Abstract
Background
Change detection is a critical component in the diagnosis and monitoring of many slowly evolving pathologies.
Objective
This article describes a semi-automatic monitoring approach using longitudinal medical images. We test the method on brain scans of meningioma patients, which experts found difficult to monitor as the tumor evolution is very slow and may be obscured by artifacts related to image acquisition.
Methods
We describe a semi-automatic procedure targeted towards identifying difficult-to-detect changes in brain tumor imaging. The tool combines input from a medical expert with state-of-the-art technology. The software is easy to calibrate and, in less than five minutes, returns the total volume of tumor change in mm3. We test the method on post-gadolinium, T1-weighted Magnetic Resonance Images of ten meningioma patients and compare our results to experts’ findings. We also perform benchmark testing with synthetic data.
Results
Our experiments indicated that experts’ visual inspections are not sensitive enough to detect subtle growth. Measurements based on experts’ manual segmentations were highly accurate but also labor intensive. The accuracy of our approach was comparable to the experts’ results. However, our approach required far less user input and generated more consistent measurements.
Conclusion
The sensitivity of experts’ visual inspection is often too low to detect subtle growth of meningiomas from longitudinal scans. Measurements based on experts’ segmentation are highly accurate but generally too labor intensive for standard clinical settings. We described an alternative metric that provides accurate and robust measurements of subtle tumor changes while requiring a minimal amount of user input.
Keywords: Automatic Change Detection, Meningioma, Growth Rate, Time Series Analysis, Statistical Modeling, Longitudinal Studies, Slowly Evolving Pathologies
Introduction
Meningiomas account for at least 25% of all primary intracranial tumors in the United States 1, 2. They are generally slowly growing lesions that can occur within the meningeal tissues on the surface of the brain, the skull base, the dural reflections, or within the ventricles. About 90% of these tumors are classified as histologically benign3. However, benign brain tumors that demonstrate continuous growth may cause neurologic deficits and eventually, if left untreated, death. Thus, knowing whether a tumor is growing or not over time is a critical decision point for the treatment of the patient. The growth patterns of benign menignioma are still an active field of research4. Even relatively large meningioma can suddenly stop growing5, 6. Many neurosurgeons therefore generally avoid operating on patients with benign meningiomas, particularly in those cases where the pathology is relatively small or difficult to access surgically and the patient is not symptomatic. Instead, close monitoring with serial imaging, usually with magnetic resonance imaging (MRI), and neurologic evaluation is used to assess for tumor progression7. Neuroradiologists and clinicians visually inspect the scans for evidence of change in tumor volume.
In clinical practice, finding evidence for subtle growth through visual inspection of serial imaging can be very difficult. This is especially true for scans taken at relatively short intervals (less than a year). Visual inspection often misses the slow evolution of many meningiomas, as the change can be obscured by variations in head position, slice position, or intensity profile between scans. In addition, very small changes in the linear dimensions seen on cross-sectional imaging can reflect appreciable volumetric change.
Surgeons and oncologists frequently analyze the evolution of meningioma by combining visual inspection of brain scans via a light box or visualization software with sophisticated measuring techniques. These techniques commonly estimate the tumor volume in each scan based on the diameter of the tumor. The medical expert then computes the change in tumor volume by comparing the measurements of consecutive scans. Based on these metrics, the change in tumor volume, however, is often difficult to detect between two sequential scans (such as in Figure 1). Instead, neuroradiologists tend to compare the most recent scan to the earliest available image to find any visible evidence for the evolution of the tumor. The resulting analysis does not reflect the current development of the tumor but rather a retrospective perspective of the tumor evolution. In addition, such methods generally do not provide a quantitative measure for the rate of volume change. Accurate quantitative measurements can aid clinicians in treatment decisions.
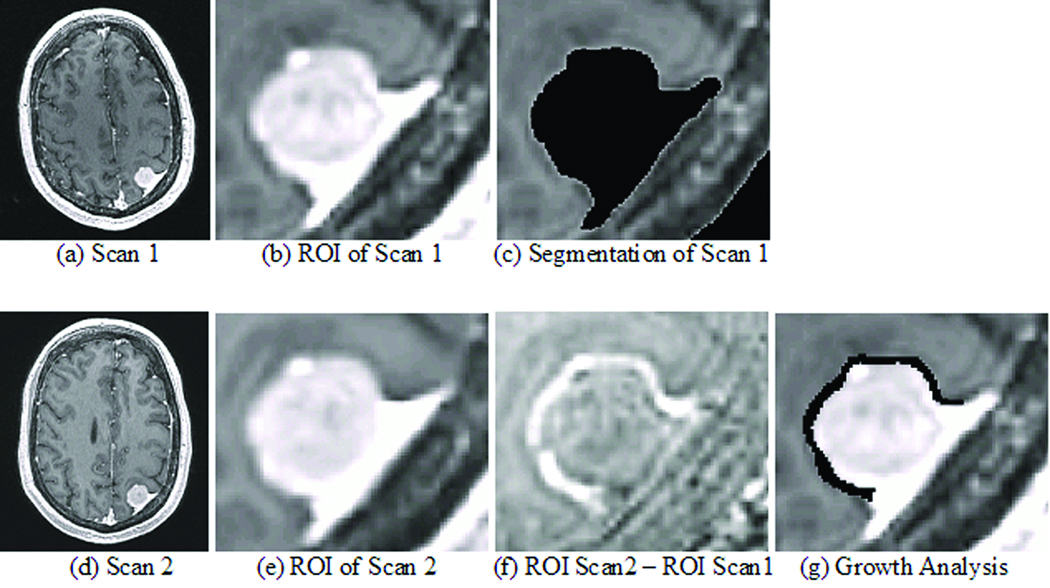
Figure 1.
The top row shows images related to the first (earlier time point) scan and the bottom row to the second (later time point) scan. The first column consists of the input of the pipeline. The second column is the result of the second step of the pipeline, which fuses both images. The third column shows the resulting segmentation of scan 1 and the image resulting from subtracting (b) from (e). Bright voxels indicate tumor growth. The image in (g) is the result of the growth analysis showing the growth in black.
