Abstract
The crystal structure of a murine monoclonal antibody, 4C3, that binds to the C-terminal lobe of the cockroach allergen Bla g 2, has been solved at 1.8 Å resolution. Binding of 4C3 involves different types of molecular interactions with its epitope compared to the monoclonal antibody 7C11, which binds to the N-terminal lobe of Bla g 2. We found that the 4C3 surface epitope on Bla g 2 includes a carbohydrate moiety attached to Asn268 and that a large number of antigen-antibody contacts are mediated by water molecules and ions, most likely zinc. Antibody binding experiments conducted with an enzymatically deglycosylated Bla g 2 and a N268Q mutant, showed that the carbohydrate contributes, without being essential, to the Bla g 2-4C3 mAb interaction. Inhibition of IgE antibody binding by the mAb 4C3 shows a correlation of the structurally defined epitope with reactivity with human IgE. Site-directed mutagenesis of the 4C3 mAb epitope confirmed that amino acids Lys251, Glu233, and Ile199 are important for the recognition of Bla g 2 by the 4C3 mAb antibody. The results show the relevance of X-ray crystallographic studies of allergen-antibody complexes to identify conformational epitopes that define the antigenic surface of Bla g 2.
Cockroach allergy is associated with development of asthma, especially in inner-cities where it affects up to 80% of asthmatic children that are sensitized and exposed to allergens produced by Blattella germanica (1,2). Bla g 2, an inactive aspartic protease with a bilobal structure (3) is the most potent allergen in terms of prevalence of sensitization among German cockroach allergic patients (50–70%) (4,5). Unlike viral aspartic proteases that have two identical subunits forming a similar bilobal structure, Bla g 2 consists of two fused lobes, with structure comparable to typical active aspartic proteases exemplified by pepsin, renin, or chymosin (6). However, amino acid substitutions in the active site render Bla g 2 inactive (7). Although both lobes are structurally similar and evolved from fusion of the single-domain subunits of an ancestral protein, presumably similar to the viral aspartic proteases (8), the amino acid sequence of the lobes differs, and thus would be expected to cause antigenic differences on their molecular surface.
Our goal has been to map the antigenic surface of Bla g 2 by solving the X-ray crystallographic structures of complexes of the allergen with specific murine monoclonal antibodies (mAb). Most of the reported epitope mapping techniques for allergens omit the identification of conformational epitopes. Such methods are based on the use of libraries of overlapping synthetic peptides, fragments from digested allergens or parts of allergens expressed as recombinant proteins. The use of synthetic peptides has proven useful to identify linear B cell epitopes, especially in foods (9,10). However, epitope mapping of globular proteins such as Bla g 2 with allergen peptides or fragments has an important limitation of missing the identification of conformational epitopes. Crystallographic techniques have proven to be an effective strategy to perform epitope mapping of globular proteins (11). This approach aims to map the antigenic surface of an allergen by solving complexes of the allergen with specific non-overlapping antibodies.
We have previously reported the X-ray crystal structure of Bla g 2 alone (12) and in complex with monoclonal antibody 7C11 that binds to the N terminus of Bla g 2 (13). Here we describe the epitope of a murine monoclonal antibody 4C3 that binds to the opposite, C-terminal, lobe of Bla g 2. Binding of 4C3 involves different types of molecular interactions than the binding of 7C11 to its epitope. We found that the 4C3 surface epitope on Bla g 2 included a carbohydrate moiety and that a large number of antigen-antibody contacts were mediated by water molecules.
Materials and Methods
Expression and purification of rBla g 2
Recombinant Bla g 2 mutants (partially deglycosylated by a substitution of either N93Q or N268Q in one of the three glycosylation sites, and the mutants of the 4C3 mAb epitope) were expressed in Pichia pastoris and purified by affinity chromatography using 7C11 monoclonal antibody, as previously described (3,12–14). Briefly, the DNA encoding for the mature form of Bla g 2 was inserted into a pGAPZα expression vector (Invitrogen). The plasmid was mutated using the QuickChange™ site-directed mutagenesis kit (Stratagene, La Jolla, CA). The linearized plasmid was electroporated into Pichia pastoris for constitutive expression of the allergen, followed by purification from culture media. The purity of rBla g 2 was >95% as judged by SDS-PAGE.
Production of Fab from the 4C3 monoclonal antibody and formation of the rBla g 2-Fab complex
Monoclonal anti-Bla g 2 antibody 4C3 (clone 4C3 H5 F6 F7) was raised against affinity-purifed natural Bla g 2 and purified from ascites by affinity chromatography through a Protein A column. Purified antibody was fragmented using papain. F(ab) fragments were purified (>95%) by Protein A chromatography and eluted in 20 mM sodium phosphate 150 mM NaCl, pH 7.2. Fab were mixed with rBla g 2-N93Q at a 1.2:1 molar ratio. The mixture was further purified over a HiPrep 16/60 Sephacryl S-100 HPLC column equilibrated with the buffer 20 mM Tris 0.2 M NaCl pH 7.2 and concentrated to ~5.2 mg/ml for crystallization.
Sequencing of the monoclonal antibody 4C3
The cell line producing the anti-Bla g 2 mAb 4C3 H5 F6 F7 was grown at the Lymphocyte Culture Center (University of Virginia, VA). Total RNA (20 μg/3×106 cells) was isolated from the 4C3 mAb cell line using an RNeasy Mini kit (Qiagen, Valencia, CA). cDNAs encoding for the light and heavy chains of the mAb were obtained by reverse transcription from RNA (SuperScript™ III, Invitrogen Corporation, Carlsbad, CA), and the DNA was PCR amplified using specific primers, sequenced and analyzed.
Two and four pairs of primers were initially tested for amplification of the light and heavy chains of the antibody, respectively. A degenerate primer for the N terminus [VHb: 5′-gag gtg cag ctg gtg ga(ag) tc-3′, encoding for EVQLVE], and the C-terminal primer [CH2: 5′-tt agg agt cag agt aat ggt gag cac atc c-3′, encoding for DVLTITLTP] amplified the heavy chain. Forward [5′-gcc aaa acg aca ccc cat c-3′, encoding for AKTTPH] and reverse [5′-a ggt cac tgt cac tgg ctc agg-3′, encoding for PEPVTVT] middle primers were used to complete heavy chain sequencing. The light chain was amplified with primers for the N terminus [Vκ4: 5′-caa att gtt ctc acc cag tct cca-3′, encoding for QIVLTQSP] and the C terminus [Cκ: 5′-gat gga tac agt tgg tgc-3′, encoding for APTVSI]. The mAb was isotyped G1 for the heavy chain and kappa (κ) for the light chain.
Structure determination
Crystals of the complex of Bla g 2 with Fab 4C3 were obtained at room temperature using the hanging-drop, vapor diffusion method. Each drop contained 2 μl of ~5 mg/ml allergen/antibody complex in 20 mM Tris buffer, 0.2 M NaCl and 2 mM DTT at pH 7.2, as well as 2 μl of reservoir solution consisting of 20% PEG8000, 8% ethylene glycol, 5 mM DTT and 0.2 mM CdCl2 in 0.1 M Tris buffer at pH 7.0. The crystals grew to the dimensions of ~0.15 × 0.2 × 0.1 mm in a week. They belong to the monoclinic space group C2 and each unit cell contains one molecule of Bla g 2, as well as one molecule of Fab.
Diffraction data extending to 1.8 Å resolution were collected from one crystal at the SER-CAT beamline 22-ID, located at the Advanced Photon Source synchrotron (Argonne, IL). Data were measured with a Mar300CCD detector and were integrated and scaled with the HKL2000 package (15). Prior to data collection the crystal was rapidly cooled to 100 K in a nitrogen stream, after being transferred to the cryo-solution containing 20% PEG 18K and 10% ethylene glycol, and 0.2 mM CdCl2 in Tris buffer at pH 7.0 (Table 1).
