Abstract
Thefood grade bacterium Lactococcus lactis is a potential vehicle for protein delivery in the gastrointestinal tract. As a model, we constructed lactococcal strains producing antigens of infectious bursal disease virus (IBDV). IBDV infects chickens and causes depletion of B-lymphoid cells in the bursa of Fabricius and subsequent immunosuppression, morbidity, or acute mortality. The two major IBDV antigens, i.e., VP2 and VP3, that form the viral capsid were expressed and targeted to the cytoplasm, the cell wall, or the extracellular compartment of L. lactis. Whereas VP3 was successfully targeted to the three compartments by the use of relevant expression and export vectors, VP2 was recalcitrant to export, thus confirming the difficulty of translocating naturally nonsecreted proteins across the bacterial membrane. This defect could be partly overcome by fusing VP2 to a naturally secreted protein (the staphylococcal nuclease Nuc) that carried VP2 through the membrane. Lactococcal strains producing Nuc-VP2 and VP3 in various bacterial compartments were administered orally to chickens. The chickens did not develop any detectable immune response against VP2 and VP3 but did exhibit an immune response against Nuc when Nuc-VP2 was anchored to the cell wall of lactococci.
Lactic acid bacteria (LAB) constitute an important group of industrial microorganisms that have long been used for fermentation and preservation of a broad range of food products. Intensive studies of the fundamental mechanisms of LAB biology in physiology and genetics have provided opportunities for the use of these bacteria in new applications (26, 41). One of them is the use of LAB as vehicles with which to deliver biologically active molecules such as enzymes, antigens, or therapeutic drugs into humans and animals.
One of the most investigated and challenging new fields of application for LAB is their use as antigen delivery vehicles for mucosal immunization (29, 41, 49). The mucosa, which provides areas of contact with the outside environment, is the entry route of many pathogens, including viruses, bacteria, and parasites. Pathogen proliferation and/or penetration on or through mucosal surfaces is prevented by (i) the physical barrier provided by the mucosal epithelium itself and by the mucosal physicochemical environment (mucus layer, peristalsis, acidity, enzymes), (ii) the normal mucosal microflora that prevents development of exogenous microorganisms, and (iii) the mucosal immune system. While the first two mechanisms are nonspecific, the mucosal immune system is highly specialized and involves different types of cells that establish specific responses against pathogens (22). The mucosal immune response is characterized by the synthesis of large amounts of class A immunoglobulins (IgA) that are secreted onto the mucosal surfaces and have a major role in the clearance of pathogens (9). Mucosal (as well as systemic) immunization can be achieved by presenting antigens at mucosal sites. LAB are well suited for this purpose. They are organisms that are generally regarded as safe and are regularly and widely ingested by humans and animals through food products, and many LAB species are members of the normal gut microflora of humans and animals.
An intermediate step for the use of LAB in new applications, such as vaccine delivery, is the development of expression systems for stable production of heterologous proteins. We have recently designed a protein-targeting system that allows the targeting of a reporter protein to the cytoplasm, the cell wall, or the extracellular medium of various LAB species (10). With this system, we are investigating the potential of the best-characterized LAB, Lactococcus lactis, to be used as a live mucosal vaccine against chicken infectious bursal disease virus (IBDV). IBDV is the causative agent of a highly contagious chicken disease known as Gumboro (42). It belongs to the family of Birnaviridae, which consists of naked viruses characterized by a bisegmented, bistranded RNA genome (11). The largest double-stranded segment (segment A) harbors an open reading frame that encodes a precursor polyprotein. Its self-processing yields viral proteins VP2, VP3, and VP4 (21). VP2 and VP3 form the viral capsid of IBDV, and VP4 is the maturation protease of the polyprotein. IBDV infects young chickens through the digestive tract and massively destroys B cells in the bursa of Fabricius, a primary lymphoid organ, causing immunosuppression and death. Surviving birds are severely immunocompromised and more susceptible to other avian pathogens (38). IBDV is therefore of major concern in the poultry industry. Vaccination methods currently used in poultry industries consist of individual subcutaneous or intramuscular injection of inactivated IBDV into birds (1). A method that incorporates the vaccine in the drinking water also exists, but this method is only effective when live (attenuated) IBDV strains are used. Infection with these vaccine strains results in bursa damage. In the case of the more potent vaccine, severe depletion of B-lymphoid cells is observed, which results in immunosuppression, leading to an impaired immune response to other vaccinations and greater vulnerability to opportunistic infections (1, 31). Alternatively, both recombinant vaccines and DNA vaccines against IBDV are under investigation (6, 27).
Here we report the expression of the major IBDV antigens VP2 and VP3 in L. lactis. VP2 and VP3 were targeted to the cytoplasm, the cell wall, and the culture medium of L. lactis. The recombinant lactococci were used for oral immunization of chickens, and the immune response was monitored. In this first report of a trial that used antigens producing LAB in production animals such as chickens, possible improvements of this vaccination strategy are discussed.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli strains were grown in Luria broth (39) supplemented with thymine at 37°C with shaking. L. lactis strains were grown in M17 medium (46) at 30°C without shaking. When appropriate, antibiotics were added as follows: for E. coli, erythromycin (150 μg/ml) and ampicillin (100 μg/ml); for L. lactis, erythromycin (5 μg/ml).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Source or reference |
|---|---|---|
| Strains | ||
| E. coli TG1 | 39 | |
| L. lactis MG1363 | Plasmid-free strain | 16 |
| Plasmids | ||
| pHB-22R | Apr; A-segment cDNA of IBDV strain D6948 cloned into pGEM-T (Promega) | 5 |
| pVE5506 | Emr; pIL252 derivative | 10 |
| pVE5523 | Apr Emr; pBS::pIL252::ttrpA::P59::spUsp45-nucA-t1t2 | 10 |
| pVE5524 | Apr Emr; pBS::pIL252::ttrpA::P59::spUsp45-nucA-cwaM6-t1t2 | 10 |
| pVE5529 | Apr Emr pBS::pIL252::ttrpA::P59::nucA-t1t2 | 10 |
| pVE5588 | Apr Emr; pBS::pIL252::ttrpA::P59::vp2-t1t2 | This work |
| pVE5539 | Apr Emr; pBS::pIL252::ttrpA::P59::spUsp45-nucA-vp2-t1t2 | This work |
| pVE5586 | Apr Emr; pBS::pIL252::ttrpA::P59::spUsp45-vp2-t1t2 | This work |
| pVE5540 | Apr Emr; pBS::pIL252::ttrpA::P59::spUsp45-nucA-vp2-cwaM6-t1t2 | This work |
| pVE5587 | Apr Emr; pBS::pIL252::ttrpA::P59::spUsp45-vp2-cwaM6-t1t2 | This work |
| pVE5541 | Apr Emr; pBS::pIL252::ttrpA::P59::spUsp45-vp3-t1t2 | This work |
| pVE5542 | Apr Emr; pBS::pIL252::ttrpA::P59::spUsp45-vp3-cwaM6-t1t2 | This work |
| pVE5543 | Apr EMr; pBS::pIL252::ttrpA::P59::vp3-t1t2 | This work |
pBS refers to pBSII-KS+ (Stratagene), ttrpA refers to the trpA operon terminator of E. coli (8), P59 refers to a lactococcal promoter (48), spUsp45 refers to the signal peptide of Usp45 (47), nucA refers to the A form of staphylococcal nuclease (30), and t1t2 refers to the rrnB operon terminators of E. coli (32).