In the remainder of this report we attempt to address these issues by describing a semi-automatic procedure specifically targeted towards identifying difficult-to-detect changes in pathology. Our approach first semi-automatically segments the tumor in the initial patient scan. It then co-registers the sequence of scans. Finally, it measures growth or shrinkage from these images based on the statistical analysis of the differences in their intensities. The software is easily calibrated and, in less than five minutes, returns the total volume of tumor change in mm3 and percent change. This approach is disseminated as part of the 3D Slicer (www.slicer.org), a publicly available software package for medical imaging processing and visualization.
We tested the method on synthetic data with known tumor growth as well as on MRI scans of ten patients with intracranial meningioma. These experiments demonstrated that our semi-automatic tool provided a reliable measurement for volume change requiring minimal user input.
Method: A Software Tool for Monitoring Slowly Growing Tumors
The goal of our semi-automatic procedure is to detect subtle changes in pathology at the level of a few voxels. It does so by completing three steps: the tool identifies the tumor in the first scan, then normalizes the series of scans, and finally detects growth based on these results.
The first step performs a user-guided segmentation of the tumor in the first scan. The tool ignores the follow up scans at this point as the variations in the acquisition across scans could negatively influence the accuracy of the approach. Manual supervision of the segmentation ensures highly accurate segmentations. The user guides the approach by specifying a region of interest around the tumor and a lower and upper bound on the intensities characterizing the pathology. Based on the input by the user, the pipeline reliably extracts the tumor volume from post-gadolinium, T1-weighted MRIs, as meningiomas are generally characterized as homogeneous, bright objects8. The software then “cleans” the segmentation by removing small islands and holes caused by noise in the MRIs. The resulting map may include false positives, as Dura, vessels, and skull can have similar intensity patterns as the tumor (see Figure 1(c)). These structures, however, should be stable over time so that their impact on the analysis can be ignored.
The second step automatically fuses the remaining scans to the first by completing four stages. It first (rigidly) aligns the remaining scans to the first one using mutual information9. As discussed in the Introduction, this procedure adjusts the global pose of the scans while preserving the size of the tumor. The tumor regions across all scans are then roughly aligned with each other so that the previously assigned region of interest defines the tumor region across all scans. The method then increases the resolution by performing interpolation within each region to address partial volume effects in the MRIs. Partial volume artifacts are caused by insufficient image resolution leading to combining multiple structures within a voxel. This often causes blurry tissue boundaries, as the intensity of these voxels is a mixture of the intensities of adjacent structures. In the third stage, the framework addresses non-linear distortion artifacts by rigidly registering only the tumor regions. This process results in a series of images where, in theory, barring temporal changes, the pathology is well aligned (see Figure 1 (b) and (e)). The final stage normalizes the intensity patterns of each scan as the uptake of the enhancement agent by the tumor generally varies across scans. The tool normalizes the intensities by first specifying the region where we do not expect any change in tumor volume. This region is a conservative estimate of stable tissue based on the segmentation of the first step. It then computes the intensity histogram for each scan and modifies the global intensity patterns of each scan via scaling until all scans have the same mean intensity inside the stable tissue region.
The final step of the pipeline measures the evolution of the tumor via a metric that detects change by analyzing differences in local intensity patterns. We relate change detection to hypothesis testing. The test is specified by the null hypothesis that local changes in intensity between two scans are caused by image artifacts. To perform the test, the tool first measures the normal variation in intensity patterns of stable tissue between the first and the remaining scans. Our tool measures the variation by subtracting each MRI scan from the first scan, computing the absolute value of this new set of images, and then recording the intensity distribution of the processed images within the stable tissue region. Due to the normalization in the previous step, the resulting images display stable tissue by dark intensities and changes between the scans by bright intensities (see Figure 1 (f)). Brighter intensities inside this region are generally the result of noise or slight misalignment between scans. We reject the null hypothesis for voxels whose intensities are brighter than the intensities of 99% of voxels inside the stable region. We call this percentage the “cut-off percentage” of our metric. These voxels are then further classified into growing or shrinking pathology depending on which scan shows bright intensities and the location of the voxel. The total tumor volume change is then computed by subtracting the volume of voxels labeled as shrinkage from the volume labeled as growth (see Figure 1 (g)).
Our software allows the modification of the cut-off percentage so that one can specify a value specific to each case. While our approach is somewhat insensitive to small changes in that percentage, smaller cut-off percentages make our metric more sensitive to noise in the image. For our study, we empirically choose 99%. We found this cut-off percentage to produce the most consistent results on our data sets when testing our metric with values ranging from 30% – 99.9%.
Data Acquisition
Real Data Set
Our experiments are based on data consisting of nine female patients and one male patient with benign meningioma who had not received surgery to remove the pathology. All patients were scanned twice by the Department of Radiology, Brigham and Women’s Hospital, Boston, MA following normal hospital procedure (axial scan direction, post-gadolinium, FOV: 240 mm, Matrix: 256 × 256 ×130, Voxel Dimension: 0.9375 mm × 0.9375 mm × 1.2 mm, Scan Time: 8 minutes). For the nine women, the mean follow up period was 13.2 months (ranging from 6 months to 21 months). The male patient received the contrast agent before the initial scan and then was scanned twice 8 minutes apart. The difference in contrast and image quality between the two scans was similar to the ones found in the other nine female cases. A radiologist, who we refer to as Expert A, and a neurosurgeon, who we refer to as Expert B, independently confirmed that the MRI scans of all ten cases were consistent with meningioma. The experts then manually outlined the tumor in each slice of the scans of the nine female cases using 3D Slicer. The tumor in the scans of the male cases was manually segmented by Expert B using only the same procedure as for the female cases.
Dataset with Known Ground Truth
We also generated two synthetic data sets to further analyze the strength and weaknesses of different metrics. For the first data set, which we refer to as dataset with known ground truth, we synthetically grew the pathology in one of the ten initial scans (see Figure 2). We initialized the computer simulation via a template consisting of a manual segmentation of the meningioma and surrounding tissue in the selected scan. The simulator then uses the template to parameterize a simple mechanical model that simulates the tumor growth in the original MR scan10. The mechanical model assumes that the tumor expands homogeneously and that the tumor’s uptake of enhancing agent in the follow up scan is similar to the original scan. Based on these assumptions, the computer simulator generates a new scan by deforming the surrounding tissue around the tumor while keeping the skull fixed. This simulator is somewhat insensitive to the shape of the tumor as defined by the template. The software only uses the template to extract the skull and to determine the tumor’s geometric center in the MR scan.
