SUMMARY
DNA methylation at the 5-position of cytosine (5mC) in the mammalian genome is a key epigenetic event critical for various cellular processes. The Ten-eleven translocation (Tet) family of 5mC-hydroxylases, which convert 5mC to 5-hydroxymethylcytosine (5hmC), offers a way for dynamic regulation of DNA methylation. Here we report that Tet1 binds unmodified C, 5mC- or 5hmC-modified CpG-rich DNA through its CXXC domain. Genome-wide mapping of Tet1 and 5hmC reveals mechanisms by which Tet1 controls 5hmC and 5mC levels in mouse embryonic stem cells (mESCs). We also uncover a comprehensive gene network influenced by Tet1. Collectively, our data suggest that Tet1 controls DNA methylation both by binding to CpG-rich regions to prevent unwanted DNA methyltransferase activity, and by converting 5mC to 5hmC through hydroxylase activity. This Tet1-mediated antagonism of CpG methylation imparts differential maintenance of DNA methylation status at Tet1 targets, ultimately contributing to mESC differentiation and the onset of embryonic development.
INTRODUCTION
DNA methylation at the 5-position of cytosine (5mC) occurs predominantly at CpG dinucleotides in the mammalian genome and is one of the most important epigenetic marks, playing critical roles in host defense, genome imprinting, and X chromosome inactivation (Suzuki and Bird, 2008). It is well established that individual CpGs located in different genomic regions are differentially methylated depending on cell or tissue type and developmental stage. Furthermore, it is evident that the GC density and gene transcriptional status also influence DNA methylation status. For example, the majority of CpG islands (CGIs) displaying a dense CpG content are hypomethylated while the rest of the genome, including CpG-rich repetitive heterochromatin regions and dispersed CpGs in gene coding regions are usually hypermethylated. Yet it is still poorly understood how genome-wide DNA methylation is differentially regulated at discrete loci and dynamically processed in different cell types and during development.
Increasing evidence suggests that DNA methylation is intimately linked to histone methylation. For instance, it is well known that high levels of DNA methylation at GC-rich repetitive genomic elements are protected first by methyl-binding proteins such as MBDs, which in turn recruit both histone deacetylases and H3K9 methyltransferases. This epigenetic signature can subsequently recruit HP1 protein and thus establish a condensed chromatin structure, which recruits more DNMTs to maintain this methylation pattern. On the other hand, unmethylated CpGs in CGIs recruit factors such as MLL1 and CFP1/SETD1, which only bind to unmethylated CpGs, to establish a unique chromatin environment with high H3K4me3 to deter DNA methyltransferases from binding. Thus, the underlying chromatin structure at CGIs, in terms of modifications and recruited binding partners, likely represents one mechanism to modulate DNMTs mediated DNA methylation.
A longstanding and fascinating question in the epigenetics field is whether there are enzymes capable of directly removing the methyl group. While such an enzyme has been elusive, human TET1 was recently identified as a 5mC hydroxylase that catalyzes the conversion of 5mC to 5-hydroxymethylcytosine (5hmC) (Tahiliani et al., 2009). The mammalian TET family contains three members, Tet1, Tet2 and Tet3, which share significant sequence homology at their C-terminal catalytic domains (Ito et al., 2010; Tahiliani et al., 2009). Similar enzymatic activities for mouse Tet family members have also been described (Ito et al., 2010; Ko et al., 2010). The discovery of this family of enzymes has provided a new potential mechanism for altering DNA methylation status. However, little is known as to what extent individual family members regulate the genome-wide 5mC/5hmC patterns and contribute in genome functions.
Tet1 is highly expressed in embryonic stem cells (ESCs) and its depletion leads to a reduction in global 5hmC levels (Koh et al., 2011). In addition to the 5mC hydroxylase domain, TET1 also contains a conserved CXXC domain (Tahiliani et al., 2009; Zhang et al., 2010), a domain employed by other proteins to bind unmethylated CpG DNA and enabling them to modify histone or DNA methylation. The family of CXXC domain-containing proteins includes factors involved in DNA methylation (DNMT1, MBD1) and histone methylation/demethylation (MLL, CFP1, KDM2A), all of which play important roles in gene regulation and contribute to embryonic development. Significantly, our recent study shows that human TET1 is a CpG DNA binding protein that promotes DNA demethylation when it is over-expressed in 293T cells and positively regulates transcription of a reporter gene in a 5mC hydroxylase activity-dependent manner (Zhang et al., 2010). These findings suggest that Tet1 regulates DNA methylation and gene expression through its ability to convert 5mC to 5hmC.
5hmC was first identified in T-even bacteriophage (Wyatt and Cohen, 1953), and later found in the vertebrate brain (Kriaucionis and Heintz, 2009; Penn et al., 1972) and several other tissues (Globisch et al., 2010). Interestingly, while 5hmC exists at high levels in mESCs, its level significantly decreases after mESC differentiation (Szwagierczak et al., 2010; Tahiliani et al., 2009), and rises again in terminally differentiated cells such as Purkinje neurons (Kriaucionis and Heintz, 2009). Despite these recent advances, the molecular basis for Tet1 and 5hmC functions in the ESC genome and epigenome is unknown, although a controversial role for Tet1 in maintaining ESC pluripotency and determining ESC differentiation has been proposed (Ito et al., 2010; Ko et al., 2010; Koh et al., 2011).
Here, we show that Tet1 is capable of binding to unmethylated as well as methylated and hydroxymethylated CpG DNA via its CXXC domain. Further, we report a complete genome-wide mapping of Tet1 binding and 5hmC in mESCs. Complemented with Tet1 depletion studies, this allows us to establish specific correlations among Tet1 occupancy, 5mC and 5hmC levels, histone modification and gene expression in mESCs and reveal an intricate role of Tet1 in its associated gene network. Thus, this study provides a foundation for understanding not only possible functions of Tet proteins and 5hmC but also molecular mechanisms by which Tet proteins, via dynamic regulation of DNA methylation, influence gene transcription and related biological functions in mESCs.
RESULTS
Tet1 is a CGI binding protein in vitro and in vivo
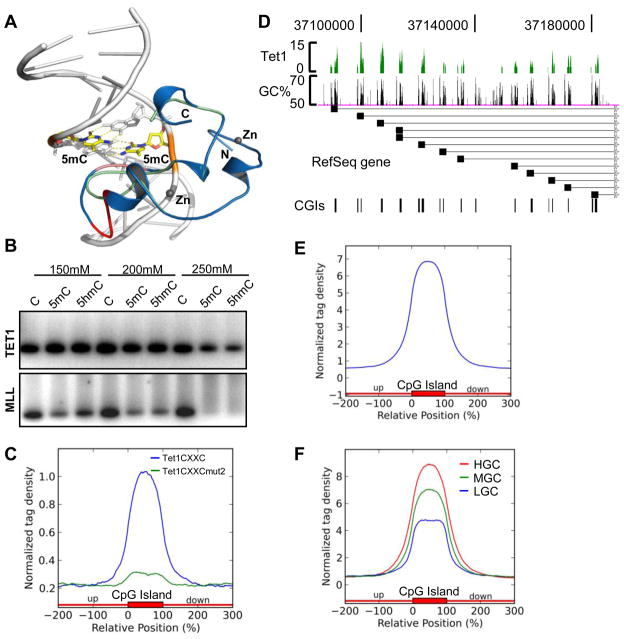
TET1 is a CXXC domain-containing 5mC hydroxylase. We have previously reported that the CXXC domain-containing N-terminus of TET1 (500–910) binds not only to unmethylated but also methylated CpG-containing DNA (Zhang et al., 2010). To extend our previous study, we first performed computer modeling based on our recent CFP1 CXXC-DNA complex crystal structure (Xu et al., 2011). In this model, like CFP1, the TET1 CXXC domain also binds CpG DNA through the CpG containing major groove; the shortened loop in the TET1 CXXC domain moves about 2 Å away from the CpG major groove, which creates enough space to allow for 5mC or 5hmC binding (Figure 1A). Hence, this model predicts that the TET1 CXXC domain may have the ability to bind both unmodified as well as 5mC- and 5hmC-modified CpG-containing DNA. Indeed, by GST pull-down assays, we demonstrate that the TET1 CXXC domain (528–674) binds to unmodified, 5mC-modified, and 5hmC-modified CpG-containing DNA, in contrast to the MLL1 CXXC domain, which is only able to recognize unmodified CpG DNA (Figure 1B, Figure S1A–S1F).
Figure 1. Tet1 binds to CpG-rich DNA via its CXXC domain.
(A) Superposition of the model of the human TET1 CXXC domain with the crystal structure of CFP1 CXXC-CpG DNA complex. The TET1 model (green) and the CFP1 crystal structure (blue) are shown in cartoon. The DNA is shown in a cartoon representation and the 5m-cytosines in the CpG motif are displayed in a stick model with its backbone colored in orange. The CXXC domain binds 2 zinc ions, which are displayed in gray balls. The CpG binding motif in the CFP1 CXXC structure and the corresponding motif in TET1 CXXC are shown in salmon color, and the extra sequence motif (DMKFGG) in the CFP1 CXXC that lacks in TET1 CXXC is colored in red.