DNA manipulation and transformation procedures.
General molecular biology techniques were performed essentially as previously described (39). Plasmid DNA was extracted as previously described for E. coli (4) and L. lactis (33). Plasmids were established in L. lactis by electroporation (24) and in E. coli by heat shock (39).
Cloning of IBDV segment A cDNA.
We used cDNA from highly virulent IBDV strain D6948 (5). Generation of the full-length cDNA of segment A was previously described (5). The cDNA was cloned into a pGEM-T vector (Promega), and the resulting pHB22 plasmid was established in E. coli (5).
Cloning strategy in L. lactis.
We previously designed vectors that allowed the targeting of Staphylococcus aureus nuclease (Nuc) to the cytoplasm, the cell wall, and the medium of L. lactis cultures (10). In those vectors, the nuc gene is flanked by unique SalI and EcoRV sites at its 5′ and 3′ ends, respectively. To clone genes encoding the IBDV antigens, we replaced nuc in the targeting vectors with vp2 and vp3 after SalI and EcoRV digestion.
Cloning of vp3.
The vp3 gene was PCR amplified from pHB22 (5) with primers VP3a (5′ CGCGACTGTCGACCGTTTTCCTCACAATCCA) and VP3b (5′ GCCAGTCGATATCTCCTCAAGGTCCTCATCAGA). The latter unraveling of the exact cleavage site between VP3 and VP4 indicated that these primers actually amplify a portion of vp4 resulting in the addition of 33 amino acids from VP4 to the N terminus of VP3 (25). VP3a bears a SalI site (in bold) at the 5′ end of the sequence derived from the vp3 gene and a tail (in italics) at its 5′ terminus. VP3b harbors an EcoRV site (in bold) adjacent to the sequence derived from the vp3 gene and a 5′ tail (in italics). The PCR product was digested with SalI and EcoRV and cloned into SalI- and EcoRV-digested pVE5523 (secretion vector), pVE5524 (cell wall anchoring vector), and pVE5529 (cytoplasmic vector) to yield plasmids pVE5541, pVE5542, and pVE5543, respectively. These plasmids were then used to transform L. lactis cells.
Cloning of vp2.
The vp2 gene was PCR amplified from plasmid pHB22 with primers VP2a (5′GGTCGGAGTCGACTGGTTAGTAGAGATCAGACAAA [the SalI site is shown in bold, and the 5′ tail is in italics]) and VP2b (5′GCCAGTCGATATCTCCCTTAGGGCCCGGATTA [the EcoRV site is in bold, and the 5′ tail is in italics]). The PCR product was restricted with SalI and EcoRV and cloned into pVE5529, pVE5523, and pVE5524 digested with the same enzymes to yield plasmids pVE5588, pVE5586, and pVE5587, respectively. To construct pVE5539, which harbors the spUsp45-nuc-vp2, plasmid pVE5586 was linearized with SalI and then treated with mung bean nuclease. A second digestion with NheI generated a 4,071-bp fragment that was ligated to the 5,656-bp EcoRV-NheI fragment of pVE5523. To construct pVE5540, which harbors the spUsp45-nuc-vp2-cwaM6 fusion, pVE5587 was linearized with SalI, treated with mung bean nuclease, and digested with BsgI. The resulting 2,643-bp fragment was ligated to the 7,618-bp EcoRV-BsgI fragment from pVE5524.
Cell fractionation, protein extraction, and Western blot analysis.
Medium, cell wall, and protoplast fractionation and protein extractions were performed as previously described (33). Briefly, 2 ml of exponential-phase cultures (optical density at 600 nm [OD600], 0.6 to 0.8) was microcentrifuged at 4°C for 3 min at 15,000 × g. The supernatant and the cell pellet were processed separately. The supernatant was filtered through 0.2-μm-pore-size filters (low protein retention; Millipore) for bacterial removal, and proteins from 1.6 ml of the filtrate were precipitated with 400 μl of ice-cold 80% (wt/vol) trichloroacetic acid (16% final concentration). The mixture was kept on ice for 20 min and then microcentrifuged at 4°C for 15 min at 15,000 × g. The resulting pellet was dissolved at 80 μl per OD600 unit in 50 ml of NaOH containing 4-(2-aminoethyl)benzenesulfonyl fluoride hydrochloride (AEBSF; 2 mM) as a protease inhibitor. The cell pellet was washed once with TES (10 mM Tris-HCl [pH 5.8], 1 mM EDTA, 25% sucrose) containing chloramphenicol (50 μg/ml) as an inhibitor of protein synthesis. The bacterial cell walls were digested with 500 μl of TES containing lysozyme (0.5 mg/ml), mutanolysin (0.1 mg/ml), RNase (0.1 mg/ml), and AEBSF (2 mM). After 1 h of incubation at 37°C, the protoplasts were recovered by a 3-min centrifugation at 15,000 × g and 4°C and then washed with TES plus chloramphenicol. The pellet was resuspended at 100 μl per OD600 unit in Tris-HCl [pH 7.4]-1 mM EDTA (TE) containing 1% sodium dodecyl sulfate (SDS) for protoplast lysis. The digested cell walls were recovered from the supernatant of the above-described centrifugation and filtered through 0.2-μm-pore-size filters, and proteins from 400 μl were precipitated with 16% trichloroacetic acid as described above. The resulting pellet was dissolved at 80 μl per OD600 unit in 50 mM NaOH containing 2 mM AEBSF. Equal volumes of 2× loading buffer were added to all samples. Extracts were subjected to SDS-polyacrylamide gel electrophoresis (12% acrylamide) (23). Electroblotting on polyvinylidene difluoride membranes (Millipore) and antibody reactions and detection (enhanced chemiluminescence) were performed in accordance with the manufacturer's recommendations. Relative amounts of VP2 and VP3 in the different fractions were determined by scanning Western blots on a PhosphorImager (Amersham). Anti-NucA polyclonal antibodies were kindly provided by James Miller. Preparation of 9.7 anti-VP3 monoclonal antibodies was done as described elsewhere (5). Anti-VP2 monoclonal antibodies were a kind gift of H. Mueller.