Table 1.
Data collection and refinement statistics
| Wavelength (Å) | 1.000 |
| Space group | C2 |
| Unit cell parameters (Å) | a=155.2, b=105.3, c=109.1, β=132.6° |
| Resolution (Å) | 50-1.8 |
| Number of reflections (unique/total) | 118,390 (441,093) |
| Completeness (last shell) | 99.3 (94.5) % |
| Rmerge (last shell) | 6.8 (49.1) % |
| No. of complexes in a.u. | 1 |
| No. of protein atoms | 5897 |
| No. of solvent molecules | 871 |
| No. of heteroatoms | 98 |
| Rcryst | 17.8% |
| Rfree (3% of data) | 20.2% |
| r.m.s. deviations from ideality | |
| Bond lengths | 0.011 Å |
| Angles | 1.4° |
The structure of Bla g 2/4C3 complex was solved by molecular replacement using the program Phaser (16). The search model for Bla g 2 was the structure of Bla g 2 complexed with Fab of 7C11 mAb. Bla g 2 and Fab were searched separately and solution of the molecular replacement was unambiguous, with the final Z-score 35.4 and LLG 2745. The structure was refined using Phenix (17) at an early stage, and then finalized with the program Refmac5 (18). The program Coot (19) was used for model building. The final model contains one Bla g 2 molecule, one Fab molecule, 3 Cd2+ ions, 6 Zn2+ ions, and 871 water molecules, and is characterized by Rfree of 20.2%. Identification of metal ions is tentative and largely based on the previous observation that Zn2+ is strongly bound to Bla g 2, and on proper behavior of the ions during refinement.
ELISA to test 4C3 mAb binding to Bla g 2 N268Q mutant
Plates were coated with 1 μg/ml of either 7C11 or 4C3 mAb, followed by a blocking step incubation with 1% BSA-PBS-0.05% Tween 20, pH 7.4 for 1h. All the steps were performed at room temperature and plates were washed three times with PBS-0.05% Tween 20, pH 7.4 between incubation steps. N268Q Bla g 2 mutant was quantified by Advanced Protein Assay (Cytoskeleton, Denver, CO) and used at 1:2 dilutions across the plate starting at 250 ng/ml. A rabbit polyclonal anti-Bla g 2 antibody was used as detection antibody followed by peroxidase-labeled goat anti-rabbit polyclonal antibody (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA). Plates were developed and read at OD 405 nm as previously described (20).
Deglycosylation of rBla g 2
Bla g 2 was deglycosylated by adding PNGase F (14 μl of the enzyme at 500,000 U/ml; New England Biolabs, Ipswich, MA) to approximately 1 ml of rBla g 2-N93Q (700 μg in PBS, pH 7.4), and incubating at 37 °C for 72 hours. Deglycosylation was confirmed by SDS-PAGE and silver staining. Deglycosylated Bla g 2 was further purified by affinity chromatography as described above. The allergen was eluted with 0.1 M glycine, 0.15 M NaCl, pH 2.5, concentrated and dialyzed against PBS pH 7.4. The allergens were quantified by measuring absorbance at 280 nm.
Comparison of rBla g 2 versus the deglycosylated allergen by ELISA
Dose-response curve experiments were performed using rBla g 2 (N93Q, glycosylated at positions N268 and N317) versus the enzymatically deglycosylated allergen (Bla g 2-DG). Microplates (96-well) were coated with 1 μg/ml of either 7C11 or 4C3 mAb in 50mM carbonate-bicarbonate buffer, pH 9.6. Plates were blocked with phosphate buffer containing 0.05% Tween 20 (PBS-T) and 1% BSA, pH 7.4. Subsequent steps and washes between steps with PBS-T were performed at room temperature. Known concentrations of allergen were added in the first well, and 1:2 serial dilutions were performed along the plate. Detection antibodies were biotinylated 4C3 mAb for plates coated with 7C11 mAb, and vice versa (at 1:1000 dilution). Peroxidase-labelled streptavidin (0.25 μg/ml) was added in the last step (Sigma, St. Louis, MO). Plates were developed and read at OD 405 nm as previously described (20).
Inhibition of IgE antibody binding by 4C3 mAb
Microplates were coated with 5 μg/ml rBla g 1-N93Q in 50mM carbonate-bicarbonate buffer, pH 9.6. Blocking step and washes were performed as described above. The antibodies tested for inhibition of IgE antibody binding were the anti-Bla g 2 mAb 4C3, and positive and negative controls. The anti-Bla g 2 mAb 4C3 was used at 0, 0.1, 1, 10 and 100 μg/ml concentrations. The rabbit polyclonal anti-Bla g 2 antibody was used as positive control (at 1:4 and 1:10 dilutions), and the anti-mite Der p 1 allergen 4C1 mAb was used as negative control (at 10 and 100 μg/ml). These antibodies were added after the blocking step, followed by addition of sera (1:5 final dilution). Sera of cockroach allergic patients with high anti-Bla g 2 IgE antibody titer were used (see next section). Incubation was at room temperature for 2.5 hours. Affinity purified peroxidase labeled goat anti-human antibody IgE (Kirkegaard and Perry Laboratories, Gaithersburg, MD) was used for detection at 1:1000. Plates were developed and read at OD 405 nm as above.
Sera from cockroach allergic patients
Sera from cockroach allergic patients were obtained from stored samples that had been collected from patients enrolled in 1988-1989 in Wilmington (Delaware) and Charlottesville (Virginia) for epidemiological studies performed at the University of Virginia (21,22). The studies performed at the University of Virginia were approved by the Human Investigation Committee and blood was drawn after informed consent was obtained from the patient. IgE antibody levels in sera were 64 and 27 ng of total IgE measured by RAST (22), and 65 and 29 ng of IgE against Bla g 2/ml, measured by mAb-based RIA (4).
Multiplex array for measurement of antibody binding to Bla g 2
Monoclonal antibody 7C11 (20 μg) was coupled to a Luminex carboxylated fluorescent microsphere bead set (Luminex Corp., Austin, TX), as previously described (23). The multiplex array assay was performed in 96 well filter plates in the dark at room temperature with incubation steps of one hour, followed by filter washing twice. After pre-wetting the filter plate with assay buffer (1% BSA-PBS-0.02% Tween 20, pH 7.4) and vacuum filtering, mAb coupled beads were added along with rBla g 2 at 400 ng/ml. The 4C3 mAb mutants were assayed in a single-plex manner. Beads and allergen were mixed and incubated. In the following step, biotinylated 4C3 mAb was added to the wells, mixed and incubated. Finally, streptavidin-phycoerythrin was added to all wells and mixed. Plate was incubated for 30 minutes and filter-washed twice. Assay buffer (100 μl) was added to wells and mixed. Plate was read in a Bio-Plex fluorescent suspension array reader (Bio-Rad Laboratories, Hercules, CA), consisting of Luminex xMAP instrumentation (Luminex Corp.) supplied with Bio-Rad proprietary software. Data were analyzed by two or one-tailed, paired, Student T-test. P values of less than 0.05 were considered significant.