(B) GST pull-down assay to determine the binding activities of human TET1 CXXC domain (upper panel) and MLL CXXC domain (lower panel) to CpG, 5mCpG and 5hmCpG containing DNA (generated by PCR, see Figure S1A, S1B) under different NaCl concentration. Data showing here is one representative from three independent assays.
(C) Normalized distribution profiles of Tet1 CXXC domain (blue) and Tet1 CXXC domain mutant2 (green) bound mESCs genomic DNA across CpG islands (CGIs). The CGIs annotation (mm9) was obtained from the UCSC website, and each CGI was normalized to 0–100%. Tag densities in the two profiles were normalized by their total reads number and plotted from 200% upstream to 200% downstream of the normalized CGI.
(D) Representative region (chr18: 37,088,056–37,188,452) to show the correlation of Tet1 binding with GC% and CGIs in mESCs.
(E–F) Normalized Tet1 tag density distribution across total CGIs (E) or with GC content classifications (F). In panel F, CGIs were sorted by GC% from high to low, and equally divided into 5 groups. The first, third, and fifth groups were chosen as HGC (red), MGC (green) and LGC (blue), respectively.
We next performed GST pull-down assays followed by deep DNA sequencing to identify the genome-wide binding profile of the mouse Tet1 CXXC domain (512–671). Purified GST-tagged Tet1 CXXC domain and mutants that contain a single C to A mutation in the core CXXC domain (Figure S1C, S1D) were incubated with sonicated genomic DNA extracted from mESCs. Protein-bound DNA was purified, sequenced and mapped onto the mouse genome (mm9). Bioinformatic analysis shows that Tet1 CXXC-bound DNA but not Tet1 CXXC mutants-bound DNA, is highly enriched for CGIs (Figure 1C, Figure S1G). Thus, these results together with the previous data demonstrate that the Tet1 CXXC domain strongly binds to CpG-rich DNA, and that the intact CXXC domain is essential for its DNA binding ability. Furthermore, these data also suggest that the CXXC domain of Tet1 is preferentially associated with CGIs.
To validate the unique DNA binding activity of Tet1 in vivo, we developed a mouse Tet1-specific Chromatin immunoprecipitation (ChIP) grade antibody and performed Tet1 ChIP-seq in mESCs where Tet1 is highly expressed. ChIP-seq quality and fidelity were verified by conventional ChIP-qPCR assays at randomly selected Tet1 bound regions (Figure S2A). In total, 9,669 Tet1 peaks were identified (P<10−5 and FDR<0.1). We find that 5166 and 7170 Refseq genes contain Tet1 peaks at promoters and gene bodies, respectively. Among them, 4944 genes show Tet1 association at both promoters and gene bodies. As exemplified by the Pcdhα gene cluster, Tet1 binding is highly correlated with the GC% and CGIs (Figure 1D). Bioinformatic analysis shows that Tet1 binding is enriched in CGIs (Figure 1E), and the binding density is positively correlated with the GC content of CGIs (Figure 1F). In fact, we find that 31.8% of all CGIs in the genome overlap with Tet1 peaks. Interestingly, we also note that 20% of methylated gene promoters (Fouse et al., 2008) and 27% of 5hmC containing promoters (based on the hMeDIP-seq results described below) in mESCs are bound by Tet1 (Figure S2B), which indicates that Tet1 protein binds to a subset of methylated and hydroxymethylated CpG DNA in vivo. As an example, Tet1 binds to promoters of both Elf5 gene isoforms (Figure S2C), which are hypermethylated in mESCs (Koh et al., 2011; Ng et al., 2008). Collectively, we conclude that Tet1 is a DNA binding protein, which preferentially binds to unmodified CpG. Importantly, it is also capable of binding to 5mCpG- and 5hmCpG-modified DNA in vitro and in vivo, which provides a potential avenue for identifying 5hmC binding proteins in vivo.
Genome-wide mapping of Tet1 reveals that it targets gene promoters and exons in mESCs
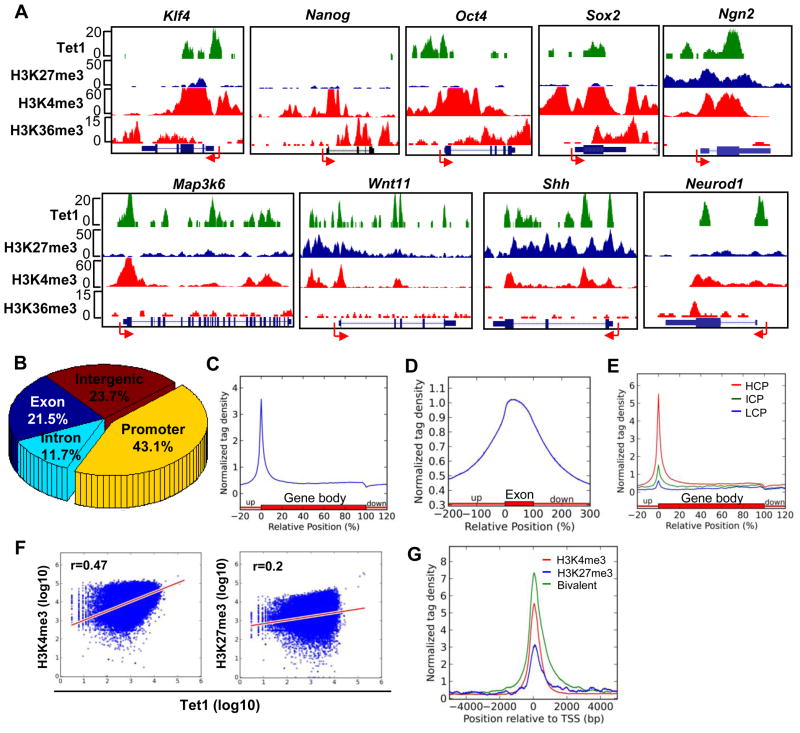
Having defined Tet1 as a CpG-rich DNA binding protein, we next analyzed Tet1 genome-wide distribution in detail. As shown in the representative Tet1 binding map (Figure 2A), Tet1 strongly binds to the promoters and exons of a variety of key genes. Bioinformatic analysis shows that among all the Tet1 peaks, almost half (43.1%) are located at promoters, while the remaining peaks (21.5% and 11.7%) are found at exons and introns, respectively (Figure 2B). The Tet1 profile across an average Refseq gene shows high density around the transcription start site (TSS) that drops dramatically upon entering the gene body (Figure 2C). Furthermore, a clear preference for exons is noted (Figure 2D). Thus, these analyses suggest that Tet1 is preferentially associated with gene promoters and exons.
Figure 2. Tet1 is enriched at gene promoters and positively correlates with promoter CpG content and H3K4me3.
(A) Representative Tet1 ChIP-seq results and associated histone modification patterns. Arrow denotes promoter orientation.
(B) Genomic distribution of Tet1-enriched regions. The genomic features (exons, introns, and intergenic regions) were defined based on RefSeq gene (mm9) annotations. Promoter was defined as −2kb to +2kb relative to TSS.
(C) Normalized Tet1 tag density distribution across the gene body. Each gene body was normalized to 0–100%. Normalized Tag density is plotted from 20% of upstream of TSSs to 20% downstream of TTSs.
(D) Normalized Tet1 tag density distribution across exons. Each exon is normalized to 0–100%. Normalized Tet1 density is plotted from 200% upstream to 200% downstream of the normalized exon.
(E) Normalized Tet1 tag density distribution across gene bodies with promoter classifications. The high CpG promoters (HCPs), intermediate CpG promoters (ICPs) and low CpG promoters (LCPs) were defined as described previously (Meissner et al., 2008; Mikkelsen et al., 2007). Each gene body region was normalized to 0–100%. Normalized tag density is shown from 20% upstream of TSSs to 20% downstream of TTSs.
(F) The correlations between Tet1 and H3K4me3 (left) or H3K27me3 (right) tag densities at gene promoters. H3K4me3 and H3K27me3 densities were obtained from the reference (Mikkelsen et al., 2007). Tet1, H3K4me3, and H3K27me3 values at promoters were calculated as log10 (tag density).
(G) Normalized Tet1 tag density distributions at ‘univalent’ H3K4me3 (red), ‘univalent’ H3K27me3 (blue) and ‘bivalent’ (green) promoters. Promoters were classified according to their histone modification patterns (Mikkelsen et al., 2007). Tet1 enrichment from −5kb to +5kb relative to TSSs is shown.