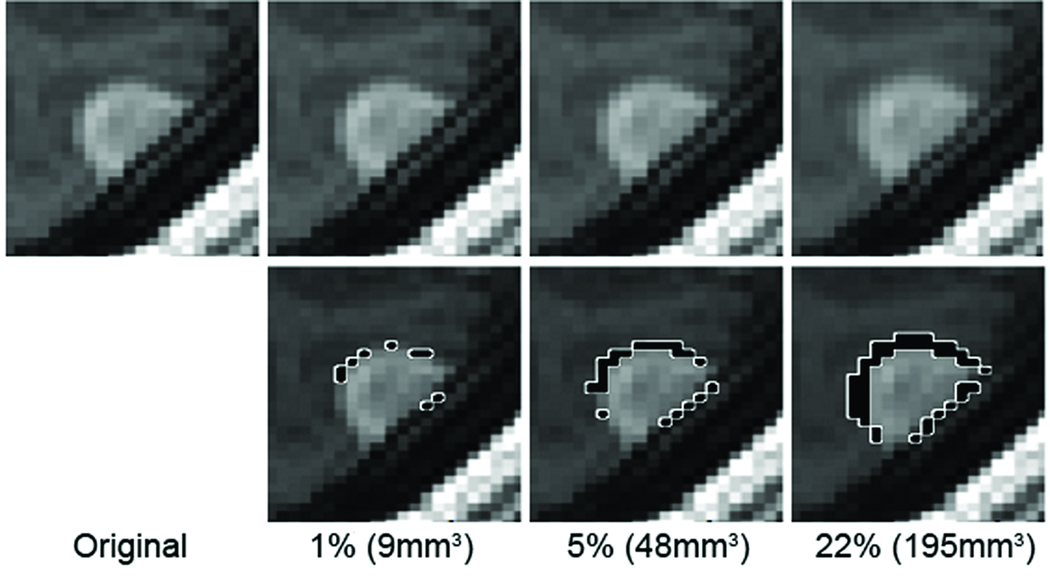
Figure 2.
The real scan (first panel) of the patient with synthetically evolving pathology of 1%, 5% and 22% volume growth (highlighted in black with white boundaries in the second row). We note that visually detecting growths of 1% and 5% is extremely difficult.
For our test sequence, the simulator generated six synthetic follow up scans with different growth values. For each synthetic scan, we computed the true growth percentage of the tumor by applying the simulator to the template instead of the MR scan, computing the volume difference of the tumor between the original and modified segmentation, and then normalizing the result by the volume of the tumor in the template. We then applied ten randomly generated rigid transformations to each scan to simulate changes in head positions between consecutive scans. The resulting test sequence consisted of 60 cases for which the exact tumor growth was known. The growth percentages were 1% (9 mm3), 3%, 5%, 11%, 16%, and 22% (195 mm3).
Dataset with Modified Scan Resolution
The second synthetic data set consists of follow up scans with varying scan resolution. We synthetically modified the slice thickness in the follow up scan of the case used in the previous data set. This resulted in four follow up scans with slice thinness of 1.2 mm (the original slice thickness), 2.4mm, 3.6mm, and 4.8 mm.
Results
We performed four experiments to determine the accuracy of different approaches for detecting subtle growth in meningioma. The first experiment tested the accuracy of experts detecting subtle growth via visual inspection, manual segmentations, and our semi-automatic approach. For this experiment, we use the synthetic data set, where the change in volume is known. The remaining three experiments compared manual and semi-automatic growth metrics on synthetic as well as real MR scans.
Synthetic Dataset with Known Tumor Growth
The first experiment assessed the accuracy of qualitative tumor change detection via visual inspection. We trained five post-doctoral students in visually detecting changes in meningioma. After all five raters reliably analyzed the training cases we recorded their accuracy in visual inspection of 30 of the 60 cases of the synthetic data set with known ground truth. Each rater first visually compared the original to the follow up scan and then classified each case as ‘stable’, ‘shrinking’ or ‘growing’ pathology. We then compared those findings with known ground truth. For each of the six growth categories (1%, 3%, 5%, 11%, 16%, 22%), we computed for each rater an accuracy score, which was the number of times the rater correctly identified growth for cases in this category divided by the number of cases in this category. Table 1 lists the average accuracy with standard deviation across the experts. We also repeated this study for three growth categories with Expert A. Expert A wrongly classified all cases with 1% growth as stable, achieved an accuracy of 20% for cases with 5% growth, and correctly classified all cases with 22% growth. The accuracy scores of Expert A were thus within one standard deviation of those of the five raters.
Table 1.
Accuracy of five experts detecting growth through visual inspection. We used a computer simulation to generate 30 synthetic scans, where the tumor grew from 1% to 22% in comparison to the original scan (see Section “Data Acquisition”). We then showed the original and the synthetic follow up scans to five experts, who independently classified each case as ‘stable’, ‘shrinking’ or ‘growing’ pathology. The table lists the average and standard deviation in % of the experts correctly identifying growth in the five cases for each growth category. The experts achieved an average accuracy of more than 50% for cases with at least 16% (or 142 mm3) growth. The relatively low accuracy of visually detecting subtle tumor growth underlines the need for more accurate metrics.
| Growth | 1% | 3% | 5% | 11% | 16% | 22% |
|---|---|---|---|---|---|---|
| Accuracy of Detection (mean ± std) |
8 ± 8 % | 6 ± 6 % | 28 ± 11% | 44 ± 9% | 52 ± 24% | 88 ± 12% |
An alternative metric for detecting change is based on manual segmentation of the tumor in the original and follow up scan. The change in pathology is then defined by the volume difference of tumor in the two segmentations. We quantify the volume difference between the initial and follow-up scans by first recording the volume of the tumor in each scan based on the corresponding manual segmentations. We then compute the change in tumor volume between scans by subtracting the tumor volume in the initial scan from the one in the follow up scan. We note that this metric is sensitive to the expert’s opinion about the shape of the tumor in the initial scan. Based on our experiments, experts often disagree about the exact shape. For the nine female patients, the average intra-rater correlation coefficient of the tumor volume in the first scan measured by Expert A and Expert B was 97% (F=34.193, df = 8). To account for the differences between the experts, we report the change in tumor volume in percent. The percentage in volume change is defined by the change in volume divided by the volume of the tumor in the first scan as measured by the expert.