Results
Description of the Bla g 2-4C3 mAb-Fab complex
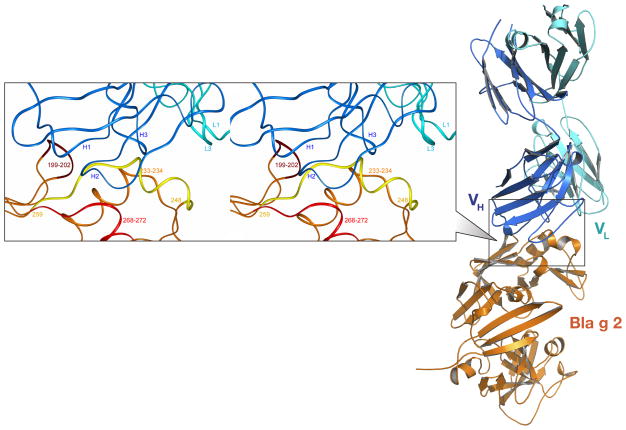
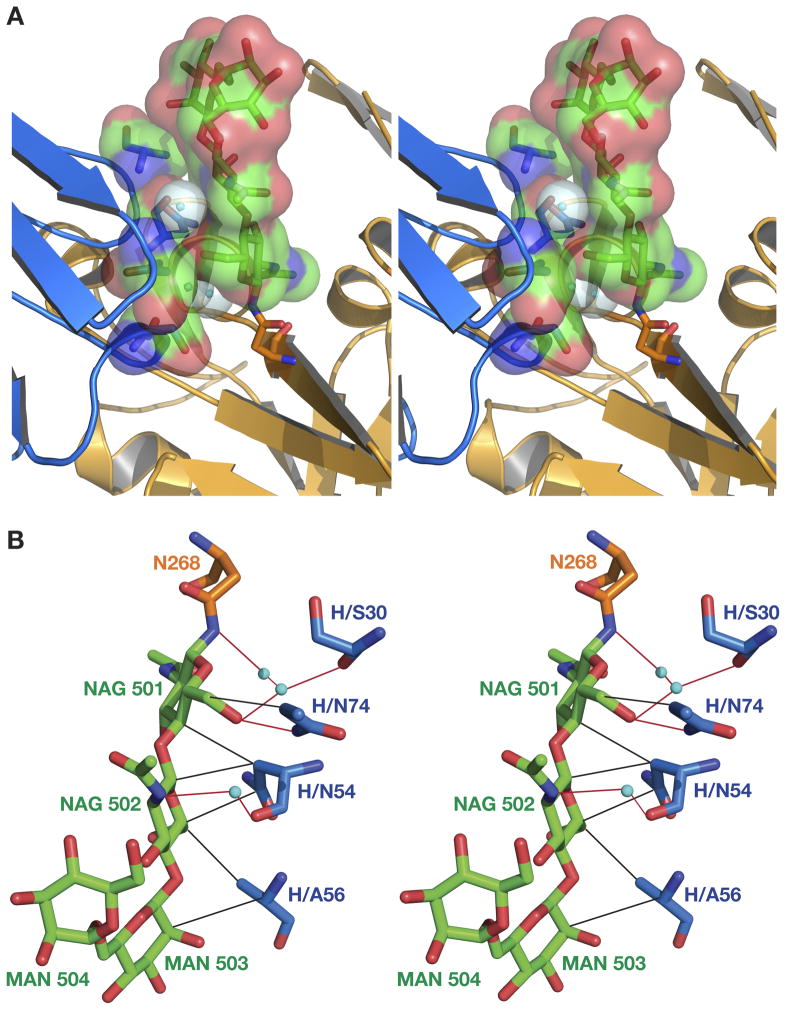
The crystal structure of the complex between Bla g 2 and monoclonal antibody 4C3 was solved at the resolution of 1.8 A (Table 1). The structure of the complex demonstrated that the antibody 4C3 binds tightly to Bla g 2 in 1:1 ratio, revealing a novel epitope, located in the C-terminal domain of the allergen molecule (Fig. 1, Supplemental Fig. 1). This epitope is comprised of the residues from three loops of the allergen (199–202, 248–259 and 268–272), as well as of the residues from the C-terminal end of the helix 225–235 (Fig. 1 inset). The loop 248–259 represents a continuous part of the epitope. This fragment of the structure of the allergen is composed of a helical segment that includes residues 248–254 which adopt the conformation of a 310 helix, with the remainder of this fragment assuming an extended conformation. The latter segment interacts with the most extended part of the CDR H1 in an antiparallel fashion maintaining the main chain – main chain contact between residues 255–257 of the allergen and SerH31-AlaH33 of the H1 CDR of the antibody. (In the text that follows, residue numbers of the heavy and light chains of the antibody are preceded with letters H and L, respectively.) The side chain of Asn268 of Bla g 2 is glycosylated, and a carbohydrate molecule attached to this residue is involved in extensive interactions with the antibody, predominantly water mediated.
Figure 1. Crystal structure of the complex between Bla g 2 and mAb 4C3.
A cartoon representation of the complex with the Bla g 2 molecule colored orange and 4C3 in two shades of blue. Inset: Stereoview of the interface between Bla g 2 and mAb 4C3 shown in ribbon representation. The chain of the antigen is colored orange, with the epitope loops brown, red, and yellow. The heavy chain of 4C3 is dark blue and the light chain is cyan, with the CDRs identified and numbered.
The allergen molecule interacts with all three CDRs of the heavy chain (H1–H3) of the 4C3 antibody, either directly or via solvent. The interactions of the allergen with the variable loops L1 and L3 of the light chain of 4C3 are, with one exception, mediated by solvent. The only direct contacts involve the side chain of Lys251, which forms electrostatic interactions with the residues from both the L1 and L3 CDRs. The remaining L2 CDR is not involved in any interactions with Bla g 2, either directly or via solvent.
The combined solvent-accessible surface buried in the Bla g 2 – 4C3 complex is 1831 Å2 (calculated with the program CNS, including both the allergen and antibody surfaces), which is within the range of 1,400–2,300 Å2 observed for the known antibody antigen complexes (24,25). In order to quantify the shape complementarity in 4C3-Bla g 2 complex, we calculated a shape correlation statistic (Sc), that estimates the geometric match for the interface (26). We found that the value of Sc is 0.62 for the Bla g 2/4C3 complex, compared to Sc = 1.0 for the interfaces with a geometrically perfect fit.
The epitope on the surface of a Bla g 2 molecule recognized by the 4C3 antibody is of conformational type and is comprised of residues originating from several secondary structure elements of the C-terminal domain of the allergen (Fig. 1 inset). Based on the type of the allergen-antibody interactions, the overall interface involves four kinds of contacts: direct contacts between residues from the counterparts, solvent-mediated interactions, interactions of mixed character, and carbohydrate-mediated interactions.
Specific interactions between Bla g 2 and 4C3 mAb
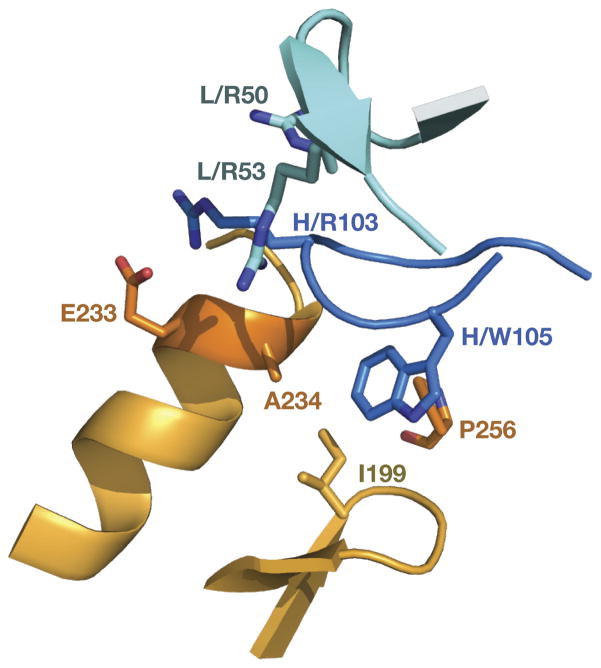
The overall views of the areas with numerous antibody-antigen interactions are shown in Supplemental Figs. 2 and 3. The most specific interactions between Bla g 2 and 4C3 are charge-charge interactions between the guanidinium group of ArgH103 from the CDR H3 and a negative charge of the C terminus of the helix comprised of residues 225–235, enhanced by the presence of properly oriented, negatively charged side chain of Glu233 (Fig. 2). In a similar manner, the positive charge of Arg103 is enhanced by the close proximity of ArgL50 and ArgL53 - two positively charged residues from the CDR L2. Although the latter two residues do not interact directly with the allergen, they contribute to antibody-allergen interactions by forming a cluster of positively charged residues on the surface of the antibody, which is able to specifically recognize its negatively charged counterpart on the surface of the allergen. These long distance, charge-driven interactions bring the side chain of TrpH105 of the variable loop H3 to the exactly right position in order to dock it into the hydrophobic pocket on the surface of allergen molecule comprised of Ala234, Ile199, and Pro256 (Fig. 2). A main chain–main chain hydrogen bond is formed between the carbonyl oxygen of Ala234 and the amide nitrogen of ArgH103 (Fig. 3).