When promoters are grouped based on their CpG content as previously described (Meissner et al., 2008; Mikkelsen et al., 2007), Tet1 binding shows significant enrichment at high CpG promoters (HCPs) compared to low (LCPs) or intermediate CpG promoters (ICPs) (Figure 2E), suggesting a positive correlation between Tet1 binding density and promoter CpG content. In addition, we observe a better positive correlation of Tet1 binding with H3K4me3 (r=0.47) than H3K27me3 (r=0.2) at promoters (Figure 2F). Consistently, 49% of Tet1 bound promoters are ‘univalent’ H3K4me3, 44% are ‘bivalent’ promoters (a total of 93% H3K4me3 positive) and only 2.6% are ‘univalent’ H3K27me3 promoters. The average Tet1 density at ‘bivalent’ promoters is significantly higher than either type of ‘univalent’ promoters (P=2.58e–316) (Figure 2G). These data suggest that Tet1 presence is positively correlated with H3K4me3 at promoters and is highly enriched at HCPs, which are mostly hypomethylated in ESCs, raising an intriguing question of how Tet1 might regulate 5hmC and 5mC levels at specific genome loci.
A genome-wide map of 5hmC illustrates a unique outlook of the 6th base in mESCs
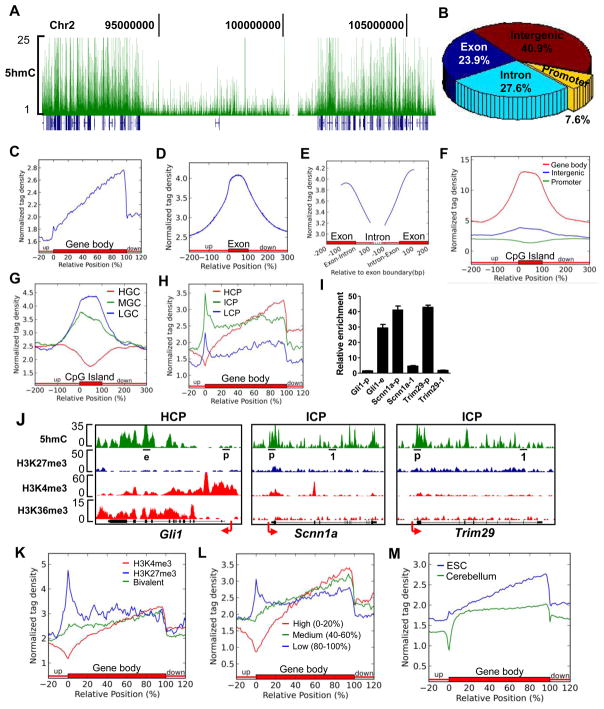
To gain insight into Tet1-mediated regulation of 5hmC levels, we first performed a genome-wide mapping of 5hmC in mESCs using a 5hmC antibody-based hydroxymethylated DNA immunoprecipitation (hMeDIP) protocol recently established in our lab (Figure S3A). We collected about 1.5G bases sequencing data and identified a total of 47,472 5hmC peaks (P=10−7, FDR≤0.01). In general, 5hmC levels are localized predominantly in gene-rich areas, with a paucity of signal in gene-poor regions (gene deserts) (Figure 3A).
Figure 3. Genome-wide distribution of 5hmC in mESCs.
(A) The distribution of 5hmC density (green) in the region of chr2: 90,353,915–106,163,465 by hMeDIP-seq. Refseq genes (blue) are shown under 5hmC peaks.
(B) Genomic distribution of 5hmC-enriched regions.
(C–E) Normalized 5hmC tag density distribution across the gene body (C), exon (D) or exon-intron boundary (E). Normalized tag density is plotted from −100bp to 100bp relative to exon-intron boundaries in panel E.
(F–G) Normalized 5hmC tag density distribution across CGIs with genomic location (F) or GC content (G) classifications. In panel F, CGIs were grouped into promoter (−2k to +2k relative to TSS, green), gene body (+2k to TTS, red), and intergenic CGIs (TTS to −2k of the downstream gene, blue) based on their locations. In panel G, CGIs were sorted by GC% from high to low, and equally divided into 5 groups. The first, third, and fifth groups were chosen as HGC (red), MGC (green) and LGC (blue), respectively.
(H) Normalized 5hmC tag density distribution across the genes with HCPs (red), ICPs (green) or LCPs (blue).
(I) hMeDIP q-PCR to detect 5hmC levels. Results are shown as mean +/− SEM (n=3). The targeting region for each primer set is underlined in panel J. Arrow denotes promoter orientation.
(J) hMeDIP-seq results of Gli1 (HCP) and Scnn1a and Trim29 (ICPs) genes.
(K) Normalized 5hmC tag density distribution across the genes with ‘univalent’ H3K4me3 (red), ‘univalent’ H3K27 me3 (blue) or ‘bivalent’ (green) promoters.
(L) Normalized 5hmC tag density distribution across genes with high (red), medium (green) or low (blue) expression levels. Genes were sorted by their expression levels identified in microarray assay from high to low, and equally divided into 5 groups. The first, third and fifth groups were chosen as genes with high-, medium- and low-expression levels, respectively.
(M) Normalized 5hmC tag density distribution relative to the average gene body in mouse ESCs (blue) and cerebellum (green) (Song et al., 2011). Tag densities in these two groups were normalized by their total reads number and shown from 20% upstream of TSSs to 20% downstream of TTSs.
5hmC is abundant in gene bodies with a specific enrichment at exons
Statistical analysis reveals that 51.5% of 5hmC peaks are located at either exons (23.9%) or introns (27.6%), while only 7.6% are at promoters (Figure 3B). The 5hmC profile across averaged Refseq genes confirms this distribution, where 5hmC density demonstrates a gradual increase from TSSs towards transcription termination sites (TTSs), dropping drastically around the TTSs (Figure 3C). Importantly, 5hmC is highly enriched in gene exons compared to introns (Figure 3D, 3E).
5hmC levels at promoters relative to CpG content and histone modifications
Given that 5hmC is derived from 5mC and the majority of 5mC occurs at CpG sites in ESCs, we analyzed the correlation between 5hmC and CpG content. Interestingly, 5hmC is uniquely enriched within gene body CGIs, but very low in CGIs at promoters and intergenic regions (Figure 3F). We also find that 5hmC is enriched in CGIs with low or medium GC content and shows a negative correlation with the GC content in CGIs (Figure 3G). Consistently, when promoters are grouped according to their CpG content, we find that weak CpG promoters (ICPs and LCPs) have relatively higher 5hmC levels than strong CpG promoters (HCPs) (Figure 3H). In addition, the average 5hmC level continues to increase towards the TTS in genes with HCPs, while it remains constant in the body of genes with ICPs and LCPs (Figure 3H). Importantly, these bioinformatic analyses can be confirmed by conventional hMeDIP-qPCR on randomly selected genes including Scann1a and Trim29, representative ICP genes, and Gli1, a representative HCP gene (Figure 3I and 3J). Finally, we find 5hmC levels to be enriched at ‘univalent’ H3K27me3 promoters, in contrast to ‘univalent’ H3K4me3 or ‘bivalent’ promoters (Figure 3K).
5hmC levels at promoter or within gene body relative to gene expression
The regulation of gene expression in ESCs is a complex process influenced by transcriptional factors, DNA methylation and histone modifications. So far, 5hmC is an unknown contributor to gene expression in ESCs. We therefore set out to determine whether gene expression levels correlate with their respective 5hmC content. Based on the microarray assay gene expression profile in E14 mESCs (described below), we find that 5hmC density is much higher at lowly expressed gene promoters compared to those of genes with medium and high expression levels, with extremely low levels around the TSS of highly expressed genes (Figure 3L), suggesting a negative correlation between the 5hmC level at gene promoters and associated gene transcription activity.
In contrast to promoters, 5hmC levels gradually increases towards TTSs of genes expressed at high and medium levels, but remain constant at lowly expressed genes (Figure 3L). Genes that are expressed at the high and medium levels have relatively higher 5hmC levels than lowly expressed genes at the 3′ end of intragenic regions (Figure 3L). Using the independent gene transcriptome data derived from J1 mESCs (described below), we observe similar 5hmC distribution profiles relative to gene expression levels (Figure S3B). Interestingly, we note that our results are different from the recently reported genome-wide mapping of 5hmC in mouse cerebellum (Song et al., 2011), particularly at promoters and the 5′ end of intragenic regions. To address this discrepancy, we compared the 5hmC distribution profiles of mouse ESCs and cerebellum tissue (Song et al., 2011). Globally, mESCs show a unique 5hmC profile around TSSs and a more overt 5hmC level increase from TSSs to TTSs (Figure 3M). In addition, 5hmC patterns at certain genomic loci are also different between mESCs and mouse cerebellum (Figure S3C). Thus, it is possible that during development or differentiation from ESCs to terminally differentiated neurons, 5hmC levels are dynamically changed at specific gene bodies and/or promoters.