Chicken immunization.
Six groups of 10 specific-pathogen-free chickens were housed separately. At the age of 28 days, each chicken in each group received orally 1 ml of M17 containing 109 viable lactococci for 5 consecutive days. At the age of 42 days, each chicken of each group again received orally 1 ml of M17 containing the same 109 viable lactococci for 5 consecutive days. Blood samples were taken from each chicken when it was 28, 42, and 49 days old. Group 1 received VE5611 (secreted NucA), group 2 received VE5612 (cell wall-anchored NucA), group 3 received VE5662 (secreted VP3), group 4 received VE5663 (cell wall-anchored VP3), group 5 received VE5670 (secreted Nuc-VP2 fusion), and group 6 received VE5671 (cell wall-anchored Nuc-VP2). Sera of the collected samples were stored at −20°C and analyzed for the presence of IgG antibodies against NucA, IBDV (IDEXX), and VP3 by enzyme-linked immunosorbent assay.
RESULTS
We have recently designed a protein-targeting system that allows the expression of a heterologous enzyme, staphylococcal Nuc, at three cellular locations in L. lactis and other LAB (10). The system comprises (i) the strong lactococcal promoter P59 (48), (ii) the signal peptide from Usp45, the major secreted protein in L. lactis (47) for the secretion of fusion proteins, and (iii) the cell wall anchor motif from Streptococcus pyogenes M6 protein (19) for sortase-mediated cell wall anchoring (28). The combination of these tools enabled the design of vectors suitable for the expression of Nuc in the cytoplasm, the cell wall, or the culture medium of LAB; the vectors are designated below as cytoplasmic, cell wall-anchoring, and secretion vectors, respectively (Fig. 1). In an attempt to target IBDV antigens in L. lactis, we cloned the genes encoding VP2 and VP3 into these vectors.
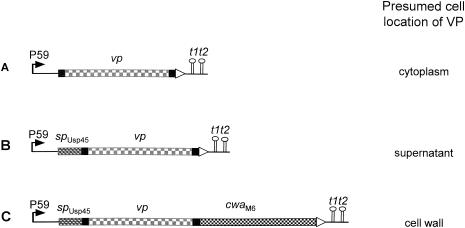
FIG. 1.
Fusion genes constructed and expressed and presumed cell localization of IBDV antigens. P59, lactococcal promoter; spUsp45, signal sequence from the Usp45 preprotein; vp, structural gene for viral protein VP2 or VP3; cwaM6, sequence specifying the cell wall anchor domain from the M6 preprotein; t1t2, transcriptional terminators.
VP3 can be targeted to three cell compartments in L. lactis.
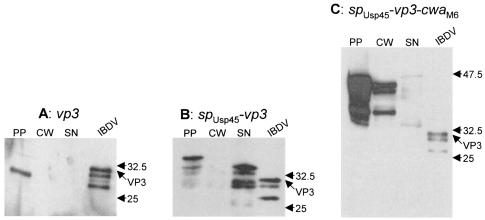
A PCR fragment specifying the vp3 gene was cloned into the cytoplasmic, the secretion, and the cell wall-anchoring vectors. The resulting plasmids were established in L. lactis cells, and the expression of fusion proteins was analyzed by Western blotting. Protoplast, cell wall, and supernatant fractions prepared from log-phase cultures were subjected to SDS-polyacrylamide gel electrophoresis and revealed with polyclonal antibodies raised against VP3.
(i) Cytoplasmic-fraction targeting.
One band with the expected size of VP3 (33 kDa) was detected in the protoplast fraction (Fig. 2A). As expected, no signal was revealed in either the cell wall or the supernatant fraction. This validates the protocol used for protein fractionation in that no cytoplasmic proteins leak into the cell wall and supernatant fractions.
FIG. 2.
Analysis of L. lactis strains expressing VP3. Proteins from log-phase growing cultures were fractionated and analyzed by Western blotting with 9.7 anti-VP3 monoclonal antibodies. spUsp45, signal peptide from the Usp45 preprotein; cwaM6, cell wall anchor domain from the M6 preprotein; PP, protoplast; CW, cell wall; SN, supernatant; IBDV, purified IBDV. The positions of full-length VP3 and molecular mass standards (kilodaltons) are indicated.
(ii) External-medium targeting.