Figure 2. The region of specific interactions between Bla g 2 and mAb 4C3.
Ionic interactions are formed between the C terminus of helix 225–235 in Bla g 2 and an arginine cluster originating from the H3 and L2 CDRs. A salt bridge is formed by the guanidinium group of ArgH103 from the CDR H3 and the side chain of Glu233, with an additional negative charge provided by the C terminus of the helix. Residues from the heavy chain of the antibody are colored dark blue, from the light chain cyan, and the antigen is gold.
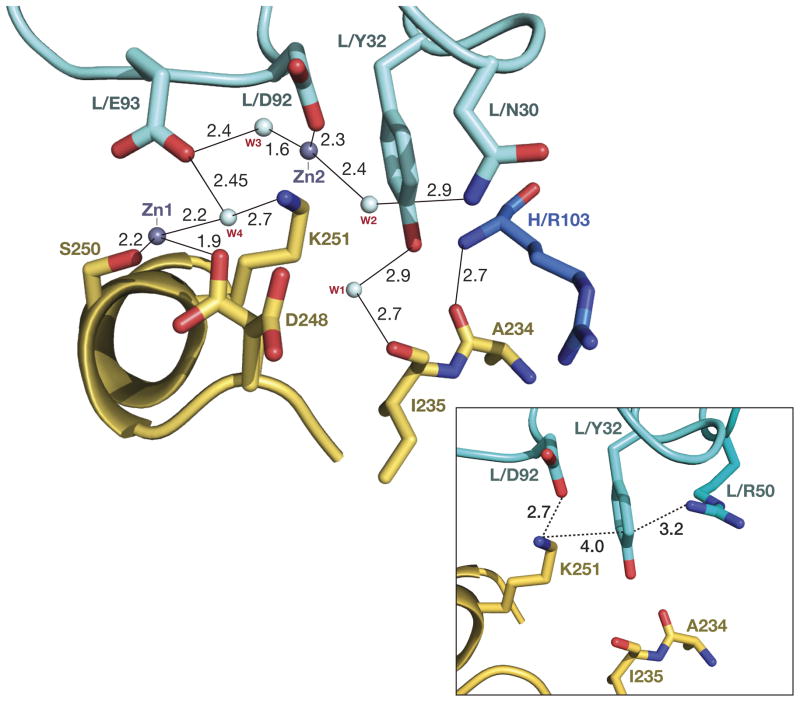
Figure 3. Interactions involving CDRs L1 and L3.
The contacts are predominantly solvent-mediated and involve residues from two fragments of the allergen epitope, comprising residues 248–252 from the loop 248–259 (the continuous part of the epitope), and Ala234-Ile235 from the helix 225–235. Residues from the heavy chain of the antibody are dark blue, from the light chain cyan, and the antigen is gold. Hydrogen bonds and metal-ligand interactions are shown as thin lines, with marked distances. Inset: ionic and cation–π interactions involving TyrL32 and AspL92 with Lys251.
An area adjacent to the one described above involves the interactions between CDRs L1 and L3 of the light chain of 4C3 and the residues from two fragments of the allergen epitope. The first of these fragments comprises residues 248–252 from the loop 248–259 (the continuous part of the epitope), and the second involves Ala234 from the helix 225–235. There are no direct contacts between Bla g 2 and the antibody in this area, with the exception of Lys251. The side chain of this residue is involved in two types of electrostatic interactions with the antibody. On one hand, it forms an ion pair with AspL92 of the variable loop L3, whereas, on the other hand, it is involved in cation-π type interactions with the side chain of TyrL32 of CDR L1 (Fig. 3 inset). The orientation of the aromatic ring of TyrL32, favorable for that interaction, is supported by another strong cation-π interaction formed with ArgL50 of L2 CDR on the opposite side of the ring (Fig. 3 inset). All other interactions in this region are mediated by water molecules. Two metal ions (presumably Zn2+) participate in this extensive hydrogen bonded network (Fig. 3). In addition to the involvement in the cation-π interactions described above, TyrL32 of CDR L1 is water bridged (W1 in Fig. 3) to the carbonyl oxygen of Ile235 of Bla g 2, whereas the side chain of AsnL30 contributes to the network of hydrogen bonds between CDR L3 and the fragment 248–252 of Bla g 2, by being hydrogen bonded to one of the two water molecules (W2) in the coordination sphere of Zn2 (Fig. 3). The metal ion is also directly coordinated by AspL92 of L3, as well by GluL93 via a water molecule (W3). The other metal ion (Zn1) is coordinated on the side of the allergen by the side chains of Ser250 and Asp248, while its contact with the antibody is mediated by the water molecule (W4). The latter water molecule is polarized by a strong hydrogen bond with Lys251 of Bla g 2, and thus able to maintain a short hydrogen bond with GluL93 of CDR L3 of the antibody. The side chain of Asp248 was modeled in two conformations in the structure of the complex, with only one of them participating in the interactions described above (Fig. 3).
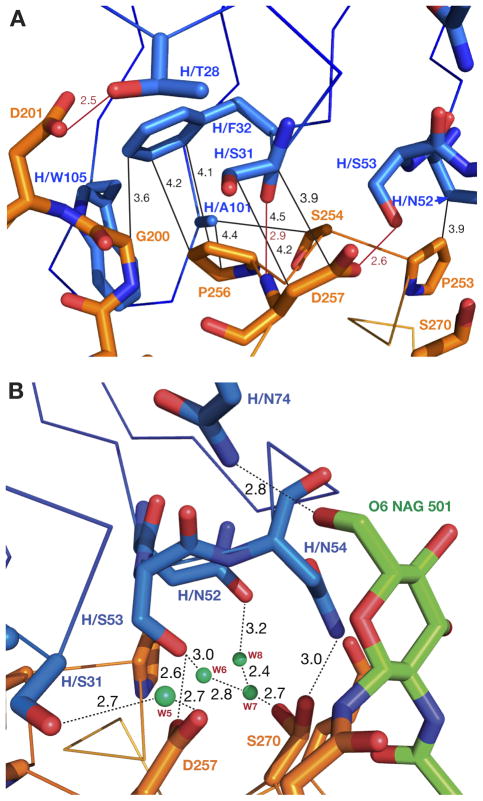
Three CDRs of the heavy chain of the antibody interact with all four loops involved in formation of the epitope recognized by 4C3 (Fig. 1 inset). Loop H1 of 4C3 is involved in direct, predominantly hydrophobic interactions with the fragments 200–201 from the loop 199–202 of Bla g 2, and 256–257 from the loop 248–259 (Fig. 4A). In addition, two residues of CDR H1, ThrH28 and SerH31, make polar contacts with the residues from the same two loops of the epitope. A short hydrogen bond is formed between the side chain of Asp201 from the first loop and the hydroxyl of the ThrH28 of H1, while the carbonyl oxygen of H1 SerH31 maintains direct hydrogen bond with the amide nitrogen of Asp257 from the second loop of Bla g 2 (Fig. 4A). In turn, the side chains of the latter two residues are also hydrogen bonded via a water molecule (W5 in Fig. 4B), i.e. hydroxyl of SerH31 side chain interacts with the OD1 atom of Asp257 of Bla g 2 epitope (Fig. 4B).