We measured the accuracy of this metric by having Expert A manually segment the tumor in the original MR scan as well as 12 synthetically generated follow up scans. The results are shown in Figure 3 with the true growth percentage (ground truth) represented by the dotted line and the outcome of Expert A by the grey dots. The true growth percentage and the percentage measured by Expert A were defined with respect to Expert A’s segmentation of the initial scan instead of the template. As in the case of the template, we computed the true growth percentage by applying the expert’s segmentation to the simulator, computing the volume difference of the tumor between the original and modified segmentation, and then normalizing the result by the volume of the tumor in the segmentation. The true growth percentages, with respect to Expert A’s initial segmentation, were 1.2%, 3.6%, 6.0%, 12.1%, 18.5%, and 24.9%.
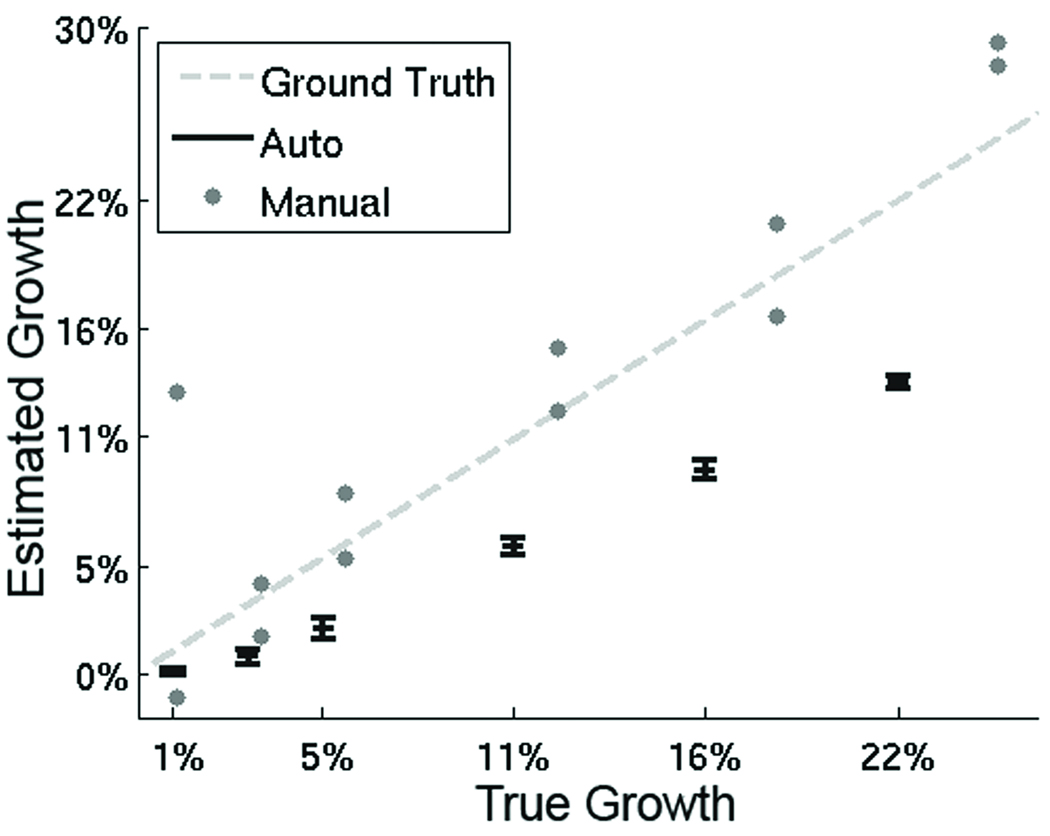
Figure 3.
The graph summarizes the measurements on the synthetic data based on our semi-automatic procedure (Auto) and manual segmentations (Manual). On this synthetic data set, the ground truth is known (dotted line). The x-axis represents the true growth while the y-axis corresponds to the growth computed by the metrics. The gray dots represent a single Manual measurement by Expert A, who did two trials for each growth category. The black bars indicate the mean and standard deviation of the measurements of Auto for each growth category. In general, Manual seems to overestimate change but seems closer to the ground truth. Auto seems to underestimate change. Furthermore, the relatively small standard deviation of Auto implies that the metric is robust to changes in head position.
Our semi-automatic tool is an alternative to visual inspection and manual segmentations. We applied our tool to all 60 cases initialized by the template of Section “Data Acquisition”. For each case, the tool completed its analysis in less then 5 minutes compared to up to 40 minutes for the measurements based on manual segmentations. Table 1 shows the mean and standard deviation of the measured percentage for each growth category (1%, 3%, 5%, 11%, 16%, 22%). For 1% (9 mm3) growth, our semi-automatic metric successfully detected growth for seven cases and labeled three cases as stable. Our metric successfully detected growth for all cases with growth larger than 1%. The mean error between the growth percentage measured by our metric and the actual growth percentage was −4.47% with a standard deviation of 2.62%. In comparison, the mean error of Expert B was 3.26% with a standard deviation of 3.71%. The unpaired t-test of the error scores of our metric versus the ones of Expert B revealed a (two-tailed) p value of less than 0.0001 (t = 9.1353, df = 72). The results of the two metrics are therefore significantly different in this study.
Dataset with Modified Scan Resolution
The second experiment studies the impact of the scan resolution on the outcome of our proposed metric. For this, we apply our approach to the synthetic data set consisting of follow up scans with varying scan resolution. The relative growth percentage based on the original scans (1.2 mm slice thickness) was 1.99%, based on the follow up scan with 2.4 mm slice thickness was 2.40%, based on the follow up scan with 3.6 mm slice thickness was 2.31%, and based on the follow up scan with 4.8 mm slice thickness was 2.29%. The growth percentage across the four cases thus only deviated by 0.41%.
