating DNA repair enzymes HHR6A and B suggests a role in spermatogenesis and chromatin modification. Dev Biol. 1996;173:119–132. doi: 10.1006/dbio.1996.0011. [DOI] [PubMed] [Google Scholar]
- Kuroda M, Oikawa K, Yoshida K, Takeuchi A, Takeuchi M, Usui M, Umezawa A, Mukai K. Effects of 3-methylcholanthrene on the transcriptional activity and mRNA accumulation of the oncogene hWAPL. Cancer Lett. 2005;221:21–28. doi: 10.1016/j.canlet.2004.08.006. [DOI] [PubMed] [Google Scholar]
- Labhart P, Morgan GT. Identification of novel genes encoding transcription elongation factor TFIIS (TCEA) in vertebrates: conservation of three distinct TFIIS isoforms in frog, mouse, and human. Genomics. 1998;52:278–288. doi: 10.1006/geno.1998.5449. [DOI] [PubMed] [Google Scholar]
- Marmorstein R. Protein modules that manipulate histone tails for chromatin regulation. Nat Rev Mol Cell Biol. 2001;2:422–432. doi: 10.1038/35073047. [DOI] [PubMed] [Google Scholar]
- Marzluff WF, Gongidi P, Woods KR, Jin J, Maltais LJ. The human and mouse replication-dependent histone genes. Genomics. 2002;80:487–498. [PubMed] [Google Scholar]
- Minsky N, Oren M. The RING domain of Mdm2 mediates histone ubiquitylation and transcriptional repression. Mol Cell. 2004;16:631–639. doi: 10.1016/j.molcel.2004.10.016. [DOI] [PubMed] [Google Scholar]
- Minsky N, Shema E, Field Y, Schuster M, Segal E, Oren M. Monoubiquitinated H2B is associated with the transcribed region of highly expressed genes in human cells. Nat Cell Biol. 2008 doi: 10.1038/ncb1712. [DOI] [PubMed] [Google Scholar]
- Nagata M, Ito T, Arimitsu N, Koyama H, Sekimizu K. Transcription arrest relief by S-II/TFIIS during gene expression in erythroblast differentiation. Genes Cells. 2009;14:371–380. doi: 10.1111/j.1365-2443.2008.01277.x. [DOI] [PubMed] [Google Scholar]
- Nakanishi T, Nakano A, Nomura K, Sekimizu K, Natori S. Purification, gene cloning, and gene disruption of the transcription elongation factor S-II in Saccharomyces cerevisiae. J Biol Chem. 1992;267:13200–13204. [PubMed] [Google Scholar]
- Nakanishi T, Shimoaraiso M, Kubo T, Natori S. Structure-function relationship of yeast S-II in terms of stimulation of RNA polymerase II, arrest relief, and suppression of 6-azauracil sensitivity. J Biol Chem. 1995;270:8991–8995. doi: 10.1074/jbc.270.15.8991. [DOI] [PubMed] [Google Scholar]
- Osley MA, Fleming AB, Kao CF. Histone ubiquitylation and the regulation of transcription. Results Probl Cell Differ. 2006;41:47–75. doi: 10.1007/400_006. [DOI] [PubMed] [Google Scholar]
- Palangat M, Renner DB, Price DH, Landick R. A negative elongation factor for human RNA polymerase II inhibits the anti-arrest transcript-cleavage factor TFIIS. Proc Natl Acad Sci U S A. 2005;102:15036–15041. doi: 10.1073/pnas.0409405102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavri R, Zhu B, Li G, Trojer P, Mandal S, Shilatifard A, Reinberg D. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell. 2006;125:703–717. doi: 10.1016/j.cell.2006.04.029. [DOI] [PubMed] [Google Scholar]
- Phelps ED, Updike DL, Bullen EC, Grammas P, Howard EW. Transcriptional and posttranscriptional regulation of angiopoietin-2 expression mediated by IGF and PDGF in vascular smooth muscle cells. Am J Physiol Cell Physiol. 2006;290:C352–361. doi: 10.1152/ajpcell.00050.2005. [DOI] [PubMed] [Google Scholar]
- Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinberg D, Roeder RG. Factors involved in specific transcription by mammalian RNA polymerase II. Transcription factor IIS stimulates elongation of RNA chains. J Biol Chem. 1987;262:3331–3337. [PubMed] [Google Scholar]
- Reines D, Conaway RC, Conaway JW. Mechanism and regulation of transcriptional elongation by RNA polymerase II. Curr Opin Cell Biol. 1999;11:342–346. doi: 10.1016/S0955-0674(99)80047-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robzyk K, Recht J, Osley MA. Rad6-dependent ubiquitination of histone H2B in yeast. Science. 2000;287:501–504. doi: 10.1126/science.287.5452.501. [DOI] [PubMed] [Google Scholar]