Figure 4. Interactions involving CDRs H1 and H2 of 4C3.
A) CDR H1 makes contacts with fragments 200–201 from the loop 199–202 of Bla g 2, as well as with residues 256–257 from the loop 248–259. Residues from the heavy chain of the antibody are dark blue and the antigen is gold. Hydrogen bonds are shown as red dotted lines, and hydrophobic interactions as black dotted lines, with the distances marked in respective colors. B) Solvent-mediated interactions between CDR H2 of 4C3 mAb and a part of the epitope on Bla g 2 that includes a glycosylation site. The side chain of SerH31 of CDR H1 is also shown. Hydrogen bonds are marked with dotted lines and their lengths are given.
Residues from the variable loop H2 form predominantly polar interactions with two loops of the allergen that carry residues Asp257 and Ser270. A short hydrogen bond is formed between the OD2 atom of Asp257 and the hydroxyl of SerH53 of H2. Ser270 is found in two conformations, maintaining different types of contacts with the antibody. One direct hydrogen bond is formed between the serine hydroxyl in the first orientation and the amino group of the side chain of AsnH54 of CDR H2, but when Ser270 adopts the second conformation, it interacts with the antibody via solvent molecules (W6-W8 in Fig. 4B). A single hydrophobic contact in this area is found between Pro253 of Bla g 2 and Cβ atom of the side chain of AsnH52 of H2 (Fig. 4A).
The contacts with H3 are rather limited in this area, flanking the continuous hydrophobic contacts of CDRs H1 and H2. The Cβ atom of Ser254 makes an important hydrophobic contact with AlaH101 of H3 (Fig. 4A), which, when combined with the interactions formed by TrpH105 (Fig. 2), completes the hydrophobic pillow in this area of the allergen-antibody interface.
Carbohydrate-mediated interactions
The epitope on the surface of Bla g 2 involved in binding mAb 4C3 includes a glycosylation site on the side chain of Asn268 of the allergen. The well-defined carbohydrate molecule attached to this residue interacts with the antibody either directly or via solvent (Fig. 5A). The interactions with the antibody significantly stabilize conformation of the oligosaccharide. Two mannose (Man) residues that follow the first two N-acetylglucosamine moieties (GlcNAc, also abbreviated here NAG1 and NAG2) can be traced in the electron density map of the complex (Supplemental Fig. 4), whereas these sugars are much more mobile and thus not visible in the electron density map of the uncomplexed allergen. An additional intramolecular contact between the side chain of Arg324 of the allergen molecule and O7 of NAG2, absent in unbound Bla g 2, supports the conformation of the sugar in the complex with the antibody, as well as its interactions with the Fab residues. Involvement of a sugar moiety in the interactions with antibody increases the total buried surface (for allergen and Fab) from 1585 to 1831 Å2. An extensive network of the interactions between the sugar and the antibody includes both hydrophobic and polar interactions (Fig. 5B). The polar interactions are mostly maintained via water molecules, with the exception of a single strong hydrogen bond between Asn74 from the third loop (which does not contain a CDR and is located between the second and third CDRs) of the heavy chain and O6 atom of NAG1. Several hydrophobic contacts are maintained between the residues from the antibody and three out of four sugar rings that could be traced in the electron density maps. Very good surface complementarity for this region is observed. To the best of our knowledge, this is the first structure that places a carbohydrate as part of the antigen epitope that is recognized by an antibody and reveals direct contacts between them.
Figure 5. Location of the carbohydrate moieties on Bla g 2.
A) Stereoview of the carbohydrate molecule bound to Asn268 of Bla g 2 (orange cartoon) shown as sticks covered by the van der Waals surface. The fragments of mAb 4C3 interacting with the carbohydrate are shown as blue cartoon. B) Interactions between the oligosaccharide (carbon atoms green) and antibody (carbon atoms blue). The residue glycosylated in Bla g 2, Asn268, is shown in orange. Hydrogen bonds are shown by red lines and the hydrophobic contacts in black.
Structural comparison of the two antibody complexes
Crystal structures of the complexes of Bla g 2 with two mAb’s, 7C11 (13) and 4C3 (this work), mark two epitopes on the surface of allergen. These epitopes are located in two different domains of the molecule. While 7C11 recognized an epitope in the N-terminal domain of Bla g 2, the second epitope, recognized by 4C3 mAb, is located in the C-terminal domain (Supplemental Fig. 5). Both epitopes have similar structural organization, which includes their continuous part as well as three short loop areas. Combined, these elements form the epitopes of conformational type for both antibodies. The residues from the continuous parts of both epitopes are involved in cation-π interactions. The epitopes are also of comparable size (1739 A2 and 1831 A2), although there are significant differences between them in the nature of the antibody-antigen interactions. The interface between Bla g 2 and 4C3 includes more solvent molecules compared to that in the complex with 7C11 (twenty one water-mediated interactions as compared to only four).
The other distinctive feature of the 4C3 epitope is the involvement of the glycosylation site at Asn268. The carbohydrate molecule interacts with the antibody directly and via solvent. When included in the calculations, sugar increases the contact area in the complex from 1586 A2 to 1831 A2.
The most profound differences in the conformations of the variable loops in two antibodies are detected for the loops H3 (with maximum deviation over 6 Å between the corresponding Cα atoms). The residues from H3 CDR in 4C3 complex are involved in most specific direct contacts with the allergen (see above), therefore the differences in the conformations of H3 CDRs reflects the specific properties of two different epitopes on the surface of the allergen molecule.
Relevance of carbohydrates that mediate the Bla g 2-4C3 mAb interaction
Binding of 4C3 mAb to Bla g 2 N268Q mutant by ELISA
In order to assess the relevance of the carbohydrate involved in 4C3 mAb binding, a recombinant Bla g 2 mutant N268Q, expressed in Pichia pastoris, was tested for antibody binding by ELISA. The mutant N268Q that retained two of the three N-glycosylation sites present in Bla g 2 wild type (Asn93 and Asn317), was able to bind the antibodies (either 7C11 or 4C3 mAb used for capture, and the rabbit polyclonal used for detection), and the dose-response curves were parallel to the ones obtained with rBla g 2-N93Q and natural Bla g 2 (data not shown). These results prove that the N268Q mutant folds similarly to the Bla g 2 wild type and that the presence of carbohydrate at position 268 is not essential for the allergen-antibody interaction.