In the protoplast fraction of cells harboring the VP3 secretion vector, one major band that corresponds in size to the SPUsp45-VP3 precursor (37 kDa) accounted for about 20% of the total signals detected (Fig. 2B). One additional faint band that corresponded in size to mature VP3 (34 kDa) released from the SPUsp45-VP3 precursor and a smear indicating additional cytoplasmic degradation of VP3 were revealed. No signal was detected in the cell wall fraction of cells expressing the secreted fusion of VP3. The two upper bands migrated more slowly than the control viral VP3, possibly because of 45 additional amino acids at the N terminus of the recombinant VP3 protein: in the SPUsp45-VP3 construct, 12 amino acids were introduced downstream of the cleavage site of the Usp45 signal peptide in order to both improve secretion efficiency and create cloning sites (10); also, 33 amino acids derived from the adjacent VP4 protein in the IBDV polyprotein were included. About 80% of the signal was present in the supernatant fraction as one major band and three lower-molecular-weight bands (Fig. 2B). The largest band that migrated at a distance similar to that of the faint band in the protoplast fraction corresponds to VP3. The second lower band detected in the supernatant fraction is most likely a degradation product of the higher band. Probably, the amino acid tails introduced at the N terminus of VP3 are highly susceptible to proteolysis because of a folding defect. Additional proteolysis targeted sites within the VP3 sequence and yielded two additional degradation products. From this hypothesis, the two higher bands that reflect the whole VP3 sequence account for about 70% of the total supernatant signal. This shows that VP3 could be efficiently secreted in L. lactis although some degradation occurred.
(iii) Cell wall targeting.
Cell wall targeting of VP3 was assessed in L. lactis expressing the spUsp45-vp3-cwaM6 fusion. CWAM6 comprises (i) 35 amino acids that are necessary for anchoring and are cleaved off upon tethering of the protein to the peptidoglycan and (ii) a 105-residue upstream sequence used as a spacer to display the protein of interest outside of the cell wall layer (10). Western blot analysis revealed bands in the cell wall fraction in the range of 35 to 47 kDa; very little VP3 protein was detected in the supernatant (Fig. 2C). The higher band in the cell wall fraction is likely to correspond to the full-length anchored VP3 fusion (theoretical molecular mass, 49 kDa); the other bands might result from proteolysis, as previously observed when Nuc was expressed at the surface of L. lactis (10). This result shows that some proportion of VP3 could be targeted to the L. lactis cell wall. Nevertheless, the majority (about 80%) of signals were present in the protoplast fraction (Fig. 2C). These bands probably correspond to the SPUsp45-VP3-CWAM6 precursor (the higher band) and to degradation products. Comparison of the secreted (Fig. 2B) and anchored (Fig. 2C) forms of VP3 indicates that a smaller proportion of total VP3 is cell wall anchored than secreted.
In summary, IBDV VP3 antigen can be targeted to three cell compartments in L. lactis. VP3 could be stably produced in the cytoplasm. When fused to suitable signals, about 80% of the VP3 could be secreted and about 20% could be anchored to the cell wall, although some proteolysis occurred in both cases. The protocol used for protein fractionation appeared efficient, as each VP3 species that was targeted to a defined cell compartment provided a typical pattern that was not found in the other fractions. This demonstrates that no contamination between the different fractions occurred.
VP2 failed to be secreted in L. lactis.
The vp2 gene was cloned in frame into cytoplasmic and secretion vectors, and its expression in L. lactis was analyzed by Western blotting of fractionated samples with antibodies raised against VP2. As expected, cells harboring the cytoplasmic vector generated a unique band in the protoplast fraction (data not shown). However, in cells harboring the secretion vector, the only detected band was in the protoplast fraction as well (Fig. 3A); it migrated at the expected position of SPUsp45-VP2 (54 kDa). No signal was present in either the cell wall or the supernatant fraction. This result suggests that VP2 could not be exported in L. lactis. As VP2 is naturally expressed in a cytoplasmic context in IBDV, one can imagine that despite the addition of SPUsp45, VP2 remained in a state incompetent for membrane translocation.
FIG. 3.
Analysis of L. lactis strains expressing VP2 fusions. Proteins from log-phase growing cultures were fractionated and analyzed by Western blotting with anti-VP2 (A) or anti-Nuc (B and C) antibodies. spUsp45, signal peptide from the Usp45 preprotein; cwaM6, cell wall anchor domain from the M6 preprotein; PP, protoplast; CW, cell wall; SN, supernatant; IBDV, purified IBDV; Nuc, S. aureus nuclease. The positions of pre-VP2 (pVP2) and VP2 and molecular mass standards (kilodaltons) are indicated.
The fusion of VP2 to staphylococcal Nuc allows its secretion and cell wall anchoring.
We suspected that fusion of VP2 to a readily secreted protein, staphylococcal Nuc, could enhance secretion efficiency. To test this possibility, we expressed an spUsp45-nuc-vp2 fusion in L. lactis. Western blot analysis with antibodies raised against Nuc showed one major band and a few faster-migrating weak bands in the supernatant (Fig. 3B). The distribution of the fusion protein was about 80 and 20% in the protoplast and supernatant fractions, respectively. This suggests that fusion of VP2 with Nuc facilitates its export. Nevertheless, the detected species in both the protoplast and supernatant fractions were smaller than the expected sizes (theoretical molecular mass of 69 kDa for Nuc-VP2), suggesting that degradation occurs. As degradation products are mainly observed in the protoplast fraction, it is likely that Nuc-VP2 is degraded before its export. To further characterize the degradation events, we blotted the same membrane with antibodies raised against VP2. No signal was detected (data not shown), suggesting that the proteolysis targets the VP2 sequence.
For assessment of VP2 targeting to the L. lactis cell wall, we constructed the spUsp45-nuc-vp2-cwaM6 fusion. Its expression product was analyzed in L. lactis by Western blotting with antibodies against Nuc. The protoplast fraction produced one strong band that corresponds to the 86-kDa precursor protein SPUsp45-Nuc-VP2-CWAM6 (Fig. 3C). The cell wall fraction displayed a strong band of the size expected for the anchored species (84 kDa), which represented ∼20% of the signals detected in the three cell compartments. This proportion is the same as that of secreted Nuc-VP2 obtained as described above, which suggests that, once translocated across the membrane, the Nuc-VP2 fusion is efficiently targeted to the cell wall of L. lactis. Interestingly, anchored Nuc-VP2 appears to be more stable than the secreted fusion since its migration corresponds to the full-size protein. Possibly, the addition of CWAM6 partly protects VP2 against proteolysis. However, some cell surface proteolysis may occur, as suggested by the presence of a low-molecular-weight compound in the supernatant fraction. Three factors suggest that this proteolysis occurs within theVP2 sequence. First, this sequence is, as shown above, highly susceptible to proteolysis. Second, the degradation compound still reacts with Nuc antibodies. Third, the topology of the Nuc-VP2-CWAM6 fusion strongly suggests that its release into the supernatant results from C-terminal degradation as it is anchored to the cell wall by its C terminus. Since little proteolysis within CWAM6 occurred in the above-described studies with VP3 (Fig. 2C), VP2 was probably the major target for proteolysis.