Growth Measurements on Cases with Scans taken at least 6 Months Apart
The next experiment evaluated the inter-rater and intra-rater variability of volumetric measurements based on the manual segmentations by Expert A and Expert B. We also compared these measurements to the ones obtained with the semi-automatic method. Table 2 summarizes the measurements across the nine female patients. The patients are listed by their average percentage of change in tumor volume across the three measurements. In the remainder of this section we classify a case as stable if the corresponding measured percentage is above −0.5% and below 0.5%. Otherwise, we categorize the case as shrinkage or growth depending on whether the percentage is negative or positive. We label two different measurements of the same patient as similar when their percentages in tumor volume change are less than 1.5% apart.
Table 2.
The table list manual and semi-automatic volume differences of the tumor in the nine cases. Based on these measurements, we found relatively large variations between the results of the two experts. Our semi-automatic procedure seems to be a conservative estimate of change (see also Figure 4).
| Case | Expert A | Expert B | Semi-Automatic | |||
|---|---|---|---|---|---|---|
| % | mm3 | % | mm3 | % | mm3 | |
| 1 | −4.98 | −276 | −6.28 | −323 | −2.54 | −227 |
| 2 | 2.54 | 99.7 | −3.72 | −68.9 | 0.0 | 0.0 |
| 3 | 2.22 | 278.4 | 0.1 | 6.3 | 0.0 | 0.0 |
| 4 | 3.50 | 27.07 | −0.44 | −2.4 | 0.0 | 0.0 |
| 5 | 0.94 | 177.0 | 5.44 | 883.8 | 1.99 | 341.9 |
| 6 | 3.94 | 544.9 | 4.81 | 596.4 | 4.36 | 593.0 |
| 7 | 16.61 | 1138 | 22.23 | 1697 | 13.30 | 1190 |
| 8 | 13.39 | 377.6 | 30.66 | 685.5 | 11.47 | 257.9 |
| 9 | 21.29 | 902.6 | 38.98 | 1165 | 22.83 | 808.2 |
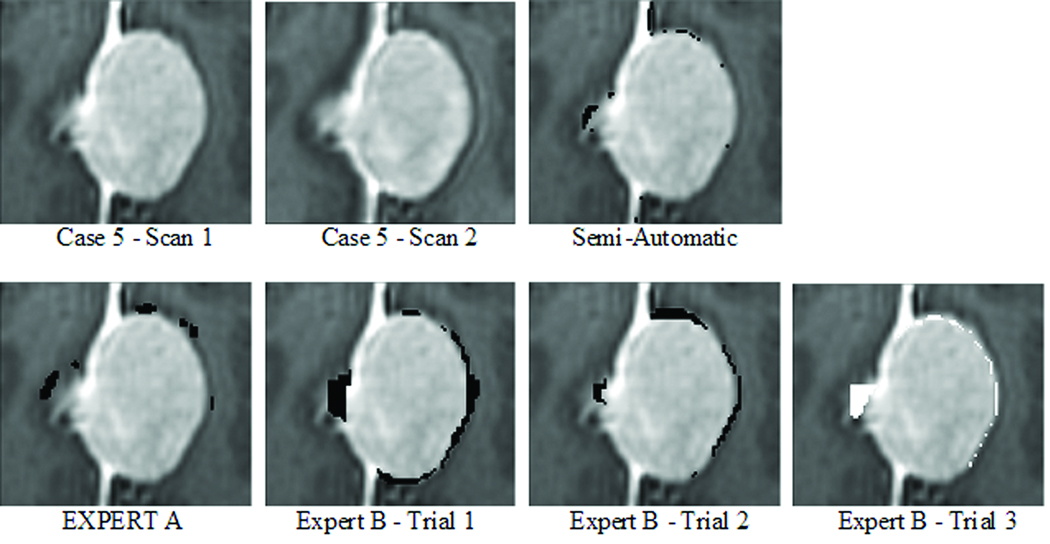
We first measured the intra-rater variability of Expert B. The expert segmented the two scans of Case 5 three times, with a delay of one week between the segmentation sessions. Case 5 was selected as the patient that had the least amount of change and both experts agreed about the type of change (growth, stable, or shrinkage). Furthermore, the initial and follow up scans only showed subtle differences, making accurate visual change detection challenging (see Figure 2). The resulting volumetric measurements in change varied by almost 1 ml or 6% in volume (first: 883.8 mm3, second: 545.8 mm3, third: −99.8 mm3).
Next, we captured the inter-rater variability of volumetric change detection of the two experts across the nine patients (see also Figure 4). Expert A measured larger volumetric change in four patients, Expert B indicated larger volumetric change in three patients, and in only two patients (Case 1 and Case 6) the growth analysis of the experts was similar. The mean difference in the percentage of volume change was −3.7%, implying that Expert B generally measured a higher percentage of volume change than Expert A.
