Abstract
A protocol is described using lipid mutants and thiol-specific chemical reagents to study lipid-dependent and host-specific membrane protein topogenesis by the substituted-cysteine accessibility method as applied to transmembrane domains (SCAMTM). SCAMTM is adapted to follow changes in membrane protein topology as a function of changes in membrane lipid composition. The strategy described can be adapted to any membrane system.
Keywords: Membrane protein, Topology, Lipid-dependent topogenesis, Phospholipid, Lactose permease, SCAMTM
1 Introduction
Membrane proteins represent at least 30% of the all currently sequenced genomes and represent more than half the drug targets pursued by pharmaceutical companies (1). Effective drug design is dependent on understanding membrane protein structure and the rules that govern the folding and assembly of native and mutant membrane proteins. A fundamental aspect of the structure of polytopic membrane proteins is membrane protein topology. Membrane protein topology describes the way a polypeptide chain is arranged in the membrane, i.e. the number of transmembrane domains (TMs) and their orientation in the membrane. Final protein topology is determined by topogenic signals in the nascent polypeptide chain that are recognized and decoded not only by the protein insertion machinery (2) but by the lipid profile (3–9).
Although the methodology for obtaining high-resolution structures for membrane proteins is improving, the need to determine low-resolution organizational information on membrane proteins in a native membrane continues. Given the enormous number of sequences that are available in genome-sequencing projects, it is not realistic to assume that the structures of all the encoded proteins will be generated by crystallographic approaches, especially for membrane proteins. Moreover purification, crystallization and structure determination of membrane proteins still remain a challenge. Crystal structures are static and may be distorted due to purification and crystallization constraints. Information on interactions with other proteins and the lipid environment are also lost during purification. During protein purification, molecular interactions with lipids are replaced by non-native detergent interactions. Heterologous expression in a host strain with a different lipid composition than the native host can also result in loss of proper lipid-protein interactions, which can affect topological organization and function. Therefore, non-crystallographic approaches have been developed to determine lower resolution topological arrangement of membrane spanning segments in whole proteins as a function of membrane lipid composition (3, 6, 8).
Dynamic aspects of protein structure as a function of the physiological state of the cell is best probed in whole cells or membranes. Escherichia coli strains genetically altered in their lipid composition (10) (Fig. 1) and thiol-specific chemical reagents have been developed to study lipid-dependent and host-specific membrane protein topogenesis by the substituted-cysteine accessibility method as applied to TMs (SCAMTM) (3, 5–8). In this approach cysteine residues replace individual amino acids that reside in the putative extracellular or intracellular loops connecting TMs of a membrane protein expressed in a strain in which lipid composition can be changed either before (3, 8) or after (3, 5, 9) membrane protein synthesis and assembly. Combining of these techniques provides a system in which to study the role of lipid-protein interactions in determining the structure, assembly and function of membrane proteins. By combining SCAMTM with mutants of E. coli in which membrane phospholipid composition can be systematically controlled, the role of phospholipids as determinants of membrane protein topological organization was established (3, 8, 9). In addition, the ability to change lipid composition post-assembly of a membrane protein demonstrated the potential for polytopic membrane proteins to change their topological organization after insertion and assembly in the membrane (3, 5, 9).
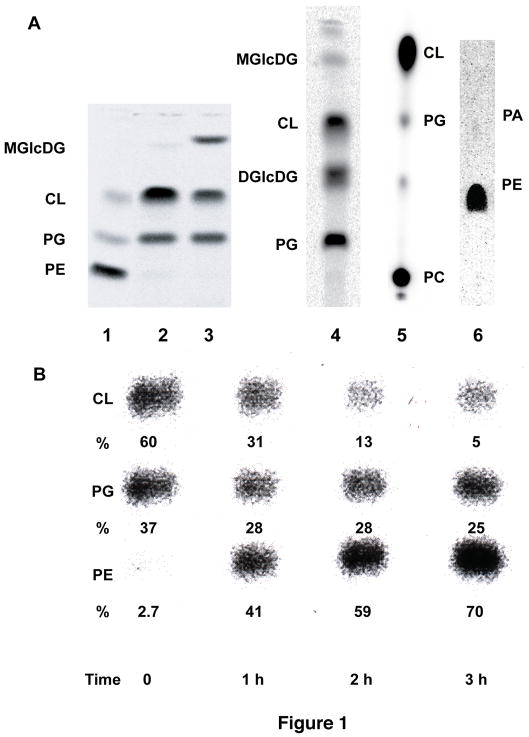
Fig. 1.