Chicken immune response to recombinant lactococci.
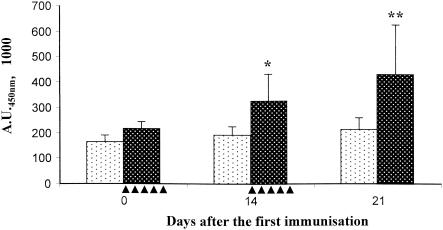
The immunogenicity of recombinant L. lactis strains expressing IBDV antigens or Nuc was evaluated after oral administration to chickens. The birds received 109 cells daily for 5 successive days, and the same administration protocol was repeated 2 weeks later. Specific serum IgG production was analyzed at days 0, 14, and 21 following the first administration. The strain expressing the cell wall-anchored Nuc-VP2 fusion induced a weak but significant IgG response against Nuc (Fig. 4). This response appeared at day 14 after the first administration (P < 0.02) and increased at day 21 postimmunization (P < 0.016), which corresponds to 1 week after the booster immunization (Fig. 4). However, we did not detect any serum IgG response against VP2 (data not shown). Also, no immune response was detected after immunization of chickens with L. lactis expressing the other antigens tested in this study (data not shown).
FIG. 4.
Serum IgG response to Nuc. Ten chickens were immunized with 109 bacteria (black dots, negative control [strain VE5505 containing pVE5502]; white dots, strain VE5671 containing pVE5540 and producing anchored Nuc-VP2) for 5 consecutive days starting at days 0 and 14 (indicated by filled triangles). Individual serum samples were collected at days 0 (before the first administration), 14, and 21 and tested by enzyme-linked immunosorbent assay for NucA-specific IgG. *, P < 0.02; **, P < 0.016 (Student's t test). A.U.450nm, units of absorption at 450 nm.
DISCUSSION
In a preceding work, we have designed a system for targeting of a reporter protein to the cytoplasm, the cell wall, or the supernatant culture of LAB (10). With this system, our ultimate objective is to study the host immune response in relation to antigen localization in the LAB vehicle. In the present work, we decided to target the two major antigens from chicken IBDV to different cell compartments of L. lactis and use the recombinant lactococci to immunize chickens by oral administration.
Targeting of IBDV antigens in L. lactis. (i) VP2 and VP3 are stable in the cytoplasm of L. lactis.
VP2 and VP3 were successfully produced in the cytoplasm and appeared as unique bands of the expected sizes, suggesting that no proteolysis occurred. We attribute this stability of VP2 and VP3 in the cytoplasm to their rapid folding, which would provide them with a structure resistant to cytoplasmic proteases. Indeed, the compact conformational structure and multimerization of VP2 and VP3 are supported by studies with yeast (45). However, as previously observed with other proteins in L. lactis, VP2 and VP3 were produced at a lower yield than their secreted and cell wall-anchored counterparts (Fig. 2 and 3) (3, 10, 12, 37). No explanation for this finding has been found.
(ii) Secretion of NNSP versus their degradation.
Naturally nonsecreted proteins (NNSP) are known to be difficult to secrete. This is attributed to their tendency to fold rapidly in the cytoplasm, which prevents their translocation across the cytoplasmic membrane (35). A first way to delay their folding and keep them in the unfolded state required for translocation is to fuse them with a signal peptide. In an attempt to drive secretion of the two NNSP VP2 and VP3, we used the signal peptide from Usp45 (the most-secreted protein in L. lactis), followed by negatively charged amino acids downstream of the cleavage site. This combination has previously been shown to be the most efficient in LAB for secretion of heterologous naturally secreted proteins (10). With this system, efficient secretion of VP3 (80%) was obtained. This is a particularly high level for an NNSP compared with other viral proteins that were secreted much less in L. lactis (12, 37). On the other hand, fusion of VP3 with SPUsp45 also induced VP3 degradation both at the precursor level in the protoplast and at the mature-protein level in the extracellular medium. As native VP3 was stable in the cytoplasm of L. lactis (Fig. 2A), we attribute the cytoplasmic degradation to the addition of SPUsp45, which would impede VP3 folding, rendering it more susceptible to proteolysis. It is likely that cytoplasmic and extracellular degradation is mediated by ClpP and HtrA, two housekeeping proteases located in the cytoplasm and on the outer side of the membrane of L. lactis, respectively (14, 34).
Fusion of VP2 to SPUsp45 failed to drive VP2 secretion (Fig. 3A). This suggests that this Usp45 signal peptide is not sufficient to prevent VP2 folding and/or multimerization in the cytoplasm of L. lactis. It has been shown in both Escherichia coli and Bacillus subtilis that the protein-folding process can be further delayed (and protein secretion can be improved) by overexpression of chaperones proteins (40, 51). We first chose another strategy, which consisted of fusing VP2 to the C terminus of naturally secreted S. aureus Nuc. Our rationale was based on the postulate that addition of Nuc might delay the folding of VP2, thus maintaining the fusion protein in an export-competent state. To a certain extent, this strategy appeared successful, allowing a secretion efficiency of 20% for the Nuc-VP2 fusion. This suggests that an NNSP such as VP2 that is totally recalcitrant to translocation in its native form can be partly carried through the secretion machinery when it is fused to a naturally secreted protein such as Nuc. This may find many applications in the development of protein delivery by LAB, as some candidate proteins, including antigens and enzymes, are not naturally secreted in their native organism. An adverse effect of the partial secretion of Nuc-VP2 was observed in protein degradation that concerned the VP2 moiety of the fusion. Altogether, these results show that there are conflicting interests in retarding protein folding; i.e., translocation may occur, but the unfolded protein is more prone to proteolysis.
(iii) Cell wall anchoring of NNSP and protection against proteolysis.