Effect of enzymatic deglycosylation on antibody binding
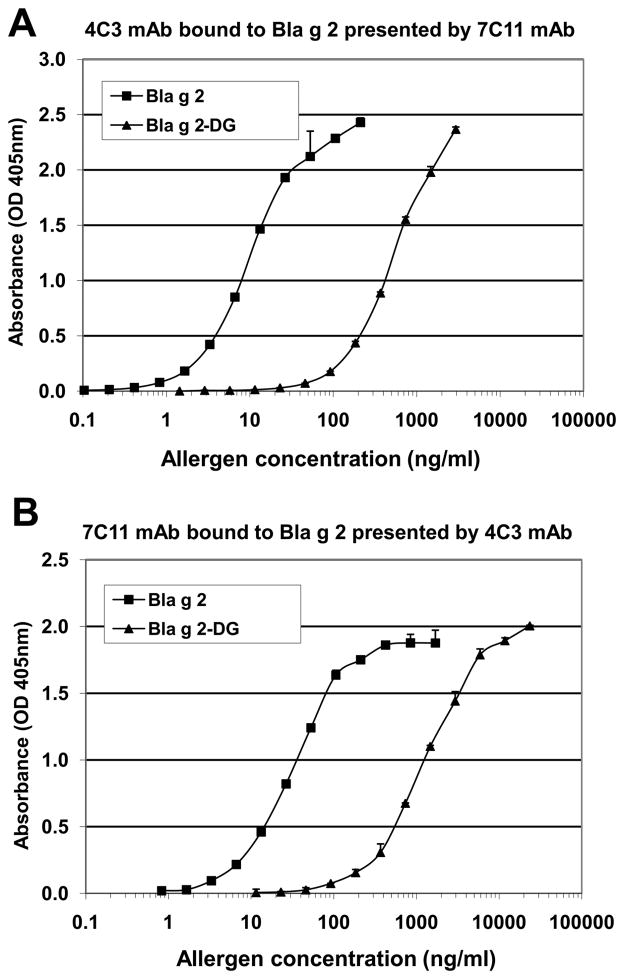
Effective deglycosylation of rBla g 2 was confirmed by a reduction of the allergen molecular weight on SDS-PAGE. Enzymatically deglycosylated recombinant Bla g 2-N93Q (Bla g 2-DG) was compared to non-deglycosylated rBla g 2-N93Q (Bla g 2) by similar ELISAs that used either 7C11 mAb as capture and biotinylated 4C3 mAb (Fig. 6A), or vice versa (Fig. 6B). ELISA experiments showed parallel dose-response curves for Bla g 2 and the enzymatically deglycosylated allergen Bla g 2-DG. In both ELISAs the dose-response curves were parallel, but the one for Bla g 2-DG displayed a ~40-fold displacement to the right, compared to the curve for the non de-glyscosylated allergen.
Figure 6. Dose-response curves of Bla g 2 deglycosylated versus non-deglycosylated allergen using sandwich mAb ELISAs.
A) Binding of biotinylated 4C3 mAb to Bla g 2 presented by 7C11 mAb. B) Binding of biotinylated 7C11 mAb to Bla g 2 presented by 4C3 mAb. Values represent average of duplicates and standard deviation from one representative experiment of 2 performed for each of the ELISA.
Inhibition of IgE antibody binding by 4C3 mAb
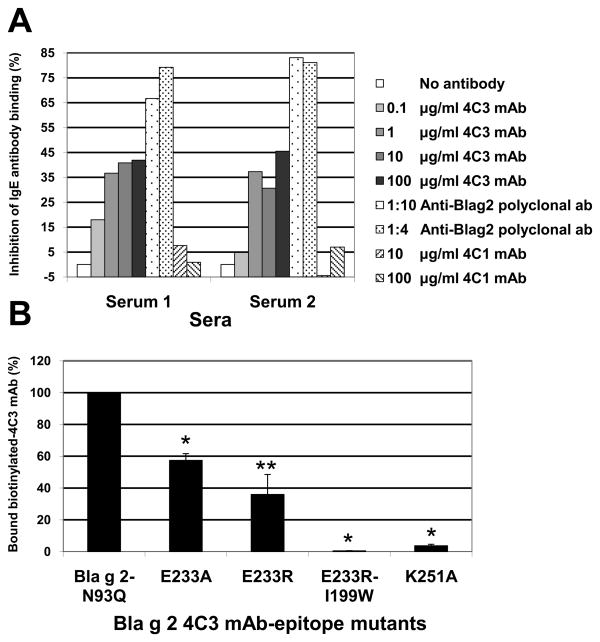
At increasing concentrations (from 0.1 to 100 μg/ml), the mAb 4C3 showed increased inhibition of IgE antibody binding of up to 45%. The positive control (an anti-Bla g 2 rabbit polyclonal antibody) showed inhibition of IgE antibody binding of up to 83%, whereas the negative control (the anti-mite allergen Der p 1 mAb 4C1), showed inhibition below 7% at the highest concentration of 100 μg/ml (Fig. 7A).
Figure 7.
A) Overlap of 4C3 mAb and IgE antibody epitopes. Inhibition of IgE antibody binding to Bla g 2 by 4C3 mAb (0.1, 1, 10, 100 μg/ml), an anti-Bla g 2 rabbit polyclonal antibody (as positive control) and an anti-Der p 1 mAb 4C1 (as negative control) at 10 and 100 μg/ml. B) Binding of biotinylated 4C3 mAb to Bla g 2-N93Q and four epitope mutants (E233A, E233R, E233R-I199W, and K251A). Average of n = 2 out of three experiments performed (p < 0.05 paired t-test compared to the wild type, two (*) and one (**) tailed distribution).
Site-directed mutagenesis analysis of the 4C3 mAb epitope
Site-directed mutagenesis of the 4C3 mAb epitope was performed based on structural analysis of the allergen-antibody interactions. Mutants K251A-N93Q, E233A-N93Q, E233R-N93Q and E233R-I199W resulted in a significant decrease of 4C3 mAb binding versus the wild type (WT-N93Q) in a validated multiplex array assay (27) (Fig. 7B). Bla g 2-specific polyclonal antibody bound Bla g 2-N93Q and the epitope mutants (presented by 7C11 mAb) in a similar manner, showing that the overall folding of the mutants was similar to the wild type (data not shown). Decrease of 4C3 mAb binding to the mutants confirmed the importance of amino acids K251, E233 and I199 in the interaction and highlighted the importance of a rational design for site-directed mutagenesis for the identification of key amino acids for the allergen-antibody interaction. Interestingly, K132A and K251A, two single mutants from Bla g 2 epitopes for 7C11 and 4C3, respectively, that significantly reduce antibody binding, have very similar interaction patterns in both complexes.
Discussion
The conformational epitope on the surface of the cockroach allergen Bla g 2 for the monoclonal antibody 4C3 has been defined by X-ray crystallography, and the biological significance of the individual contacts within the interface was evaluated by site-directed mutagenesis. The key residues responsible for specific recognition of the epitope by the 4C3 antibody were identified. A carbohydrate moiety (N-acetylglucosamine) attached to the allergen appears to play a role in antibody recognition. Finding positive contribution of carbohydrates to allergen-antibody interactions contrasts with many previous studies that report “masking” role of carbohydrates covering the parts of the epitope and thus interfering with the antibody recognition and impairing antibody binding (28,29). The masking of epitopes by carbohydrates could be compared to the blockage of IgE antibody binding observed by the pro-part of the mite cysteine protease allergen Der p 1 (30). The structure of Bla g 2 complexed to 4C3 mAb suggests a possibility of another role of such glycosylation.
Whereas a number of structures of antibodies complexed to their carbohydrate antigens have been published (exemplified by the complexes between a trisaccharide and scFv based on Se155-4 (31), between human antibody 2G12 and an oligosaccharide, Man9GlcNAc2 (32), or between hu3S193 binding to Lewis Y tumor antigen (33,34)), few if any structures show antibodies interacting simultaneously with both the protein and the carbohydrate. However, a recent structure of the broadly neutralizing human antibody CR6261 Fab complexed with the major surface antigen from the H5N1 avian influenza virus (35) also describes an epitope on the antigen surface with a sugar molecule located at its border. Since some atoms in the sugar molecule attached to Asn154 in HA2 are in close proximity to the antibody, (i.e. ~4 Å), we can hypothesize that, similarly to our structure, a solvent-mediated antigen-antibody interaction which could include sugar moieties might be present in that case, although few water molecules were actually seen in this comparatively low-resolution structure. The involvement of carbohydrates in the interactions, as seen in the structure of Bla g 2 complexed to 4C3 mAb, suggests a possibility of a dual role of such glycosylation sites, which can either mask the surface of the epitope preventing direct interactions with the antibody, or facilitate the contacts for the parts of the antibody that cannot otherwise interact with the antigen.