Lipid profiles of E. coli mutants with altered lipid compositions. (a) Lane 1. AL95 (pss93::kanR lacY::Tn9)/pDD72 (pssA+ camr) has wild type phospholipid composition (80 mole % PE and 20 mole % PG plus CL) due to complementation by a plasmid (pDD72) copy of the null allele of the pssA gene that encodes the committed step to PE biosynthesis (3). Lane 2. AL95 (pss93::kanR lacY::Tn9) is PE-lacking due to the a null allele of the pssA gene and contains mainly CL and PG (3). Lane 3. Introduction of plasmid pTMG3 (ALmgs ampR) into strain AL95 results in 35 mole % monoglucosyldiacylglycerol (MGlcDAG) due to the expression of the A. laidlawii MGlcDAG synthase gene. The remaining lipids are primarily PG (35 mole %) and CL (25 mole%) (8). Lane 4. Introduction into AL95 of the genes from A. laidlawii that synthesize MGlcDAG and diglucosyldiacylglycerol (DGlcDAG) results in about 30–40 mole % DGlcDAG with less than 1 mole % MGlcDAG (18). Lane 5. Introduction of the pcs gene (placed under OParaB regulatory control) from Legionella pneumophila (30) that confers the ability to synthesize PC results in about 70 mole % PC with the remainder being PG (2.5 mole %) plus CL (26 mole %) and other minor lipids (P. N. Heacock and W. Dowhan, unpublished). Lane 6. UE54 (pgsA::FRT-kan-FRT lpp-2 Δara714 rcsF::mini-Tn10cam) carries a null allele of the pgsA gene encoding the committed step to PG and CL biosynthesis making it devoid of PG and CL and containing about 90 mole % PE, 4.0 mole % phosphatidic acid (PA) and 3.2% CDP-diacylglycerol. Figure and compositional results taken from (31). (b) Phospholipid composition as a function of pssA gene induction. Strain AT2033 was grown first in the absence of aTc (time 0) followed by growth in the presence of aTc for 3 h. When grown without aTc, AT2033 has a greatly reduced level of PE and contains elevated levels of PG and CL. Growth in the presence of aTc results a progressive increase in PE to normal levels (5). Lipid composition is shown as mole % of the total major phospholipid species.
The following protocol describes (1) preparation of derivatives of the target protein, (2) description and use of hosts with varied lipid composition and (3) use of SCAMTM to map the topological organization of the target protein.
2 Materials
2.1 Transformation Protocol
10X RbCl/CaCl2 Transformation Salts for the preparation of transformation competent bacterial cells (MP Biomedicals, USA).
Centrifuge 5417R (Eppendorf).
2.2 Growth of E. coli Strains
Luria–Bertani (LB) medium.
2.5 M MgCl2.
Ampicillin (amp) (100 mg/ml).
500 mM isopropyl-β-D-thiogalactoside (IPTG).
Anhydrotetracycline (aTc) (Spectrum) (10 mg/ml).
2.3 SCAMTM
Buffer A: 100 mM HEPES/KOH buffer, 250 mM sucrose, 25 mM MgCl2, 0.1 mM KCl, adjusted to pH 7 or 10.5.
10 mM 3-(N-maleimidylpropionyl) biocytin (MPB) (Invitrogen-Molecular Probes) freshly dissolved in dimethyl sulfoxide (DMSO).
2 M β-mercaptoethanol.
Digital Sonifier (Branson, USA).
Beckman Coulter TLA-100 ultracentrifuge equipped with TLA-55 rotor.
Microfuge Polyallomer Tubes (natural tint, capacity 1.5 ml) (Beckman Coulter).
Centrifuge 5417R (Eppendorf).
2.4 Membrane Protein Solubilization Buffer
Solubilization buffer: 50 mM Tris-HCl buffer (pH 8.1) 2% SDS, 1 mM EDTA.
Fisher Vortex Genie 2 equipped with microtube foam rack for multiple polyallomer tubes (Fisher Scientific).
2.5 SDS-PAGE
2X SDS gel loading (sample) buffer: 10 mM Tris-HCl (pH 6.8), 5.6% SDS, 200 mM dithiothreitol, 10% glycerol, 0.01% bromophenol blue.
READY GEL: Tris-HCl precast gel for SDS Polyacrylamide Electrophoresis, 12.5% polyacrylamide (BIO-RAD Laboratories).
2.6 Immunoprecipitation (IP) buffer
50 mM Tris-HCl (pH 8.1), 0.15 M NaCl, 2% Lubrol-PX, 0.4% SDS, 1 mM EDTA (see note 1).