- Scotto L, Narayan G, Nandula SV, Arias-Pulido H, Subramaniyam S, Schneider A, Kaufmann AM, Wright JD, Pothuri B, Mansukhani M, Murty VV. Identification of copy number gain and overexpressed genes on chromosome arm 20q by an integrative genomic approach in cervical cancer: potential role in progression. Genes Chromosomes Cancer. 2008;47:755–765. doi: 10.1002/gcc.20577. [DOI] [PubMed] [Google Scholar]
- Selth LA, Sigurdsson S, Svejstrup JQ. Transcript Elongation by RNA Polymerase II. Annu Rev Biochem. 2010;79:271–293. doi: 10.1146/annurev.biochem.78.062807.091425. [DOI] [PubMed] [Google Scholar]
- Shema E, Tirosh I, Aylon Y, Huang J, Ye C, Moskovits N, Raver-Shapira N, Minsky N, Pirngruber J, Tarcic G, et al. The histone H2B-specific ubiquitin ligase RNF20/hBRE1 acts as a putative tumor suppressor through selective regulation of gene expression. Genes Dev. 2008;22:2664–2676. doi: 10.1101/gad.1703008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigurdsson S, Dirac-Svejstrup AB, Svejstrup JQ. Evidence that transcript cleavage is essential for RNA polymerase II transcription and cell viability. Mol Cell. 2010;38:202–210. doi: 10.1016/j.molcel.2010.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanny JC, Erdjument-Bromage H, Tempst P, Allis CD. Ubiquitylation of histone H2B controls RNA polymerase II transcription elongation independently of histone H3 methylation. Genes Dev. 2007;21:835–847. doi: 10.1101/gad.1516207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubukata T, Shimizu T, Adachi N, Sekimizu K, Nakanishi T. Cleavage, but not read-through, stimulation activity is responsible for three biologic functions of transcription elongation factor S-II. J Biol Chem. 2003;278:8580–8585. doi: 10.1074/jbc.M211384200. [DOI] [PubMed] [Google Scholar]
- Weake VM, Workman JL. Histone ubiquitination: triggering gene activity. Mol Cell. 2008;29:653–663. doi: 10.1016/j.molcel.2008.02.014. [DOI] [PubMed] [Google Scholar]
- Wyce A, X. T, Whelan KA, Kosman C, Walter W, Eick D, Hughes TR, Krogan NJ, Strahl BD, Berger SL. H2B ubiquitylation acts as a barrier to Ctk1 nucleosomal recruitment prior to removal by Ubp8 within a SAGA-related complex. Mol Cell. 2007;27:275–288. doi: 10.1016/j.molcel.2007.01.035. [DOI] [PubMed] [Google Scholar]
- Xiao T, Kao CF, Krogan NJ, Sun ZW, Greenblatt JF, Osley MA, Strahl BD. Histone H2B ubiquitylation is associated with elongating RNA polymerase II. Mol Cell Biol. 2005;25:637–651. doi: 10.1128/MCB.25.2.637-651.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Nakanishi T, Sekimizu K, Natori S. Cloning and identification of testis-specific transcription elongation factor S-II. J Biol Chem. 1994;269:3100–3103. [PubMed] [Google Scholar]
- Zhang XY, Varthi M, Sykes SM, Phillips C, Warzecha C, Zhu W, Wyce A, Thorne AW, Berger SL, McMahon SB. The putative cancer stem cell marker USP22 is a subunit of the human SAGA complex required for activated transcription and cell-cycle progression. Mol Cell. 2008;29:102–111. doi: 10.1016/j.molcel.2007.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Lang G, Ito S, Bonnet J, Metzger E, Sawatsubashi S, Suzuki E, Le Guezennec X, Stunnenberg HG, Krasnov A, et al. A TFTC/STAGA module mediates histone H2A and H2B deubiquitination, coactivates nuclear receptors, and counteracts heterochromatin silencing. Mol Cell. 2008;29:92–101. doi: 10.1016/j.molcel.2007.12.011. [DOI] [PubMed] [Google Scholar]
- Zhu B, Zheng Y, Pham AD, Mandal SS, Erdjument-Bromage H, Tempst P, Reinberg D. Monoubiquitination of human histone H2B: the factors involved and their roles in HOX gene regulation. Mol Cell. 2005;20:601–611. doi: 10.1016/j.molcel.2005.09.025. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.