The role of the carbohydrates will depend on particular orientation of the antibody towards the epitope, which is dictated by the majority of specific interactions within the antibody-antigen interface. When carbohydrates cover the essential part of the epitope on the surface of the antigen which carries a majority of the residues specifically recognized by the antibody, then their role can be considered to “mask” antibody interactions. In a case like ours, when the presence of the sugars at the periphery of the epitope does not obstruct the specific antibody-antigen interactions within the major part of the epitope, involvement of carbohydrates in the interactions (either direct or via solvent) with the antibody may contribute to the stability of the complex. While taking into account the non-specific nature of such interactions (predominantly via solvent molecules), combined with the generally high B-factors of the sugar moieties, we would not like to overemphasize their significance, but our data on deglycosylated allergen indicate that such interactions might be helpful for increasing antibody binding.
N-glycans involved in IgE antibody recognition have been reported for plant and insect allergens and are named cross-reactive carbohydrate determinants (CCD) (36). N-glycans play an important role in IgE antibody binding for some allergens such as latex Heb v 4 and yellow jacket Ves m 2. For both allergens, a total reduction of IgE antibody recognition of non-glycosylated recombinant allergen versus native or allergen expressed in P. pastoris has been reported (37,38). However, the clinical relevance of CCDs is controversial (39–41). In this study we compared two rBla g 2 (enzymatically deglycosylated versus non-deglycosylated Bla g 2 expressed in Pichia pastoris) that retain a similar fold, as proven by antibody binding. The selection of PNGase F as deglycosylating enzyme was based on its capacity to remove the trimannosyl core which is involved in the 4C3 mAb interaction with Bla g 2 and is the minimal identical structure common to all N-glycans, regardless of the species in which they have been synthesized. We showed a displacement of the dose-response curves to the right for the deglycosylated versus the non-deglycosylated allergen using two similar ELISAs that involved 4C3 mAb either as detection or as capture antibody. Since carbohydrates are not involved in the 7C11 mAb-Bla g 2 interaction (13), the displacement to the right indicates that the 4C3 mAb binds less to the deglycosylated allergen. These results show that the carbohydrates at position Asn268 contribute to the allergen-4C3 mAb interaction. However, the carbohydrate at this position is not essential for the interaction since 4C3 mAb binds to the mutant N268Q. An overlap between the 4C3 mAb epitope and IgE antibody binding site was proven by high levels of inhition (up to 45%) of IgE antibody binding to Bla g 2 by 4C3 mAb, compared to a negative control (anti-mite Der p 1 mAb 4C1). We have also reported an overlap between 4C3 mAb and IgE antibody epitopes by mutagenesis analysis, as well as a contribution of carbohydrates and certain amino acids in the 4C3 mAb epitope to the IgE-allergen interaction (27). These results show the clinical relevance of the 4C3 mAb epitope described in this study. The present study highlights the importance of X-ray crystallographic studies on allergen-antibody complexes to map the conformational epitopes on the surface of inhaled allergens such as Bla g 2 and the molecular mechanisms that contribute to allergen/antibody interactions.
Supplementary Material
Acknowledgments
We would like to thank Bryan Smith and Dr. Eva King for their support in the multiplex array studies, and Leah Stohr for technical assistance. We acknowledge the use of beamline 22-ID of the Southeast Regional Collaborative Access Team (SER-CAT), located at the Advanced Photon Source, Argonne National Laboratory. Use of the APS was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. W-31-109-Eng-38.
AP and MDC were funded in part by grant R01AI077653 from the National Institute of Allergy and Infectious Diseases. This project was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research, and in part with Federal funds from the National Cancer Institute, NIH, under Contract No. HHSN261200800001E. The content of this publication is solely the responsibility of the authors and does not necessarily represent the official views or policies of the Department of Health and Human Services, nor does the mention of trade names, commercial products, or organizations imply endorsement by the U. S. Government.
Footnotes
The coordinates and structure factors have been deposited in the Protein Data Bank with the accession code 3LIZ.
Reference List
- 1.Rosenstreich DL, Eggleston P, Kattan M, Baker D, Slavin RG, Gergen P, Mitchell H, McNiff-Mortimer K, Lynn H, Ownby D, Malveaux F. The role of cockroach allergy and exposure to cockroach allergen in causing morbidity among inner-city children with asthma. N Engl J Med. 1997;336:1356–1363. doi: 10.1056/NEJM199705083361904. [DOI] [PubMed] [Google Scholar]
- 2.Gruchalla RS, Pongracic J, Plaut M, Evans R, III, Visness CM, Walter M, Crain EF, Kattan M, Morgan WJ, Steinbach S, Stout J, Malindzak G, Smartt E, Mitchell H. Inner City Asthma Study: relationships among sensitivity, allergen exposure, and asthma morbidity. J Allergy Clin Immunol. 2005;115:478–485. doi: 10.1016/j.jaci.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 3.Pomés A, Chapman MD, Vailes LD, Blundell TL, Dhanaraj V. Cockroach allergen Bla g 2: structure, function, and implications for allergic sensitization. Am J Respir Crit Care Med. 2002;165:391–397. doi: 10.1164/ajrccm.165.3.2104027. [DOI] [PubMed] [Google Scholar]
- 4.Arruda LK, Vailes LD, Mann BJ, Shannon J, Fox JW, Vedvick TS, Hayden ML, Chapman MD. Molecular cloning of a major cockroach (Blattella germanica) allergen, Bla g 2. Sequence homology to the aspartic proteases. J Biol Chem. 1995;270:19563–19568. doi: 10.1074/jbc.270.33.19563. [DOI] [PubMed] [Google Scholar]
- 5.Satinover SM, Reefer AJ, Pomés A, Chapman MD, Platts-Mills TA, Woodfolk JA. Specific IgE and IgG antibody-binding patterns to recombinant cockroach allergens. J Allergy Clin Immunol. 2005;115:803–809. doi: 10.1016/j.jaci.2005.01.018. [DOI] [PubMed] [Google Scholar]
- 6.Dunn BM, Goodenow MM, Gustchina A, Wlodawer A. Retroviral proteases. Genome Biol. 2002;3:REVIEWS3006. doi: 10.1186/gb-2002-3-4-reviews3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wünschmann S, Gustchina A, Chapman MD, Pomés A. Cockroach allergen Bla g 2: an unusual aspartic proteinase. J Allergy Clin Immunol. 2005;116:140–145. doi: 10.1016/j.jaci.2005.04.024. [DOI] [PubMed] [Google Scholar]
- 8.Tang J, James MNG, Hsu IN, Jenkins JA, Blundell TL. Structural evidence for gene duplication in the evolution of the acid proteases. Nature. 1978;271:618–621. doi: 10.1038/271618a0. [DOI] [PubMed] [Google Scholar]
- 9.Burks AW, Shin D, Cockrell G, Stanley JS, Helm RM, Bannon GA. Mapping and mutational analysis of the IgE-binding epitopes on Ara h 1, a legume vicilin protein and a major allergen in peanut hypersensitivity. Eur J Biochem. 1997;245:334–339. doi: 10.1111/j.1432-1033.1997.t01-1-00334.x. [DOI] [PubMed] [Google Scholar]