Taken together, our genome-wide mapping of 5hmC together with the finding from mouse cerebellum suggests that high 5hmC enrichment in gene bodies is likely a common feature for most 5hmC-associated genes. However, unique to mESCs, our study reveals that 5hmC is generally absent or present at very low levels at gene promoters that are associated with high CpG density, high H3K4me3 and/or high expression levels, whereas it is enriched at gene promoters with ‘univalent’ H3K27me3, ICPs, or low expression levels. Importantly, although 5hmC levels are low at gene promoters in comparison with gene bodies, the 5hmC level within gene bodies is not a simple reflection of associated gene expression. For example, we find that the 5hmC level is very low both at the promoter and the body of a set of genes with constantly high expression levels, such as house keeping genes (Figure S3D). These findings highlight the central question of how 5hmC is regulated as well as where it fits within the network of epigenetic regulation and transcription in ESCs.
Tet1 regulates 5hmC levels at targeted gene promoters and exons
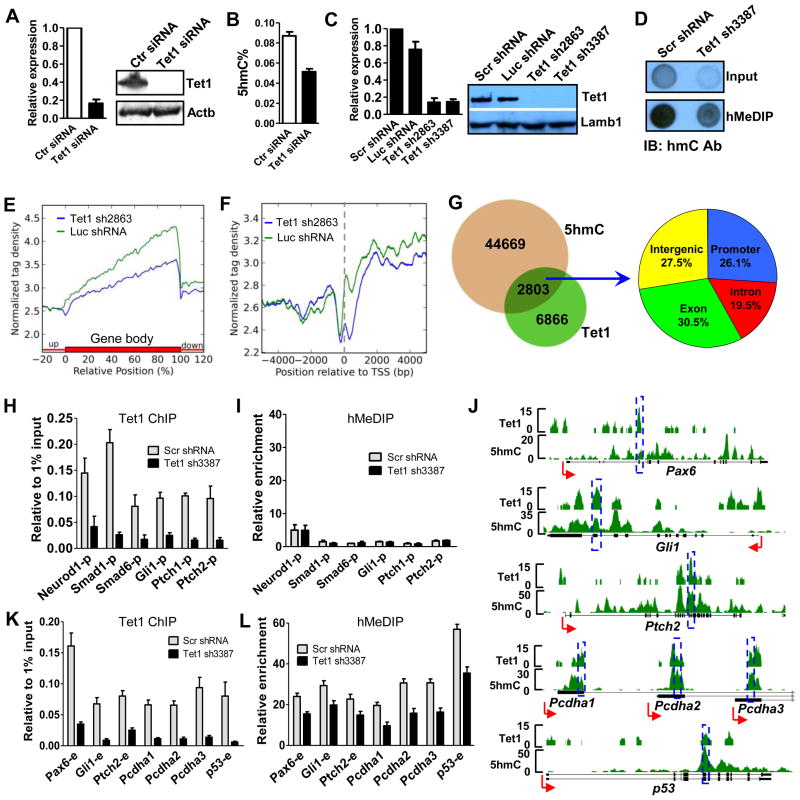
The C-terminus of Tet1 converts 5mC to 5hmC, while the CXXC domain likely targets the protein to specific CpG-rich regions at gene promoters and exons. Furthermore, Tet1 is highly expressed in mESCs and depletion of Tet1 results in a 30% decrease of global 5hmC level in mESCs (Koh et al., 2011). Thus, it is possible that the unique DNA binding and enzymatic activities of Tet1 may be coordinated to regulate 5hmC generation at specific genome loci. We employed RNAi to address this hypothesis and observed depleted Tet1 expression at the mRNA and protein levels after siRNA treatment in mESCs (Figure 4A), as well as a 35% decrease in global 5hmC by HPLC (Figure 4B). Using shRNA-mediated Tet1 depletion (Figure 4C), we further confirm a similar global 5hmC level decrease by dot-blot assays (Figure 4D). To define loci-specific 5hmC regulation by Tet1, we carried out hMeDIP-seq combined with shRNA-mediated Tet1 depletion in mESCs. In comparison with the control shRNA-treated ESCs, Tet1-depleted ESCs show a dramatic decrease of 5hmC levels within gene bodies, particularly at the 3′ end of genes (Figure 4E). Even though 5hmC level at gene promoters is relatively low, we also observe a significant 5hmC level reduction around TSSs after Tet1 depletion (Figure 4E and 4F).
Figure 4. Tet1 regulates 5hmC levels in its targeted gene promoters and exons.
(A) siRNA-mediated Tet1 depletion at mRNA (left, by RT-qPCR) and protein (right, by western blot) levels. RT-qPCR data are presented as mean +/− SEM (n=3). Actb was used as loading control.
(B) Global 5hmC level decrease caused by siRNA-mediated Tet1 depletion by HPLC assay. Results are shown as mean +/− SEM (n=3).
(C) Specific shRNAs-mediated Tet1 depletion at both mRNA (left, by RT-qPCR) and protein (right, by western blot) levels. RT-qPCR results are shown as mean +/− SEM (n=4). Lamb1 was used as loading control.
(D) Dot blot assay showing the global 5hmC level decrease after shRNA-mediated Tet1 depletion. The same amount of genome DNA from Scr shRNA or Tet1 shRNA3387 treated mESCs was processed for hMeDIP. The same quantity of input DNA and same volume of hMeDIPed DNA were blotted onto NC membrane and performed dot-blot assay using 5hmC antibody.
(E–F) The differential average 5hmC levels in Luc shRNA (green, control) and Tet1 shRNA2863 (blue) treated mESCs though out the gene (E) or at gene promoters (F). Two group 5hmC tag densities were normalized by their total reads number for comparison. The TSS is noted by a dash line in panel F.
(G) Tet1 peaks overlapped with 5hmC and their distributions.
(H–I) Tet1 occupancy (H) and 5hmC levels (I) changes at Tet1 targeted promoters after Tet1 depletion by ChIP-qPCR and hMeDIP-qPCR, respectively. Data are presented as mean +/− SEM (n=3).
(J) Representative genes show good 5hmC and Tet1 correlation in Tet1-targeted exons. The targeting regions of primers used in panel K and L are noted by blue rectangles.
(K–L) Tet1 occupancy (K) and 5hmC levels (L) changes at Tet1 bound exons after Tet1 depletion by ChIP-qPCR and hMeDIP-qPCR, respectively. Data are presented as mean +/− SEM (n=3).
Given the disproportionate number of 5hmC-containing loci (47,472; ~50% within gene bodies) compared with Tet1 sites (9,669; 43% at promoters), we hypothesize that Tet1 likely regulates 5hmC in a loci-specific fashion. To identify specific genes or gene elements regulated by Tet1, we sorted out Tet1 peaks that overlap with 5hmC. This amounts to roughly 30% (2803 of 9669) of all peaks, distributed among promoters (26.1%), exons (30.5%), introns (19.5%) and intergenic regions (27.5%) (Figure 4G). Of note, we find that these promoters and exons strongly represent genes involved in various developmental processes, particularly neural development (Figure S4).
To validate this analysis and assess the regulatory effects of Tet1 on 5hmC levels at these genomic elements, we performed Tet1 ChIP-qPCR and hMeDIP-qPCR using an independent set of control and Tet1 shRNAs. Tet1 depletion results in a marked reduction of Tet1 occupancy at targeted promoters and exons (Figure 4H, 4J and 4K). Concurrently, we detect significant 5hmC level decrease at 5hmC enriched, Tet1-targeted exons (Figure 4J and 4L) and promoters (Figure S5A, S5B) but not at Tet1-targeted promoters with low 5hmC level (Figure 4I). Importantly, 5hmC reduction is not observed at Tet1 non-targeted regions (Figure S5C, S5D), suggesting specific regulation of 5hmC production by Tet1. Taken together, we conclude that Tet1 regulates 5hmC levels in targeted key gene elements, such as gene promoters and exons.