Both Nuc-VP2 and VP3 could be anchored to the cell wall of L. lactis. Although the proportion of precursor processed to the cell wall (20%) was the same for both antigens, we believe that the respective bottlenecks hampering cell wall anchoring of VP2 and VP3 are different. In the case of Nuc-VP2, we observed the same level of processing in the case of secretion and cell wall anchoring. As membrane translocation is a prerequisite for cell wall anchoring, we believe that, in the case of Nuc-VP2, the limiting step for cell wall anchoring is translocation of the precursor through the membrane. In contrast, in the case of VP3, the secretion level was 80% and the cell wall-anchoring level was 20%. This is reminiscent of the observation previously made with Nuc (10). In this case, we showed that the defect in cell wall anchoring is due not to a translocation defect but rather to too low an activity of sortase, the transpeptidase tethering the protein to the cell wall. We believe that the same phenomenon explains why some VP3 cannot anchor to the cell wall. Probably, the sortase level in L. lactis is sufficient to anchor the low level of exported Nuc-VP2 but insufficient to anchor VP3, which is exported at a higher level.
Interestingly, cell wall anchoring of Nuc-VP2 allowed its partial protection against proteolysis. Whereas all of the Nuc-VP2 secreted had the VP2 portion truncated, most of the cell wall-anchored fusion appeared as the entire Nuc-VP2 fusion. Although we have no explanation for this observation, it could be valuable to stabilize proteins that are highly susceptible to proteolysis when they are exported outside the cell.
Serum antibody response to oral administration of IBDV antigen-producing L. lactis.
IBDV antigen-producing lactococci were used to investigate the potential of L. lactis for vaccine delivery into chickens. This represents a first step toward the development of a new strategy for vaccination against IBDV. Such a vaccination strategy, with LAB as the vehicle for oral administration of the vaccine through food or water, would be highly attractive because of its low cost, its safety, and the absence of immunosuppression.
We chose VP2 and VP3 as antigens to be expressed in L. lactis as these proteins are constituents of the IBDV capsid and represent 90% of the total IBDV proteins (11). VP2 contains the major epitopes that elicit neutralizing antibodies (13), while VP3 has been reported to bear minor neutralizing sites (20).
The lactococcal strains designed and characterized as described above were used for oral vaccination of 28-day-old specific-pathogen-free chickens. It is noteworthy that strains expressing exported forms (secreted or cell wall anchored) of VP2 and VP3 also accumulated significant amounts of antigen in their protoplast. Consequently, antigens were, in fact, located in two cell compartments. Following oral vaccination of chickens, only bacteria producing an anchored Nuc-VP2 fusion induced a systemic and specific response against Nuc but not against VP2. This is the first demonstration that oral administration of antigen-producing LAB can promote an immune response in production animals such as chickens, as the few studies performed in this field have been conducted with mice (29). However, these results raise two questions. (i) Why did only Nuc fused to VP2 and cell wall anchored induce an immune response, and (ii) why did only Nuc, and not VP2 or VP3, induce an immune response? It has been shown previously that cell wall-anchored antigens are more immunogenic in mice than are their cytoplasmic or secreted counterparts (36, 50). A hypothesis is that the expression level of the designed fusion genes is situated at the threshold required to elicit an immune response. Therefore, only the most immunogenic form, i.e., the cell wall-anchored form, of Nuc-VP2 would have allowed an immunological response of the chickens. A parameter other than the expression level did occur as lactococci expressing cell wall-anchored Nuc on its own with the same expression system were not immunogenic. Possibly, this may be due to the topology of the Nuc-VP2 fusion at the cell wall. As the fusion is anchored by its C terminus, VP2 could have a spacer role allowing the Nuc moiety to be displayed outside the peptidoglycan. This would allow better contact with immune cells. In this case, it would also explain why no response against VP2 and VP3 was observed as these antigens would be buried in the bacterial peptidoglycan layer.
Another possible explanation is that the absence of serum antibody reflects differential mucosal and systemic immune responses. This hypothesis is based on recent observations in mice after oral administration of L. lactis cells expressing bovine β-lactoglobulin (7). Significant levels of specific anti β-lactoglobulin IgA were found in feces, while all of the Ig classes tested (IgA, IgG1, IgG2a, and IgE) were absent in serum. A similar imbalance toward an IgA response might account for our results.
In the emerging field of antigen delivery by LAB, further work is needed to better understand and improve the mechanisms governing chicken immune responses. Several lines of improvement exist, including (i) antigen stabilization in LAB by down regulation of housekeeping protease genes (15, 34), (ii) better protein export by overexpression of chaperones, and (iii) improvement of protein anchoring to the cell wall by sortase overexpression. Also, improvements in the interactions between the bacteria and the host immune system will consist of (i) coexpression of both antigens and adjuvants in LAB as L. lactis is able to produce fully active cytokines (2, 43, 44) and (ii) the use of LAB able to colonize the chicken gut (17, 18). Probably, a combination of improvements along these lines will be necessary to make LAB serious candidates for live-vaccine development.
Acknowledgments
We thank B. Delmas, A. Gruss, P. Langella, A. ter Huurne, and W. Boersma for stimulating discussions. We are grateful to J. Miller and H. Mueller for the gifts of antibodies.
This work was supported by a Van Gogh program. Y.D. was the recipient of a joint grant from ID-DLO, INRA, and Fondation pour la Recherche Médicale (Paris, France).
REFERENCES
- 1.Baxendale, W. 1996. Current methods of delivery of poultry vaccines. Carfax Publishing Co., Abington, United Kingdom.