- 10.Shreffler WG, Beyer K, Chu TH, Burks AW, Sampson HA. Microarray immunoassay: association of clinical history, in vitro IgE function, and heterogeneity of allergenic peanut epitopes. J Allergy Clin Immunol. 2004;113:776–782. doi: 10.1016/j.jaci.2003.12.588. [DOI] [PubMed] [Google Scholar]
- 11.Padlan EA. X-ray crystallography of antibodies. Adv Protein Chem. 1996;49:57–133. doi: 10.1016/s0065-3233(08)60488-x. [DOI] [PubMed] [Google Scholar]
- 12.Gustchina A, Li M, Wünschmann S, Chapman MD, Pomés A, Wlodawer A. Crystal structure of cockroach allergen Bla g 2, an unusual zinc binding aspartic protease with a novel mode of self-inhibition. J Mol Biol. 2005;348:433–444. doi: 10.1016/j.jmb.2005.02.062. [DOI] [PubMed] [Google Scholar]
- 13.Li M, Gustchina A, Alexandratos J, Wlodawer A, Wunschmann S, Kepley CL, Chapman MD, Pomes A. Crystal structure of a dimerized cockroach allergen Bla g 2 complexed with a monoclonal antibody. J Biol Chem. 2008;283:22806–22814. doi: 10.1074/jbc.M800937200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pomes A, Vailes LD, Helm RM, Chapman MD. IgE reactivity of tandem repeats derived from cockroach allergen, Bla g 1. Eur J Biochem. 2002;269:3086–3092. doi: 10.1046/j.1432-1033.2002.02990.x. [DOI] [PubMed] [Google Scholar]
- 15.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 16.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallograhic software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. 2002;D58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 18.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. 1997;D53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 19.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 20.Pollart SM, Smith TF, Morris EC, Gelber LE, Platts-Mills TA, Chapman MD. Environmental exposure to cockroach allergens: analysis with monoclonal antibody-based enzyme immunoassays. J Allergy Clin Immunol. 1991;87:505–510. doi: 10.1016/0091-6749(91)90009-d. [DOI] [PubMed] [Google Scholar]
- 21.Gelber LE, Seltzer LH, Bouzoukis JK, Pollart SM, Chapman MD, Platts-Mills TA. Sensitization and exposure to indoor allergens as risk factors for asthma among patients presenting to hospital. Am Rev Respir Dis. 1993;147:573–578. doi: 10.1164/ajrccm/147.3.573. [DOI] [PubMed] [Google Scholar]
- 22.Pollart SM, Chapman MD, Fiocco GP, Rose G, Platts-Mills TA. Epidemiology of acute asthma: IgE antibodies to common inhalant allergens as a risk factor for emergency room visits. J Allergy Clin Immunol. 1989;83:875–882. doi: 10.1016/0091-6749(89)90100-0. [DOI] [PubMed] [Google Scholar]
- 23.King EM, Vailes LD, Tsay A, Satinover SM, Chapman MD. Simultaneous detection of total and allergen-specific IgE by using purified allergens in a fluorescent multiplex array. J Allergy Clin Immunol. 2007;120:1126–1131. doi: 10.1016/j.jaci.2007.06.043. [DOI] [PubMed] [Google Scholar]
- 24.Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 25.Sundberg EJ, Mariuzza RA. Advances in Protein Chemistry. Vol. 61. Elsevier Science; USA: 2003. Molecular Recognition in Anibody-Antigen Complexes; pp. 119–160. [DOI] [PubMed] [Google Scholar]
- 26.Lawrence MC, Colman PM. Shape complementarity at protein/protein interfaces. J Mol Biol. 1993;234:946–950. doi: 10.1006/jmbi.1993.1648. [DOI] [PubMed] [Google Scholar]
- 27.Pomés A, Li M, Glesner J, Wunschmann S, King EM, Chapman MD, Wlodawer A, Gustchina A. The X-ray crystal structure of two complexes of the cockroach allergen Bla g 2 with fragments of monocloncal antibodies defines two non-overlapping epitopes. J Allergy Clin Immunol. 2009;123:S229. [Google Scholar]
- 28.Munk K, Pritzer E, Kretzschmar E, Gutte B, Garten W, Klenk HD. Carbohydrate masking of an antigenic epitope of influenza virus haemagglutinin independent of oligosaccharide size. Glycobiology. 1992;2:233–240. doi: 10.1093/glycob/2.3.233. [DOI] [PubMed] [Google Scholar]
- 29.Helle F, Goffard A, Morel V, Duverlie G, McKeating J, Keck ZY, Foung S, Penin F, Dubuisson J, Voisset C. The neutralizing activity of anti-hepatitis C virus antibodies is modulated by specific glycans on the E2 envelope protein. J Virol. 2007;81:8101–8111. doi: 10.1128/JVI.00127-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takai T, Kato T, Yasueda H, Okumura K, Ogawa H. Analysis of the structure and allergenicity of recombinant pro- and mature Der p 1 and Der f 1: major conformational IgE epitopes blocked by prodomains. J Allergy Clin Immunol. 2005;115:555–563. doi: 10.1016/j.jaci.2004.11.024. [DOI] [PubMed] [Google Scholar]
- 31.Zdanov A, Li Y, Bundle DR, Deng SJ, MacKenzie CR, Narang SA, Young NM, Cygler M. Structure of a single-chain antibody variable domain (Fv) fragment complexed with a carbohydrate antigen at 1.7-A resolution. Proc Natl Acad Sci U S A. 1994;91:6423–6427. doi: 10.1073/pnas.91.14.6423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Calarese DA, Scanlan CN, Zwick MB, Deechongkit S, Mimura Y, Kunert R, Zhu P, Wormald MR, Stanfield RL, Roux KH, Kelly JW, Rudd PM, Dwek RA, Katinger H, Burton DR, Wilson IA. Antibody domain exchange is an immunological solution to carbohydrate cluster recognition. Science. 2003;300:2065–2071. doi: 10.1126/science.1083182. [DOI] [PubMed] [Google Scholar]
- 33.Ramsland PA, Farrugia W, Bradford TM, Mark HP, Scott AM. Structural convergence of antibody binding of carbohydrate determinants in Lewis Y tumor antigens. J Mol Biol. 2004;340:809–818. doi: 10.1016/j.jmb.2004.05.037. [DOI] [PubMed] [Google Scholar]
- 34.Farrugia W, Scott AM, Ramsland PA. A possible role for metallic ions in the carbohydrate cluster recognition displayed by a Lewis Y specific antibody. PLoS One. 2009;4:e7777. doi: 10.1371/journal.pone.0007777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ekiert DC, Bhabha G, Elsliger MA, Friesen RH, Jongeneelen M, Throsby M, Goudsmit J, Wilson IA. Antibody recognition of a highly conserved influenza virus epitope. Science. 2009;324:246–251. doi: 10.1126/science.1171491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aalberse RC. Clinical relevance of carbohydrate allergen epitopes. Allergy. 1998;53:54–57. doi: 10.1111/j.1398-9995.1998.tb04940.x. [DOI] [PubMed] [Google Scholar]
- 37.Seppala U, Selby D, Monsalve R, King TP, Ebner C, Roepstorff P, Bohle B. Structural and immunological characterization of the N-glycans from the major yellow jacket allergen Ves v 2: the N-glycan structures are needed for the human antibody recognition. Mol Immunol. 2009;46:2014–2021. doi: 10.1016/j.molimm.2009.03.005. [DOI] [PubMed] [Google Scholar]
- 38.Malik A, Arif SA, Ahmad S, Sunderasan E. A molecular and in silico characterization of Hev b 4, a glycosylated latex allergen. Int J Biol Macromol. 2008;42:185–190. doi: 10.1016/j.ijbiomac.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 39.van der Veen MJ, van Ree R, Aalberse RC, Akkerdaas J, Koppelman SJ, Jansen HM, van der Zee JS. Poor biologic activity of cross-reactive IgE directed to carbohydrate determinants of glycoproteins. J Allergy Clin Immunol. 1997;100:327–334. doi: 10.1016/s0091-6749(97)70245-8. [DOI] [PubMed] [Google Scholar]
- 40.Foetisch K, Westphal S, Lauer I, Retzek M, Altmann F, Kolarich D, Scheurer S, Vieths S. Biological activity of IgE specific for cross-reactive carbohydrate determinants. J Allergy Clin Immunol. 2003;111:889–896. doi: 10.1067/mai.2003.173. [DOI] [PubMed] [Google Scholar]
- 41.Altmann F. The role of protein glycosylation in allergy. Int Arch Allergy Immunol. 2007;142:99–115. doi: 10.1159/000096114. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.