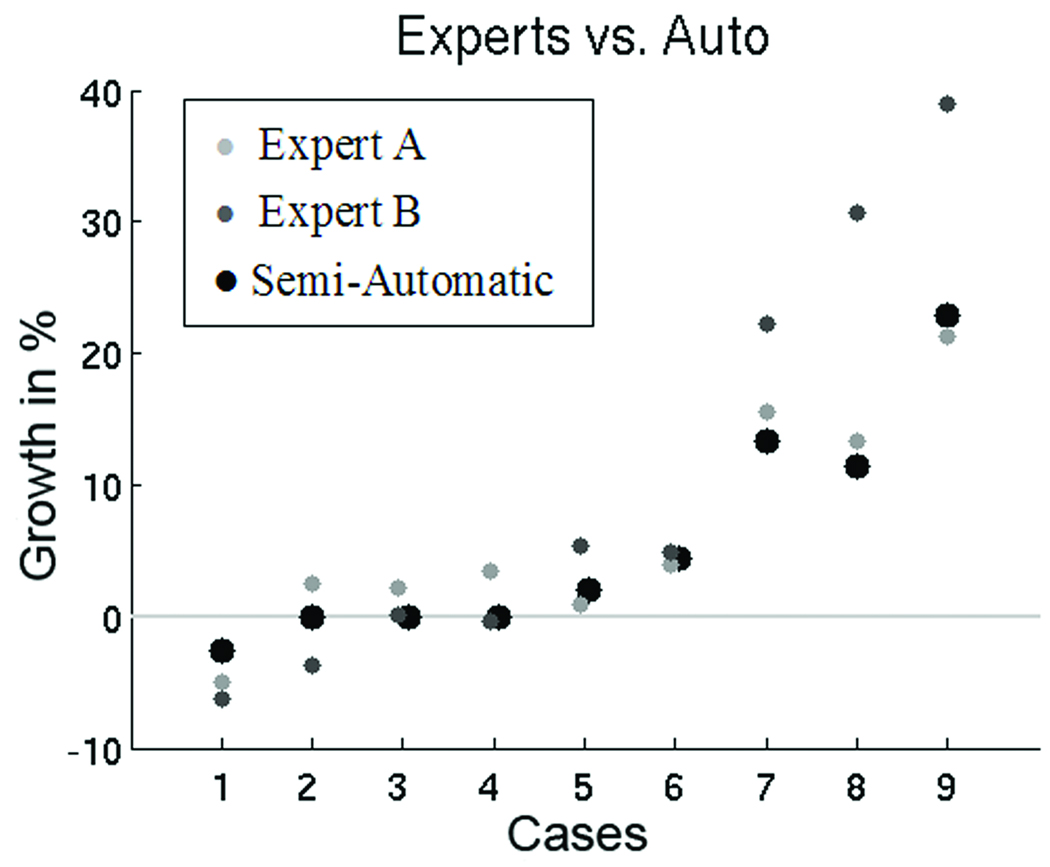
Figure 4.
Growth in percent of Expert A (light gray circle), Expert B (dark gray circle), and our semi-automatic approach (black circle). We note, that the experts only agree in six cases about shrinkage, stable, or growth of the tumor. The semi-automatic measure seems to be a more conservative metric compared to results of the experts.
We also measured the volume change in the nine female patients with our semi-automatic procedure. In six cases, the semi-automatic measurements were similar to the more conservative estimate of the two experts. A measurement X is more conservative than measurement Y, if the absolute value of X is lower than the absolute value Y. Across the nine cases, the semi-automatic results more closely correlated with Expert A’s finding (0.78 ±2.14%) than the two experts agreed with one another (−3.7 ±2.14%).
Growth Measurements on Clinical MRI Scans Taken 8 Minutes Apart
The last experiment analyzes the accuracy of the volumetric measurement as well as our semi-automatic metric in establishing stability of meningioma. Expert B segmented the tumor in the two scans of the male patient. The scans were taken 8 minutes apart so that, theoretically, no change should be detected. The relative tumor growth with respect to these segmentations was −0.52%, where the change in volume measured by our metric was −0.26%.
Discussion
As mentioned in the Introduction, many neurosurgeons avoid operating on patients with benign meningiomas by closely monitoring the tumor progression through visual inspection of serial scans of the patients. Based on our synthetic data experiment with known ground truth, our experts achieved an average accuracy above 50% for cases that showed more than 16% (or 142 mm3) in tumor growth (see also Table 1). From our clinical experience, yearly follow up scans of patients affected by meningioma often show growth of less than this. In those cases, our experiment indicates that visual inspection generally is not sensitive enough to accurately track changes in the tumor volume. Alternative metrics for capturing change are therefore necessary.
One such metric is proposed by the WHO (World Health Organization) response criteria11 and MacDonald criteria. These approaches infer the size of a tumor through two orthogonal diameters, which are the tumor’s largest diameter and perpendicular diameter12. To increase efficiency and reproducibility, the Response Evaluation Criteria in Solid Tumors (RECIST)13 is based only on the largest diameter. Alternatively, Zeideman et al.14 suggest three measurements along the perpendicular axes of the tumor. These three metrics, however, ignore small growth deviating from the largest diameter directions15, 16.
Volumetric measurements based on manual segmentations are considered more accurate,17–19 especially with respect to visual inspection. This statement is supported by our experiment on the synthetic data set with known ground truth. Using manual segmentation, the 1% (or 9 mm3) growth case was labeled as shrinking in one case and as growing in the other. All cases with higher growth percentage were correctly identified as growing using manual volumetric segmentation. On this data set, the volumetric measurement was more sensitive compared to visual inspection. Furthermore, it provided the clinician with a quantitative measurement for change in pathology.
On the downside, the volumetric measurements are sensitive to variations in head position indicated by the relatively large difference in measurements for each growth category. They are also considered sensitive to inter- and intra rater variability (see 20), which was also reflected in the outcome of our experiments based on the real MRI scans. We analyzed the impact of intra-rater variability of Expert B on the volumetric measurement by having Expert B repeat the measurement three times for one case. The first two measurements clearly indicated tumor growth, while the last measurement suggested shrinkage. The large differences between the measurements are also visible in the sample slice shown in Figure 5. The growth detected through volumetric change detection is visualized in black while white represents shrinkage. Finally, the three measurements varied by almost 1 ml or 6% in volume, which is quite large given that the measured change in pathology for each of the nine patients with serial imaging was less than 1 ml.
Figure 5.
An example slice of Case 5 with corresponding change detection of different metrics. Growth is shown by black while shrinkage is displayed in white. The change in tumor volume is quite subtle in this case and difficult to detect through visual inspection. This might explain the relatively large differences in change detection between the different metrics.
We analyzed the impact of the inter-rater variability on volumetric measurements by comparing the outcome of Expert A and Expert B on the nine serial cases. For six of the nine cases in our data set, the tumor changed by less than 10% in volume according to the two experts. The standard deviation between the two measurements of the two experts across the nine cases was 8.76%, which was relatively large given that most of the cases grew by less than 10%. Figure 5 visually confirms these findings, as the images show relatively large differences between the changes detected by both experts. Furthermore, the experts only agreed for six of the nine patients about shrinkage, stable, or growth of the tumor. Another drawback of the volumetric metric is the amount of labor needed to generate the measurements. In the case of the real MRI scans, the two experts required on average 40 minutes to generate a measurement. The labor associated with the measurement makes the method generally unsuitable for the clinical setting.