Tet1 regulates DNA methylation status of its target genes
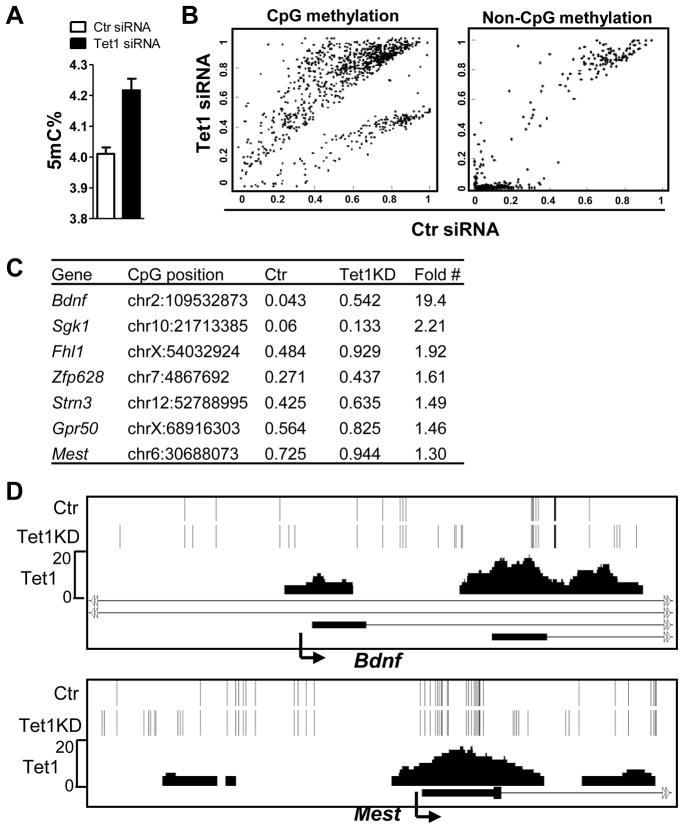
5hmC is converted from 5mC, thus we complement the above study by examining the effect of Tet1 depletion on 5mC levels at Tet1 target genes. By HPLC, we observed a moderate but consistent (5–10%) global 5mC level increase following siRNA- or shRNA-mediated Tet1 depletion in mESCs (Figure 5A and data not shown). Because of the low sensitivity of MeDIP and low DNA methylation levels at a majority of Tet1-occupied promoters in mESCs, we also utilized targeted bisulfite sequencing (Deng et al., 2009) to determine 5mC changes caused by Tet1 depletion. Even though bisulfite sequencing cannot distinguish between 5mC and 5hmC (Huang et al., 2010), we reasoned that any increased signal caused by Tet1 depletion likely reflects the 5mC increase, given that Tet1 depletion results in 5hmC level decrease but not increase. We interrogated 1,603 candidate genes (see Methods) and found differential CpG methylation in 981 out of 11,608 sites (P<0.001) (Figure 5B left panel, 5C, Figure S5E). Importantly, roughly 40% of these sites overlap with Tet1 peaks and a large portion of the others are flanking Tet1 peaks (as exemplified in Figure 5D). In contrast, only 0.3% (449 of 133,635) of non-CpG methylation sites show significant differences after Tet1 depletion (Figure 5B, right panel). Taken together, we conclude that Tet1 functions at its target genes to regulate cytosine methylation.
Figure 5. Tet1 regulates the DNA methylation status at target gene promoters.
(A) Global 5mC level increase after siRNA-mediated Tet1 depletion by HPLC assay. Results are shown as mean +/− SEM (n=3).
(B) Global analysis of DNA methylation after siRNA-mediated Tet1 depletion by targeted bisulfite sequencing. The differential CpG methylation (left) and non-CpG methylation (right) are presented. CpG or non-CpG methylation levels were extracted from mapped reads. The resulting values are real numbers ranging from 0 to 1, corresponding to completely unmethylated to completely methylated.
(C) Examples of increased CpG methylation after Tet1 knockdown (KD) and associated genes. The CpG methylation levels are shown as numbers from 0 to 1, corresponding to completely unmethylated to completely methylated. “#”: the methylation level differences at all listed CpG sites are statistically significant (see Figure S5E).
(D) Representative regions in Bdnf and Mest genes show the significantly increased CpG methylation caused by Tet1 KD. Each bar represents single CpG methylation. Note that unique bars in Tet1 KD sample show the significant CpG methylation increase (from undetectable in control to detectable) and that most of those sites are overlapped with Tet1 peaks or flanking around Tet1 peaks, indicating the role of Tet1 in regulating CpG methylation at those sites. Arrow denotes promoter orientation.
Tet1-mediated gene expression program in mESCs
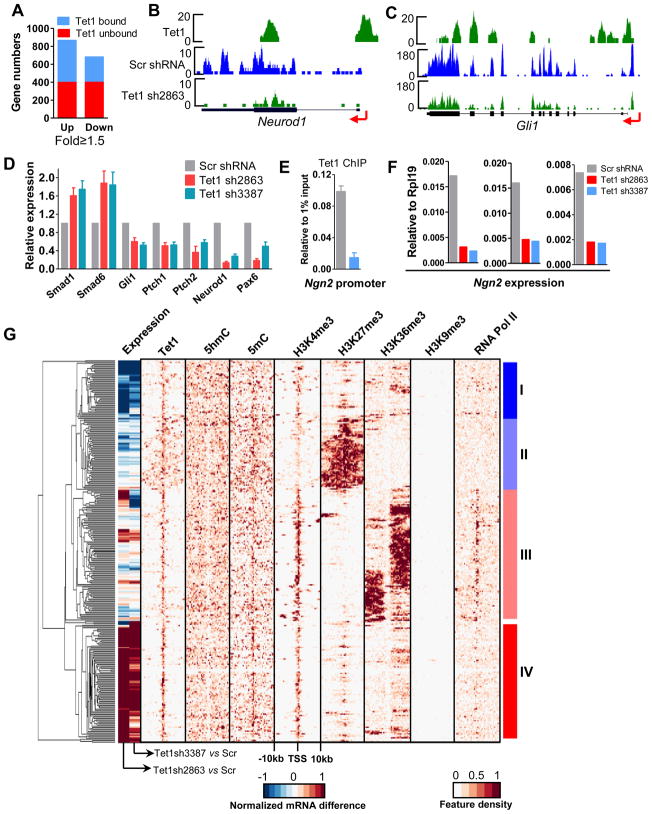
We next carried out microarray studies in J1 mESCs to explore the role of Tet1 in regulating gene expression, given its high enrichment at promoters and gene bodies. Using a criterion of 1.5-fold expression change, we identified 867 up- and 682 down-regulated genes following siRNA-mediated Tet1 depletion (Figure 6A). Among these genes, 54% of the up- and 41% of the down-regulated genes are Tet1 targets (Figure 6A), suggesting that Tet1 has both positive and negative effects on target gene expression.
Figure 6. Tet1 plays both positive and negative roles in target gene regulation in mESCs.
(A) Using 1.5-fold as a cut-off value, all differentially expressed genes in the microarray assay were selected and further classified into Tet1 bound and unbound genes.
(B–C) Representative mRNA-seq results. Arrow denotes promoter orientation.
(D) RT-qPCR confirms the differentially expressed genes after Tet1 depletion. Results are shown as mean +/− SEM (n=4).
(E) Tet1 ChIP-qPCR reveals that Tet1 binds to the Ngn2 promoter and Tet1 depletion causes the significantly decreased Tet1 occupancy at the Ngn2 promoter in mESCs. Data are presented as mean +/− SEM (n=3).
(F) Tet1 depletion delayed the Ngn2 induction during neural differentiation. ESCs were infected with control or Tet1 shRNA containing lentivirus, selected with puromycin and differentiated by LIF withdraw for 3 days. The formed embryonic bodies were treated with 1μM RA for 3 days. Ngn2 expression was examined by RT-qPCR. Results of three independent experiments are shown.
(G) Hierarchical clustering of epigenetic and transcriptional features of Tet1 target genes. The histone methylation, DNA methylation and RNA Pol II profiles in mESCs were obtained from the references (Meissner et al., 2008; Mikkelsen et al., 2007). Top 300 Tet1 target genes based on the variance of expression and epigenetic modifications density score were chosen to make the cluster. The tag densities of Tet1 binding, 5hmC, 5mC, H3K4me3, H3K27me3, H3k36me3, H3K9me3 and RNA Pol II were profiled through −10kb to +10kb relative to the TSS of each gene with bin of 500bp. The middle color bar indicates the difference of mRNA levels between Scr shRNA- and Tet1 shRNA2863- or Tet1 shRNA3387-treated mESCs.
To eliminate any potential artificial effects and confirm the above results, we also examined gene expression changes in E14 mESCs following lentivirus-mediated shRNA depletion of Tet1, using both microarray and recently developed mRNA-seq technologies. We developed and tested 5 independent Tet1 shRNAs, and selected shRNA2863 and shRNA3387, as these showed consistent and stable Tet1 depletion and negligible morphological and alkaline phosphatase activity changes in the Tet1-depleted ESCs (Figure 4C, Figure S6A). Using a criterion of P<0.05 and FDR<0.05, we generated genome-wide expression maps of both normal and Tet1-depleted ESCs by mRNA-seq. After comparing the results of mRNA-seq and two independent microarray assays, we identified a set of genes that demonstrate consistent differential expression as high confidence Tet1-regulated genes. Interestingly, among the Tet1 target genes showing increased CpG methylation after Tet1 depletion (Figure 5C), many genes also exhibit gene expression decreases (Figure S6C). These data suggest that Tet1 dynamically regulates DNA methylation level at its functional sites, which may partly account for the Tet1 influence on gene expression.