- 2.Bermudez-Humaran, L. G., P. Langella, N. G. Cortes-Perez, A. Gruss, R. S. Tamez-Guerra, S. C. Oliveira, O. S. Cardenas, R. Montes De Oca-Luna, and Y. Le Loir. 2003. Intranasal immunization with recombinant Lactococcus lactis secreting murine interleukin-12 enhances antigen-specific Th1 cytokine production. Infect. Immun. 71:1887-1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bermudez-Humaran, L. G., P. Langella, A. Miyoshi, A. Gruss, R. T. Guerra, R. Montes de Oca-Luna, and Y. Le Loir. 2002. Production of human papillomavirus type 16 E7 protein in Lactococcus lactis. Appl. Environ. Microbiol. 68:917-922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Birnboim, H. C., and J. Doly. 1979. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 7:1513-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boot, H. J., A. A. ter Huurne, A. J. Hoekman, B. P. Peeters, and A. L. Gielkens. 2000. Rescue of very virulent and mosaic infectious bursal disease virus from cloned cDNA: VP2 is not the sole determinant of the very virulent phenotype. J. Virol. 74:6701-6711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang, H. C., T. L. Lin, and C. C. Wu. 2003. DNA vaccination with plasmids containing various fragments of large segment genome of infectious bursal disease virus. Vaccine 21:507-513. [DOI] [PubMed] [Google Scholar]
- 7.Chatel, J. M., P. Langella, K. Adel-Patient, J. Commissaire, J. M. Wal, and G. Corthier. 2001. Induction of mucosal immune response after intranasal or oral inoculation of mice with Lactococcus lactis producing bovine β-lactoglobulin. Clin. Diagn. Lab. Immunol. 8:545-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christie, G. E., P. J. Farnham, and T. Platt. 1981. Synthetic sites for transcription termination and a functional comparison with tryptophan operon termination sites in vitro. Proc. Natl. Acad. Sci. USA 78:4180-4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corthesy, B., and J. P. Kraehenbuhl. 1999. Antibody-mediated protection of mucosal surfaces. Curr. Top. Microbiol. Immunol. 236:93-111. [DOI] [PubMed] [Google Scholar]
- 10.Dieye, Y., S. Usai, F. Clier, A. Gruss, and J.-C. Piard. 2001. Design of a protein-targeting system for lactic acid bacteria. J. Bacteriol. 183:4157-4166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dobos, P., B. J. Hill, R. Hallett, D. T. Kells, H. Becht, and D. Teninges. 1979. Biophysical and biochemical characterization of five animal viruses with bisegmented double-stranded RNA genomes. J. Virol. 32:593-605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Enouf, V., P. Langella, J. Commissaire, J. Cohen, and G. Corthier. 2001. Bovine rotavirus nonstructural protein 4 produced by Lactococcus lactis is antigenic and immunogenic. Appl. Environ. Microbiol. 67:1423-1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fahey, K. J., K. Erny, and J. Crooks. 1989. A conformational immunogen on VP-2 of infectious bursal disease virus that induces virus-neutralizing antibodies that passively protect chickens. J. Gen. Virol. 70:1473-1481. [DOI] [PubMed] [Google Scholar]
- 14.Frees, D., and H. Ingmer. 1999. ClpP participates in the degradation of misfolded protein in Lactococcus lactis. Mol. Microbiol. 31:79-87. [DOI] [PubMed] [Google Scholar]
- 15.Frees, D., P. Varmanen, and H. Ingmer. 2001. Inactivation of a gene that is highly conserved in gram-positive bacteria stimulates degradation of non-native proteins and concomitantly increases stress tolerance in Lactococcus lactis. Mol. Microbiol. 41:93-103. [DOI] [PubMed] [Google Scholar]
- 16.Gasson, M. J. 1983. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J. Bacteriol. 154:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gusils, C., S. Cuozzo, F. Sesma, and S. Gonzalez. 2002. Examination of adhesive determinants in three species of Lactobacillus isolated from chicken. Can. J. Microbiol. 48:34-42. [DOI] [PubMed] [Google Scholar]
- 18.Gusils, C., J. Palacios, S. Gonzalez, and G. Oliver. 1999. Lectin-like protein fractions in lactic acid bacteria isolated from chickens. Biol. Pharm. Bull. 22:11-15. [DOI] [PubMed] [Google Scholar]
- 19.Hollingshead, S. K., V. A. Fischetti, and J. R. Scott. 1986. Complete nucleotide sequence of type 6 M protein of the group A Streptococcus: repetitive structure and membrane anchor. J. Biol. Chem. 261:1677-1686. [PubMed] [Google Scholar]
- 20.Jagadish, M. N., and A. A. Azad. 1991. Localization of a VP3 epitope of infectious bursal disease virus. Virology 184:805-807. [DOI] [PubMed] [Google Scholar]
- 21.Jagadish, M. N., V. J. Staton, P. J. Hudson, and A. A. Azad. 1988. Birnavirus precursor polyprotein is processed in Escherichia coli by its own virus-encoded polypeptide. J. Virol. 62:1084-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kraehenbuhl, J. P., and M. R. Neutra. 1992. Molecular and cellular basis of immune protection of mucosal surfaces. Physiol. Rev. 72:853-879. [DOI] [PubMed] [Google Scholar]
- 23.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 24.Langella, P., Y. Le Loir, S. D. Ehrlich, and A. Gruss. 1993. Efficient plasmid mobilization by pIP501 in Lactococcus lactis subsp. lactis. J. Bacteriol. 175:5806-5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lejal, N., B. Da Costa, J. C. Huet, and B. Delmas. 2000. Role of Ser-652 and Lys-692 in the protease activity of infectious bursal disease virus VP4 and identification of its substrate cleavage sites. J. Gen. Virol. 81:983-992. [DOI] [PubMed] [Google Scholar]
- 26.Marteau, P., and J. C. Rambaud. 1993. Potential of using lactic acid bacteria for therapy and immunomodulation in man. FEMS Microbiol. Rev. 12:207-220. [DOI] [PubMed] [Google Scholar]