Computer scientists try to address this issue by developing automatic segmentation methods 21, 22. These methods outline the pathology in a scan by combining the image data with general information about the visual appearance of healthy tissue and pathology. To quantify the change in tumor volume, metrics based on segmentations first determine the tumor volume in each scan from the corresponding segmentation and then measure the difference in tumor volume across scans. This type of quantitative measure is adversely impacted by variations in image acquisition, such as changes in head position or intensity profile, as these metrics independently compute the volume of the tumor for each scan. To date, these methods are therefore not widely used by clinicians.
Another type of automatic growth analyzes of lesions simultaneously processes the sequence of scans23–25. These tools first fuse the sequence of images and then flag unusual patterns across the scan sequence as changes in pathology. Rey et al. 26 apply this concept to patients with multiple sclerosis where lesion progression is clearly visible. They first align each scan to a fixed coordinate system using highly flexible and region-specific transformations. The authors then show that the transformations in regions with growing lesions are inherently different from regions with stable tissue. Angelini et al.27 propose an alternative approach for low grade gliomas with a growth rate above 30%. They fuse the scans by first aligning the head in each scan to a fixed pose using rigid, global transformations. Afterwards, they equalize the intensity patterns across the scans. They then relate tumor growth to gross regional differences in intensity patterns across the scans. To the best of our knowledge, current state-of-the-art software targeted towards change detection in pathology has been exclusively tested on scans with visibly apparent tumor or lesion growth. The accuracy of these methods on benign meningioma cases and more generally in very slowly evolving pathologies is unclear, especially in those cases where the change is visually difficult to detect.
We attempt to address this issue in this article by describing a semi-automatic procedure specifically targeted towards identifying difficult-to-detect changes in pathology. We are specifically interested in applying our metric to cases where current methods generally fail. In the synthetic data experiments (see also Figure 3) our semi-automatic metric was more robust for cases with smaller growth value compared to the measurements based on manual segmentations of Expert A. The metric was generally a conservative estimate of change for all cases with growth larger than 1%. Furthermore, the relatively small standard deviation of our metric implied that our approach was minimally impacted by changes in head position unlike the measurements by Expert A. Additionally, the data set consisting of follow up scans with varying slice thickness revealed that the impact of the slice thickness on our measurements is relatively low. Our semi-automatic method was less accurate in cases with large changes compared to ones with small changes in meningioma. Large changes in a tumor are often captured by different properties than subtle changes. While the shape of the meningioma generally stays consistent between scans of a case with small changes, the shape can greatly differ across scans in cases where the changes are large. Based on the synthetic data experiment, volumetric measurement based on manual segmentations better capture the properties of cases with large changes than our semi-automatic approach. In those cases, however, clinicians properly would identify growth through visual inspection, as it is reliable according to our synthetic data experiment, faster, and less labor intensive than our proposed and manual segmentation based metric.
Similar to the synthetic data set, the measurements of the semi-automatic procedure seemed to be more conservative compared to the experts’ findings on the real MRI scans. The measurements also were more reliable than the experts’ findings. In the three cases where the experts’ findings were inconclusive (Case 2–4), our semi-automatic method classified these cases as stable. The case of the patient who was scanned twice 8 minutes apart furthermore supported that our metric was much more reliable than the expert findings. Expecting no change, our metric classified the case as stable. However, the volumetric measurement of Expert A indicated slight shrinkage. A more conclusive analysis of our method is difficult due to the missing ground truth and the small size of the test data set. However, the overall findings on the real data were consistent with the findings from the synthetic data where ground truth was available. We also note that our metric was less labor intensive than the manual volumetric measurements as each measurement took less than 5 minutes. This suggests that our semi-automatic procedure is more suitable for standard clinical practice than manual segmentations.
Conclusion
We have discussed a new approach that successfully measures the volume change of slowly evolving pathology from successive MRI scans. The high correlation with expert findings emphasizes the potential of the approach in standard clinical practice. Compared to visual change detection, the tool is highly sensitive to subtle change in pathology. Compared to measurements based on manual segmentations, the metric is less impacted by intra and inter-rater variability, more robust to changes in head position and scan resolution, and requires less user input. To the best of our knowledge, our semi-automatic approach is the first metric that allows, in a clinical setting, accurate and relatively fast measurements of subtle changes of slow growing tumors such as meningioma from longitudinal medical scans.