To validate these findings, we selected a subset of Tet1 target genes, including consistently changed genes such as Gli1, Ptch1, Ptch2, Smad1, Smad6, Neurod1 and Pax6, as well as unchanged genes such as Klf4, Oct4, Sox2 and Ngn2 for RT-qPCR examination. We find consistent expression changes similar to those observed in microarray and mRNA-seq assays (Figure 6B-6D, Figure S6B). In agreement with a previous report (Koh et al., 2011), we find that some ESC pluripotency genes, such as Sox2, Klf4 and Oct4, display strong Tet1 association, but do not demonstrate expression changes following Tet1 depletion (Figure S6B). Interestingly, while Tet1 depletion causes a dramatically decreased occupancy at the Ngn2 promoter (Figure 6E), it does not alter the expression of the Ngn2 gene, a key neural development gene silenced in ESCs. However, Tet1 depletion in mESCs results in a significant delay in Ngn2 induction during neural differentiation after RA treatment (Figure 6F), suggesting a potential role of Tet1 in the epigenetic regulation of neural differentiation. Taken together, these gene expression analyses strongly suggest that Tet1 may be not critical for maintaining the transcriptional status of ESC self-renewal or pluripotency genes in ESCs, whereas it certainly affects the transcriptional status of a subset of genes involved in neurogenesis, as well as Shh and TGF-β signaling pathways.
5hmC and Tet1 define a new layer of epigenetic regulation in mESCs
To understand the overall correlation of Tet1 function with DNA modification, histone methylation and gene expression, we performed a hierarchical clustering analysis of DNA modifications (5mC and 5hmC), histone modifications (H3K4me3, H3K27me3 and H3K36me3), RNA Pol II occupancy and gene expression changes for the Tet1 associated genes. We focused on 300 genes whose promoters are bound by Tet1 with the largest profile variance for more detailed analysis (Figure 6G). This reveals 4 categories of Tet1 target genes. Group I genes (n=43) contain Tet1 binding mainly around the TSS but with a modest spread flanking TSSs, with no obvious 5mC or 5hmC depletion or enrichment around the TSS compared to proximal regions. Most of these genes contain H3K4me3 and H3K27me3 around the TSS, and medium to low H3K36me3 and RNA Pol II association, suggesting that these genes contain ‘bivalent’ promoters, and are expressed at low levels. For these genes, removal of Tet1 predominantly results in down-regulated expression. Gene Ontology (GO) term analysis shows that these genes are mainly associated with proteins and lipid metabolic processes as well as cell proliferation.
Group II genes (n=54) demonstrate Tet1 binding at the TSS, but also widespread Tet1 association within the gene body. In these genes, 5hmC is absent from promoters, but enriched in gene bodies; 5mC is high at the TSS and flanking regions, similar to Tet1. Most of these genes exhibit low H3K4me3 but high H3K27me3 levels, and consistently, no obvious H3K36me3 enrichment or RNA Pol II association, suggesting that these genes are silenced. In this case, Tet1 depletion has little or no effect on gene expression. GO term and KEGG pathway analyses show an impressive functional association of this group of genes in transcriptional regulation and key signaling pathways (such as Wnt, Notch and Shh). Indeed, 74% of these genes are transcription factors, of which, 56% are homeobox containing-genes that are important for embryogenesis, cell lineage differentiation and tissue specification.
Among the group III genes (n=99), Tet1 is enriched at the TSS, in contrast to 5hmC, which is absent around the TSS. These genes exhibit high H3K4me3, H3K36me3, and RNA Pol II occupancy, but lack H3K27me3, suggesting high expression in ESCs. However, similar to the group II genes, Tet1 depletion does not cause significant gene expression changes. GO term and KEGG pathway analyses indicate that this group of genes participate in transcriptional regulation, cell cycle control and signaling pathways that are important for both ES cell biology and embryogenesis.
In the last group of genes (n=87), group IV, Tet1 is highly enriched at promoters, while 5hmC is enriched in gene bodies, and 5mC is enriched mostly around the TSS. Most of these genes, as in group I, contain ‘bivalent’ promoters, and low but obvious H3K36me3 enrichment and RNA Pol II association, suggesting low expression levels. For these genes, Tet1 depletion results in a significant increase in expression, suggesting that Tet1 negatively regulates their transcription. GO term analysis indicates that these genes are strongly associated with the regulation of transcription and protein phosphorylation.
Taken together, this integrative bioinformatic analysis reveals that Tet1 genome association is dependent on GC content and availability of CpG binding sites, but not the CpG methylation state or gene transcription activity. Although Tet1 binds to ‘univalent’ H3K4me3 (group III) or H3K27me3 (group II) promoters, it does not directly influence their transcription of in mESCs. On the other hand, there exist two types of ‘bivalent’ promoters (groups I and IV) whose transcriptional activities are clearly dependent on the presence of Tet1. For those promoters that have depleted 5hmC and high 5mC (group IV), Tet1 plays a repressive role. For those promoters that contain high 5hmC and average 5mC levels (group I), Tet1 positively regulates the associated gene expression. Thus, Tet1 and 5hmC may provide an additional layer of epigenetic regulation which, like other epigenetic controls, has an intricate role in fine-tuning the transcriptional program of ESCs.
DISCUSSION
In this study we first define Tet1 as a CGI binding factor in mESCs. We show that the CXXC domain of Tet1 is a DNA binding module that preferentially binds to CpG-rich DNA in vitro and in vivo. Significantly, Tet1 also binds to 5mC or 5hmC modified DNA in vitro and in vivo, which provides a molecular basis for Tet1 function in vivo and is distinct from most other CXXC domain containing proteins. Furthermore, we complete genome-wide mapping of 5hmC and Tet1 in mESCs, and provide comparative maps of 5hmC and gene expression profiling from Tet1-depleted ESCs. Finally, through integrative analysis of these data, we have established specific correlations among Tet1 occupancy, 5mC, 5hmC, histone modifications and gene expression in mESCs, revealing complex modes of Tet1 action in genome-wide regulation of 5hmC, DNA methylation and gene transcription in mESCs. Thus, this study sheds important light on 5mC/5hmC regulation by Tet1 and provides a foundation for understanding the functional role of Tet proteins and 5hmC in the regulation of ESC epigenome and gene transcription.
A working model for dynamic control of 5hmC and 5mC by Tet1 in mESCs
Genome-wide mapping of 5hmC and Tet1 suggests that only a portion of 5hmC in mESC is catalyzed by Tet1 protein, which is intriguing but not entirely surprising. Firstly, since Tet2 is also abundantly expressed in mESCs, we suspect that many 5hmC regions may be occupied by Tet2 or other unknown hydroxylases. Secondly, although some 5hmC-enriched regions are bound by Tet1, Tet1 may dissociate from its target regions after hydroxylation. Finally, due to the system differences between ChIP-seq and hMeDIP-seq, we do not exclude the possibility that either Tet1-bound peaks may be underestimated or that 5hmC peaks may be overestimated in our studies.
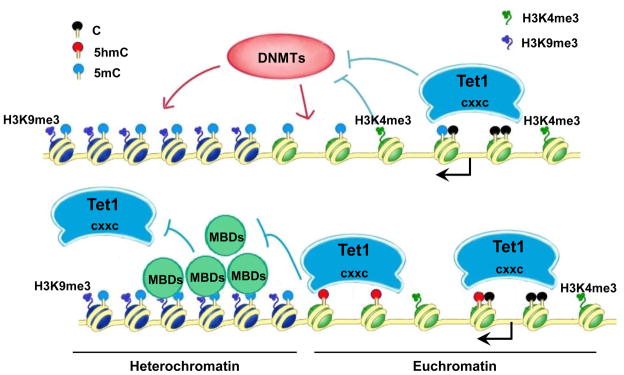
We propose that Tet1 may employ various molecular mechanisms to function at these discrete loci. To illustrate this, we propose a working model (Figure 7) whereby strong Tet1 binding to unmethylated CpG rich DNA via its CXXC DNA binding domain provides an additional layer of protection to limit the accessibility of DNMTs. Conversely, in CpG-rich regions that are already methylated (such as the pericentromeric region), the densely-methylated CpGs become an epigenetic beacon to recruit methyl-binding proteins (MBDs) such as MeCP2 and MBD4, which subsequently recruits repressive histone modifiers such as H3K9me3 methyltransferases and HP1s to establish a constitutive heterochromatin state, making Tet1 inaccessible to these hypermethylated sites and preventing the conversion from 5mC to 5hmC. Indeed, we find highly enriched 5mC but not 5hmC in heterochromatin regions in mESCs (Figure S7). However, when dispersed CpGs are methylated in euchromatin, the versatile DNA binding ability of Tet1 CXXC domain allows Tet1 to access those sites and convert 5mC to 5hmC. It has been reported that the 5mC hydroxylation product 5hmC cannot be recognized by MBDs such as MeCP2 (Valinluck et al., 2004). Therefore, the newly generated 5hmC will prevent the binding of MBDs or access of DNMTs, thus allowing 5hmC-marked regions to escape from being heterochromatinized. Furthermore, since Tet1 can also bind 5hmCpG DNA, it can then remain at those sites after the 5mC to 5hmC conversion and thereby further limit the accessibility of DNMTs or MBDs. Overall, through these separate or coordinated mechanisms, Tet1 and 5hmC provide an important layer of control to the dynamic regulation of DNA methylation in mESCs.