- 27.Martinez-Torrecuadrada, J. L., N. Saubi, A. Pages-Mante, J. R. Caston, E. Espuna, and J. I. Casal. 2003. Structure-dependent efficacy of infectious bursal disease virus (IBDV) recombinant vaccines. Vaccine 21:3342-3350. [DOI] [PubMed] [Google Scholar]
- 28.Mazmanian, S. K., H. Ton-That, and O. Schneewind. 2001. Sortase-catalysed anchoring of surface proteins to the cell wall of Staphylococcus aureus. Mol. Microbiol. 40:1049-1057. [DOI] [PubMed] [Google Scholar]
- 29.Mercenier, A., H. Muller-Alouf, and C. Grangette. 2000. Lactic acid bacteria as live vaccines. Curr. Issues Mol. Biol. 2:17-25. [PubMed] [Google Scholar]
- 30.Miller, J. R., S. Kovacevic, and L. E. Veal. 1987. Secretion and processing of staphylococcal nuclease by Bacillus subtilis. J. Bacteriol. 169:3508-3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nagarajan, M. M., and F. S. Kibenge. 1997. Infectious bursal disease virus: a review of molecular basis for variations in antigenicity and virulence. Can. J. Vet. Res. 61:81-88. [PMC free article] [PubMed] [Google Scholar]
- 32.Peschke, U., V. Beuck, H. Bujard, R. Gentz, and S. Le Grice. 1985. Efficient utilization of Escherichia coli transcriptional signals in Bacillus subtilis. J. Mol. Biol. 186:547-555. [DOI] [PubMed] [Google Scholar]
- 33.Piard, J.-C., I. Hautefort, V. A. Fischetti, S. D. Ehrlich, M. Fons, and A. Gruss. 1997. Cell wall anchoring of the Streptococcus pyogenes M6 protein in various lactic acid bacteria. J. Bacteriol. 179:3068-3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Poquet, I., V. Saint, E. Seznec, N. Simoes, A. Bolotin, and A. Gruss. 2000. HtrA is the unique surface housekeeping protease in Lactococcus lactis and is required for natural protein processing. Mol. Microbiol. 35:1042-1051. [DOI] [PubMed] [Google Scholar]
- 35.Randall, L. L., and S. J. Hardy. 1986. Correlation of competence for export with lack of tertiary structure of the mature species: a study in vivo of maltose-binding protein in E. coli. Cell 46:921-928. [DOI] [PubMed] [Google Scholar]
- 36.Reveneau, N., M. C. Geoffroy, C. Locht, P. Chagnaud, and A. Mercenier. 2002. Comparison of the immune responses induced by local immunizations with recombinant Lactobacillus plantarum producing tetanus toxin fragment C in different cellular locations. Vaccine 20:1769-1777. [DOI] [PubMed] [Google Scholar]
- 37.Ribeiro, L. A., V. Azevedo, Y. Le Loir, S. C. Oliveira, Y. Dieye, J.-C. Piard, A. Gruss, and P. Langella. 2002. Production and targeting of the Brucella abortus antigen L7/L12 in Lactococcus lactis: a first step towards food-grade live vaccines against brucellosis. Appl. Environ. Microbiol. 68:910-916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rosenberger, J. K., S. Klopp, R. J. Eckroade, and W. C. Krauss. 1975. The roles of the infectious bursal agent and several avian adenoviruses in the hemorrhagic-aplastic-anemia syndrome and gangrenous dermatitis. Avian Dis. 19:717-729. [PubMed] [Google Scholar]
- 39.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 40.Schäffner, J., J. Winter, R. Rudolph, and E. Schwarz. 2001. Cosecretion of chaperones and low-molecular-size medium additives increases the yield of recombinant disulfide-bridged proteins. Appl. Environ. Microbiol. 67:3994-4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seegers, J. F. 2002. Lactobacilli as live vaccine delivery vectors: progress and prospects. Trends Biotechnol. 20:508-515. [DOI] [PubMed] [Google Scholar]
- 42.Sharma, J. M., I. J. Kim, S. Rautenschlein, and H. Y. Yeh. 2000. Infectious bursal disease virus of chickens: pathogenesis and immunosuppression. Dev. Comp. Immunol. 24:223-235. [DOI] [PubMed] [Google Scholar]
- 43.Steidler, L., W. Hans, L. Schotte, S. Neirynck, F. Obermeier, W. Falk, W. Fiers, and E. Remaut. 2000. Treatment of murine colitis by Lactococcus lactis secreting interleukin-10. Science 289:1352-1355. [DOI] [PubMed] [Google Scholar]
- 44.Steidler, L., K. Robinson, L. Chamberlain, K. M. Schofield, E. Remaut, R. W. Le Page, and J. M. Wells. 1998. Mucosal delivery of murine interleukin-2 (IL-2) and IL-6 by recombinant strains of Lactococcus lactis coexpressing antigen and cytokine. Infect. Immun. 66:3183-3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tacken, M. G., P. J. Rottier, A. L. Gielkens, and B. P. Peeters. 2000. Interactions in vivo between the proteins of infectious bursal disease virus: capsid protein VP3 interacts with the RNA-dependent RNA polymerase, VP1. J. Gen. Virol. 81:209-218. [DOI] [PubMed] [Google Scholar]
- 46.Terzaghi, B., and W. Sandine. 1975. Improved medium for lactic streptococci and their bacteriophages. Appl. Environ. Microbiol. 29:807-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van Asseldonk, M., G. Rutten, M. Oteman, R. J. Siezen, W. M. de Vos, and G. Simons. 1990. Cloning of usp45, a gene encoding a secreted protein from Lactococcus lactis subsp. lactis MG1363. Gene 95:155-160. [DOI] [PubMed] [Google Scholar]
- 48.van der Vossen, J. M., D. van der Lelie, and G. Venema. 1987. Isolation and characterization of Streptococcus cremoris Wg2-specific promoters. Appl. Environ. Microbiol. 53:2452-2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wells, J. M., K. Robinson, L. M. Chamberlain, K. M. Schofield, and R. W. Le Page. 1996. Lactic acid bacteria as vaccine delivery vehicles. Antonie Van Leeuwenhoek 70:317-330. [DOI] [PubMed] [Google Scholar]
- 50.Wells, J. M., P. W. Wilson, P. M. Norton, M. J. Gasson, and R. W. Le Page. 1993. Lactococcus lactis: high-level expression of tetanus toxin fragment C and protection against lethal challenge. Mol. Microbiol. 8:1155-1162. [DOI] [PubMed] [Google Scholar]
- 51.Wu, S. C., R. Ye, X. C. Wu, S. C. Ng, and S. L. Wong. 1998. Enhanced secretory production of a single-chain antibody fragment from Bacillus subtilis by coproduction of molecular chaperones. J. Bacteriol. 180:2830-2835. [DOI] [PMC free article] [PubMed] [Google Scholar]