Acknowledgement
This work has been partly supported by the European Health-e-Child project (IST-2004-027749), the CompuTumor associated team, the Brain Science Foundation, the ARRA supplement to NIH NCRR P41 RR13218, and the NIH grants U54EB005149 and U41RR019703. We also would like to thank Dr. Zamani, Department of Radiology, Brigham and Women’s Hospital, for help in patient identification and image acquisition.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.CBTRUS 2007–2008: Primary brain tumors in the United States Statistical Report 2000–2004. Central Brain Tumor Registry of the United States; [Google Scholar]
- 2.Claus EB, Bondy ML, Schildkraut JM, Wiemels JL, Wrensch M, Black PM. Epidemiology of Intracranial Meningioma. Neurosurgery. 2005;57(6) doi: 10.1227/01.neu.0000188281.91351.b9. [DOI] [PubMed] [Google Scholar]
- 3.Perry A, Louis D, Scheithauer B, Budka H, von Diemling A. Meningiomas. Lyon: International Agency for Research on Cancer; 2007. [Google Scholar]
- 4.Nakasu S, Fukami T, Nakajima M, Watanabe K, Ichikawa M, Matsuda M. Growth Pattern Changes of Meningiomas: Long-term Analysis. Neurosurgery. 2005;56(5):946–955. [PubMed] [Google Scholar]
- 5.Bindal R, Goodman JM, Kawasaki A, Purvin V, Kuzma B. The natural history of untreated skull base meningiomas. Surgical Neurology. 2003;59(2):87–92. doi: 10.1016/s0090-3019(02)00995-3. [DOI] [PubMed] [Google Scholar]
- 6.Van Havenbergh T, Carvalho G, Tatagiba M, Plets C, Samii M. Natural History of Petroclival Meningiomas. Neurosurgery. 2003;52(1):55–64. doi: 10.1097/00006123-200301000-00006. [DOI] [PubMed] [Google Scholar]
- 7.Yano S, Kuratsu J-i. Indications for surgery in patients with asymptomatic meningiomas based on an extensive experience. Journal of Neurosurgery. 2006;105(4):538–543. doi: 10.3171/jns.2006.105.4.538. [DOI] [PubMed] [Google Scholar]
- 8.O'Leary S, Adams WM, Parrish RW, Mukonoweshuro W. A typical imaging appearances of intracranial meningiomas. Clinical Radiology. 2007;62:10–17. doi: 10.1016/j.crad.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 9.Viola P, Wells WM. Alignment by maximization of mutual information. International Journal of Computer Vision. 1997;24:137–154. [Google Scholar]
- 10.Clatz O, Sermesant M, Bondiau PY, et al. Realistic simulation of the 3{D} growth of brain tumors in {MR} images coupling diffusion and biomechanical deformation. IEEE Transactions on Medical Imaging %L clatz05. 2005;Vol 24:1334–1346. doi: 10.1109/TMI.2005.857217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK. World Health Organization classification of tumors of the Central Nervous System. Lyon: 2007. [DOI] [PubMed] [Google Scholar]
- 12.James K, Eisenhauer E, Christian M, et al. Measuring Response in Solid Tumors: Unidimensional Versus Bidimensional Measurement. J. Natl. Cancer Inst. 1999 March 17;91(6):523–528. doi: 10.1093/jnci/91.6.523. 1999. [DOI] [PubMed] [Google Scholar]
- 13.Therasse P, Arbuck SG, Eisenhauer EA, et al. New Guidelines to Evaluate the Response to Treatment in Solid Tumors. J. Natl. Cancer Inst. 2000 February 2;92(3):205–216. doi: 10.1093/jnci/92.3.205. 2000. [DOI] [PubMed] [Google Scholar]
- 14.Zeidman L, Ankenbrandt W, Du H, Paleologos N, Vick N. Growth rate of non-operated meningiomas. J Neurolology. 2008;255:891–895. doi: 10.1007/s00415-008-0801-2. [DOI] [PubMed] [Google Scholar]
- 15.Commins DL, Atkinson RD, Burnett ME. Review of meningioma histopathology. Neurosurg Focus. 2007;23(4):E3. doi: 10.3171/FOC-07/10/E3. [DOI] [PubMed] [Google Scholar]
- 16.McHugh K, Kao S. Response evaluation criteria in solid tumours (RECIST): problems and need for modifications in paediatric oncology? Br J Radiol. 2003 July 1;76(907):433–436. doi: 10.1259/bjr/15521966. 2003. [DOI] [PubMed] [Google Scholar]
- 17.Hashiba T, Hashimoto N, Izumoto S, et al. Serial volumetric assessment of the natural history and growth pattern of incidentally discovered meningiomas. J Neurosurg. 2008 doi: 10.3171/2008.8.JNS08481. [DOI] [PubMed] [Google Scholar]
- 18.Sorensen AG, Patel S, Harmath C, et al. Comparison of Diameter and Perimeter Methods for Tumor Volume Calculation. J Clin Oncol. 2001 January 15;19(2):551–557. doi: 10.1200/JCO.2001.19.2.551. 2001. [DOI] [PubMed] [Google Scholar]
- 19.Warren KE, Patronas N, Aikin AA, Albert PS, Balis FM. Comparison of One-, Two-, and Three-Dimensional Measurements of Childhood Brain Tumors. J. Natl. Cancer Inst. 2001 September 19;93(18):1401–1405. doi: 10.1093/jnci/93.18.1401. [DOI] [PubMed] [Google Scholar]
- 20.Hopper K, Kasales C, Van Slyke M, Schwartz T, TenHave T, Jozefiak J. Analysis of interobserver and intraobserver variability in CT tumor measurements. Am. J. Roentgenol. 1996 October 1;167(4):851–854. doi: 10.2214/ajr.167.4.8819370. [DOI] [PubMed] [Google Scholar]
- 21.Liu J, Udupa J, Odhner D, Hackney D, Moonis G. A system for brain tumor volume estimation via {MR} imaging and fuzzy connectedness. Computerized Medical Imaging and Graphics %L liu05. 2005;Vol 29:21–34. doi: 10.1016/j.compmedimag.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 22.Prastawa M, Bullitt E, Moon N, Leemput KV, Gerig G. Automatic brain tumor segmentation by subject specific modification of atlas priors. Academic Radiology %L prastawa03. 2003;Vol 10:1431–1438. doi: 10.1016/s1076-6332(03)00506-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gerig G, Welti D, Guttmann CRG, Colchester ACF, Szekely G. Exploring the discrimination power of the time domain for segmentation and characterization of active lesions in serial MR data. Medical Image Analysis. 2000;4:31–42. doi: 10.1016/s1361-8415(00)00005-0. [DOI] [PubMed] [Google Scholar]
- 24.Meier DS, Weiner HL, Guttmann CRG. MR Imaging Intensity Modeling of Damage and Repair In Multiple Sclerosis: Relationship of Short-Term Lesion Recovery to Progression and Disability. AJNR Am J Neuroradiol. 2007 November 1;28(10):1956–1963. doi: 10.3174/ajnr.A0701. 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thompson PM, Hayashi KM, Sowell ER, et al. Mapping Cortical Change in Alzheimer's Disease, Brain Development, and Schizophrenia. NeuroImage %L thompson04. 2004;Vol 23:2–18. doi: 10.1016/j.neuroimage.2004.07.071. [DOI] [PubMed] [Google Scholar]
- 26.Rey D, Subsol G, Delingette H, Ayache N. Automatic detection and segmentation of evolving processes in 3D medical images: Application to multiple sclerosis. Medical Image Analysis %L rey02. 2002;Vol 6:163–179. doi: 10.1016/s1361-8415(02)00056-7. [DOI] [PubMed] [Google Scholar]
- 27.Angelini ED, Atif J, Delon J, Mandonnet E, Duffau H, angelini LCL. Detection of glioma evolution on longitudinal {MRI} studies. IEEE International Symposium on Biomedical Imaging. 2007:49–52. [Google Scholar]