Figure 7. A Model of Tet1 functions in regulating euchromatin CpG methylation.
Tet1 strongly binds to unmethylated-CpG rich regions (such as gene promoters, CGIs) via its CXXC domain, limiting the accessibility of DNMTs. Tet1 also binds to dispersedly methylated-CpGs in euchromatin (shown as green nucleosomes) and converts 5mC to 5hmC. The newly generated 5hmC may further limit the binding of methyl-binding proteins (MBDs) or DNMTs. However, densely-methylated CpGs can recruit MBDs, which subsequently recruits repressive histone modifiers such as H3K9me3 methyltransferases to establish a heterochromatin state (shown as dark blue nucleosomes), making Tet1 inaccessible to these hypermethylated sites and preventing the conversion from 5mC to 5hmC. Arrow denotes the gene promoter. Please refer to the related text for more details.
The unique 5hmC patterns suggest an additional layer of epigenetic landscape in mESCs
Since 5hmC is the hydroxylation product of 5mC, not surprisingly, both modified bases share certain common distribution features in mESC genome. For example, like 5mC (Meissner et al., 2008; Mikkelsen et al., 2008; Weber et al., 2007), 5hmC levels are extremely low at high GC content CGIs, HCPs, ‘univalent’ H3K4me3 promoters, but are frequently associated with ICPs, LCPs and ‘univalent’ H3K27me3 promoters. In addition, our genome-wide mapping reveals 5hmC enrichment in gene bodies, particularly in gene exons, which is similar to the related finding on 5mCpG in mESCs (Chodavarapu et al., 2010). However, our finding that 5mC but not 5hmC is highly enriched in heterochromatin regions (Figure S7) suggests differential distribution patterns between 5mC and 5hmC in mESCs.
ESCs can undergo indefinite cycles of self-renewal while maintaining pluripotency, which is dependent on the network of Oct4, Nanog and Sox2 mediated transcriptional circuitry and epigenetic regulation mediated by DNA methylation, Trithorax and Polycomb group proteins (Boyer et al., 2006; Young, 2011). In this report, we have generated a genome-wide map of 5hmC, adding an additional layer of regulation to the emerging ESC epigenetic landscape. One interesting finding we report is that while 5hmC only accounts for about 0.1% of total bases in mESCs (4% for 5mC), 52% of 5hmC are located in gene bodies with an enrichment in gene exons. Although it is known that H3K36me3 is an epigenetic mark for actively transcribed gene bodies, and that nucleosome position strongly correlates with gene exons (Tilgner et al., 2009), our data suggest that 5hmC represents a potential epigenetic landmark for gene exons in mESCs, independent of transcriptional status.
In addition, the correlation between gene expression and 5hmC levels at promoters and gene bodies in mESCs is different from that in mouse cerebellum (Song et al., 2011), suggesting that 5hmC likely plays distinct roles in gene transcription in different cell or tissue types. Furthermore, by comparing genome-wide 5hmC distribution in mESCs and the terminally differentiated cerebellum tissue, our data also highlight differential 5hmC distribution profiles at specific genomic loci in different cell types. Therefore, these data suggest that not only the global 5hmC level (Szwagierczak et al., 2010; Tahiliani et al., 2009) but also the loci-specific distribution of 5hmC can be regulated during cell differentiation at various developmental stages.
An intricate role of Tet1 and 5hmC in gene regulation in mESCs
A large body of evidence has demonstrated that DNA methylation plays important roles in the expression program of several developmentally regulated genes during ESC differentiation, such as ESC pluripotency genes Nanog, Oct4, Sox2 and lineage specific gene Elf5. However, in the ESC state, promoter DNA methylation does not directly correlate with gene expression (Meissner et al., 2008; Weber et al., 2007). It has also been shown that 36% of genes remain expressed despite methylation in the proximal promoter in mESCs (Fouse et al., 2008). In addition, the DNMTs triple knockout (TKO) mESCs possess negligible DNA methylation, yet display only limited alteration in gene expression compared to normal mESCs (Fouse et al., 2008). Thus, it is generally accepted that the influence of DNA methylation on transcriptional regulation is very complex in ESCs. Therefore, it has been proposed that multiple layers of epigenetic mechanism control the transcriptional program of ESCs, and that the effects of a single layer may be minimized by other layers.
We propose that Tet1 and 5hmC provide an important layer of epigenetic regulation in mESCs. Indeed, while our present study clearly demonstrates that Tet1 plays a significant role in dynamic regulation of 5mC and 5hmC at specific genome loci in mESCs, it is worth noting that our global comparative analyses of mESCs gene expression profiling after Tet1 depletion reveal that Tet1 and 5hmC likely have only a limited impact on the transcription of their directly associated genes in mESCs. For example, we observe expression changes in a subset of target genes such as Gli1, Smad6 and Pax6, but little or no effect on many other target genes, such as the ESC pluripotency genes Sox2, Oct4 and Klf4. There are several interpretations for these various transcriptional outcomes caused by Tet1 depletion in mESCs. For instance, histone modifications may compensate for the dynamic changes of 5mC and/or 5hmC on gene expression after Tet1 depletion. In addition, the effects of master transcriptional repressors or activators on a subset of Tet1 target genes may override the regulatory effects of Tet1 in mESCs. Thus, the influence of Tet1 and 5hmC on its target gene expression is intricate, similar to the effects of DNMTs and 5mC on the gene transcriptional program in mESCs.
Interestingly, while Tet1 depletion does not alter the expression of the Ngn2 gene in mESCs (which is silenced in mESCs), Tet1-depleted mESCs show significantly delayed Ngn2 induction during neural differentiation, after RA treatment (Figure 6F). A recent report also shows that Tet1 depletion does not result in the expression change of Elf5 in mESCs; however after culturing in trophoblast stem cell condition for 2 weeks, Tet1-depleted mESC clones show significantly stronger Elf5 induction (Koh et al., 2011). In addition, it was shown that Tet1 plays an important role in mESC lineage specification (Ito et al., 2010; Koh et al., 2011). Together with the previous findings that DNMTs TKO mESCs, which exhibit global hypomethylation, can maintain self-renewal and an undifferentiated ESC state (Tsumura et al., 2006) but are defective in ESC differentiation (Jackson et al., 2004), this suggests that 5mC and Tet1/5hmC may have limited effects on mESC self-renewal and pluripotency, yet are still required for the epigenetic reprogramming during mESC differentiation.
A potential role of Tet1-modulated 5hmC in neurogenesis
Our present genome-wide study of Tet1 and 5hmC clearly reveal that Tet1 binds to a set of key developmental genes in which 5hmC is also enriched. In particular, GO analysis of genes associated with both 5hmC and Tet1 demonstrates significant enrichment in terms of developmental processes, neurogenesis and cell differentiation (Figure S4). Moreover, Tet1 regulates the expression of a set of key neural development genes, such as Pax6 and Neurod1 and governs the DNA methylation state of a group of genes that are important for normal neural function, such as Sgk1 and Bdnf, in mESCs. Tet1 also regulates several key signaling pathways, such as Shh and TGF-β pathways that are essential for neurogenesis, and the expression of Ngn2 during neural induction. Therefore, our results together with the previous findings that 5hmC is present in brain at high levels (Kriaucionis and Heintz, 2009) and that the 5hmC level is increased during mouse cerebellum maturation (Song et al., 2011), strongly suggest that 5hmC and Tet1 may have important roles in neural development and function.
We envision that the epigenetic network established and maintained by the Tet family and 5hmC represents a previously un-appreciated mechanism to ensure the establishment of a precise pattern of DNA methylation, which is dynamically regulated from fertilization, through the zygote and ESC stage and ultimately during full development of vertebrates. Further investigation is warranted to determine how Tet1 recruits or is recruited by cellular complexes involving epigenetic regulation to influence gene transcriptional status, and how 5hmC signals downstream gene expression events.
EXPERIMENTAL PROCEDURES
GST pull-down assay
Proteins were expressed in Rosetta E. Coli cells and purified by Glutathione Agarose beads. GST pull-down assays were performed in different NaCl concentrations as previously described (Zhang et al., 2010).
ChIP-seq
ChIP was performed as previously described using formaldehyde cross-linked ESCs chromatin (Fang et al., 2010). The antibody against Tet1 (1749–1902) was generated and used for ChIP. ChIP-Seq was further performed according to standard Illumina protocols. Read sequences were mapped to the mouse genome (mm9) using ELAND v2 in the CASAVA (Illumina, v1.6) package. Significantly enriched regions were determined by Model-based Analysis of ChIP-Seq (MACS) package (Zhang et al., 2008).
hMeDIP-seq
ESC genomic DNA was purified and sonicated. Illumina adapters were ligated before hMeDIP. 5μg of adapter-ligated DNA was denatured and incubated with 5μl of 5hmC antibody (Active Motif) at 4°C overnight. Antibody-DNA complexes were captured by protein A/G beads. The immunoprecipitated DNA was purified and sequenced followed by standard Illumina protocols.
Targeted bisulfite sequencing
The genomic DNA was purified from control or Tet1 siRNA transfected mESCs by DNeasy kit (Qiagen). Targeted bisulfite sequencing assay was performed as previous report (Deng et al., 2009).
Microarray assay
Two independent sets of microarray assays were performed in J1 and E14 mESCs. Firstly, J1 mESCs were transfected with control siRNA or Tet1 siRNA and harvested 4 days after transfection. Secondly, E14 ESCs were infected with Scr shRNA, Tet1 shRNA2863 or Tet1 shRNA3387 containing lentivirus and harvested after 4-days selection with puromycin. Total RNA was purified by RNeasy kit (Qiagen). The microarray assays were performed using Affymetrix GeneChip mouse genome 430 2.0 array. Data analysis was performed using the bioinformatics toolbox in MATLAB (MathWorks, R2009b).
mRNA-seq
The mRNA-seq was performed according to the previous report (Gan et al., 2010). The sequencing reads were mapped to mouse genome (mm9) using TopHat package (v1.1.2) (Trapnell et al., 2009). RPKM (reads per kilobase per million mapped reads) values were calculated using the Cufflinks package (v0.9.2) (Trapnell et al., 2010) and the differential expressed genes were identified by Cuffdiff package.
Supplementary Material
Acknowledgments
We thank Michelle Mulcahey for the HPLC assay and Di Hu for drawing the model. We thank Stephen Sugrue, Paco Kang and Erin Clark for their critical reading of the manuscript. This work was supported by NIH grants GM078458 and DK077036 to Y.G.S, DA025779 to K.Z., and partly by the “985” Program from the Chinese Ministry of Education and “973” State Key Development Program of Basic Research of China (2009CB825602, 2009CB825603). Y.G.S. is a PEW scholar. J.D. was sponsored by a CIRM post-doctoral fellowship.
Footnotes
Note added in proof
While this manuscript was under reviewed, three related papers appeared in Nature (Ficz et al., 2011; Wu et al., 2011) and Genes & Development (Wu et al., 2011).
ACCESSION NUMBERS
The Tet1 CXXC pull down-seq, microarray, hMeDIP-seq, targeted bisulfite seq, ChIP-seq and mRNA-seq data have been deposited in the GEO database under the accession number GSE28500.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Boyer LA, Mathur D, Jaenisch R. Molecular control of pluripotency. Curr Opin Genet Dev. 2006;16:455–462. doi: 10.1016/j.gde.2006.08.009. [DOI] [PubMed] [Google Scholar]
- Chodavarapu RK, Feng S, Bernatavichute YV, Chen PY, Stroud H, Yu Y, Hetzel JA, Kuo F, Kim J, Cokus SJ, et al. Relationship between nucleosome positioning and DNA methylation. Nature. 2010;466:388–392. doi: 10.1038/nature09147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng J, Shoemaker R, Xie B, Gore A, LeProust EM, Antosiewicz-Bourget J, Egli D, Maherali N, Park IH, Yu J, et al. Targeted bisulfite sequencing reveals changes in DNA methylation associated with nuclear reprogramming. Nat Biotech. 2009;27:353–360. doi: 10.1038/nbt.1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang R, Barbera AJ, Xu Y, Rutenberg M, Leonor T, Bi Q, Lan F, Mei P, Yuan GC, Lian C, et al. Human LSD2/KDM1b/AOF1 regulates gene transcription by modulating intragenic H3K4me2 methylation. Mol Cell. 2010;39:222–233. doi: 10.1016/j.molcel.2010.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouse SD, Shen Y, Pellegrini M, Cole S, Meissner A, Van Neste L, Jaenisch R, Fan G. Promoter CpG methylation contributes to ES cell gene regulation in parallel with Oct4/Nanog, PcG complex, and histone H3 K4/K27 trimethylation. Cell Stem Cell. 2008;2:160–169. doi: 10.1016/j.stem.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan Q, Chepelev I, Wei G, Tarayrah L, Cui K, Zhao K, Chen X. Dynamic regulation of alternative splicing and chromatin structure in Drosophila gonads revealed by RNA-seq. Cell Res. 2010;20:763–783. doi: 10.1038/cr.2010.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Globisch D, Munzel M, Muller M, Michalakis S, Wagner M, Koch S, Bruckl T, Biel M, Carell T. Tissue distribution of 5-hydroxymethylcytosine and search for active demethylation intermediates. PLoS One. 2010;5:e15367. doi: 10.1371/journal.pone.0015367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Pastor WA, Shen Y, Tahiliani M, Liu DR, Rao A. The behaviour of 5-hydroxymethylcytosine in bisulfite sequencing. PLoS One. 2010;5:e8888. doi: 10.1371/journal.pone.0008888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, D’Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson M, Krassowska A, Gilbert N, Chevassut T, Forrester L, Ansell J, Ramsahoye B. Severe global DNA hypomethylation blocks differentiation and induces histone hyperacetylation in embryonic stem cells. Mol Cell Biol. 2004;24:8862–8871. doi: 10.1128/MCB.24.20.8862-8871.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko M, Huang Y, Jankowska AM, Pape UJ, Tahiliani M, Bandukwala HS, An J, Lamperti ED, Koh KP, Ganetzky R, et al. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature. 2010;468:839–843. doi: 10.1038/nature09586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh KP, Yabuuchi A, Rao S, Huang Y, Cunniff K, Nardone J, Laiho A, Tahiliani M, Sommer CA, Mostoslavsky G, et al. Tet1 and tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell. 2011;8:200–213. doi: 10.1016/j.stem.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Hanna J, Zhang X, Ku M, Wernig M, Schorderet P, Bernstein BE, Jaenisch R, Lander ES, Meissner A. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454:49–55. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng RK, Dean W, Dawson C, Lucifero D, Madeja Z, Reik W, Hemberger M. Epigenetic restriction of embryonic cell lineage fate by methylation of Elf5. Nat Cell Biol. 2008;10:1280–1290. doi: 10.1038/ncb1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penn NW, Suwalski R, O’Riley C, Bojanowski K, Yura R. The presence of 5-hydroxymethylcytosine in animal deoxyribonucleic acid. Biochem J. 1972;126:781–790. doi: 10.1042/bj1260781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song CX, Szulwach KE, Fu Y, Dai Q, Yi C, Li X, Li Y, Chen CH, Zhang W, Jian X, et al. Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nat Biotechnol. 2011;29:68–72. doi: 10.1038/nbt.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- Szwagierczak A, Bultmann S, Schmidt CS, Spada F, Leonhardt H. Sensitive enzymatic quantification of 5-hydroxymethylcytosine in genomic DNA. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilgner H, Nikolaou C, Althammer S, Sammeth M, Beato M, Valcarcel J, Guigo R. Nucleosome positioning as a determinant of exon recognition. Nat Struct Mol Biol. 2009;16:996–1001. doi: 10.1038/nsmb.1658. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotech. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsumura A, Hayakawa T, Kumaki Y, Takebayashi S-i, Sakaue M, Matsuoka C, Shimotohno K, Ishikawa F, Li E, Ueda HR, et al. Maintenance of self-renewal ability of mouse embryonic stem cells in the absence of DNA methyltransferases Dnmt1, Dnmt3a and Dnmt3b. Genes to Cells. 2006;11:805–814. doi: 10.1111/j.1365-2443.2006.00984.x. [DOI] [PubMed] [Google Scholar]
- Valinluck V, Tsai HH, Rogstad DK, Burdzy A, Bird A, Sowers LC. Oxidative damage to methyl-CpG sequences inhibits the binding of the methyl-CpG binding domain (MBD) of methyl-CpG binding protein 2 (MeCP2) Nucleic Acids Res. 2004;32:4100–4108. doi: 10.1093/nar/gkh739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber M, Hellmann I, Stadler MB, Ramos L, Paabo S, Rebhan M, Schubeler D. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet. 2007;39:457–466. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- Wyatt GR, Cohen SS. The bases of the nucleic acids of some bacterial and animal viruses: the occurrence of 5-hydroxymethylcytosine. Biochem J. 1953;55:774–782. doi: 10.1042/bj0550774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Bian C, Lam R, Dong A, Min J. The structural basis for selective binding of non-methylated CpG islands by the CFP1 CXXC domain. Nat Commun. 2011;2:227. doi: 10.1038/ncomms1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young RA. Control of the embryonic stem cell state. Cell. 2011;144:940–954. doi: 10.1016/j.cell.2011.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Zhang X, Clark E, Mulcahey M, Huang S, Shi YG. TET1 is a DNA-binding protein that modulates DNA methylation and gene transcription via hydroxylation of 5-methylcytosine. Cell Res. 2010 doi: 10.1038/cr.2010.156. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu XS. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.