2.7 Immunoblot Analysis (Western Blotting)
0.45 μm Protran BA 85 Nitrocellulose transfer membranes (Whatman-Schlicher and Schuell)
Blocking buffer: 5% bovine serum albumin (BSA) (Fraction V, heat-shock treated) (Fisher Scientific) in Tris Buffered Saline (TBS) (10 mM Tris-HCl, pH 7.4, 0.9 % NaCl).
ImmunoPureR Avidin Horseradish Peroxidase (Avidin-HRP): Avidin linked to horse radish peroxidase (reconstituted to concentration of 2 mg/ml according manufacture suggestion) (Thermo Scientific).
SuperSignal West Pico chemiluminescent substrates for detection of HRP (Thermo Scientific).
Labconco Semi-Dry blotting system (W.E.P, Company Seattle, Washington, USA).
Anode buffer No.1: 0.3 M Tris (pH 10.4) in 10% methanol.
Anode buffer No. 2: 25 mM Tris (pH 10.4) in 10% methanol.
Cathode buffer: 25 mM Tris, 40mM glycine (pH 9.4) in 20% methanol.
Wash Buffer: 10 mM Tris-HCl (pH 7.4), 0.9 % NaCl containing, 0.05% Nonidet P40 (Igepal TM CA-630) (USB Corporation, Cleveland, OH, USA).
Chromatography Paper: 3 mm Cr (Whatman).
2.8 Alkali Treatment Solution
0.2 N NaOH.
Beckman Coulter TLA-100 ultracentrifuge equipped with TLA-55 rotor.
2.9 Image Acquisition and Data Processing
Fluor-S MaxTM MultiImager (Bio-Rad) equipped with a CCD camera and a Nikon 50mm 1:1.4 AD (F 1.4) lens at the ultrasensitive chemiluminescence setting which cools the camera to −33°C.
3 Methods
3.1 Lipid Mutants as Biological Reagents
The ability to regulate membrane lipid composition under steady state conditions (10) (Fig. 1a) coupled with determination of membrane protein orientation with respect to the plane of the membrane bilayer is a powerful approach to establish membrane protein topology or observe changes in topology as a function of membrane lipid environment (3, 8, 9) and the amino acid sequence of membrane proteins (5). The utilization of strains in which lipid composition is under control of a tightly inducible promoter (Fig. 1b) reveals surprising topological dynamics of a polypeptide after stable membrane insertion (3, 5, 9).
3.2 SCAMTM
This approach is based on introduction of cysteine residues one at a time into putative extracellular or intracellular loops of a cysteine-less membrane protein of interest followed by chemical modification with a membrane impermeable thiol-specific probe either before or after compromising cell membrane integrity to determine cysteine membrane sidedness. Accessibility in whole cells establishes extracellular location while accessibility only after cell disruption establishes intracellular location (see notes 6, 7 and 9). The accessibility of extramembrane domains flanking a TM then establishes orientation of the TM with respect to the plane of the membrane bilayer (6).
3.2.1 Construction of Plasmids Expressing Single Cysteine Derivatives
Cysteine is a relatively hydrophobic, small amino acid, and its introduction at most positions in a membrane protein is likely to be tolerated. Furthermore, cysteine has little preference for a particular secondary structure. As an example an AmpR plasmid encoding a derivative of LacY (lactose permease of E. coli) in which endogenous cysteines are replaced by serine or alanine (cysteine-less LacY) is constructed using the site-directed mutagenesis Quickchange XL kit from Stratagene (11). Using the same method single cysteine replacements of amino acids in putative extramembrane domains are constructed (5, 8, 12). All amino acid substitutions are verified by DNA sequencing. Functional analysis and expression level by Western blotting of each derivative should be carried out if possible. Ideally gene expression should be under control of an inducible promoter such as OPtac to minimize continuous expression of potentially disruptive gene products especial in an altered lipid environment.
The process of choosing suitable residues for replacement by cysteine is often empirically determined, and the rationale for deciding which residues to alter is aided by the following considerations. Secondary structure predicted by computer-aided hydropathy analysis (reviewed (13) and thus far 60–70% reliable) is an initial starting point for the likelihood that a particular residue is in an extramembrane domain. The native cysteine residues are usually changed into alanine or serine residues which are small, commonly found in membrane proteins and appear to be tolerated at most positions thus rendering an active protein. Replacement of charged residues is generally not advised because these have a high probability of being topogenic signals or may be involved in long-range interactions. Consideration should be given to whether the replacement will be well-tolerated based on structural and functional information about the protein. Therefore, a prerequisite for each cysteine replacement is retention of function that provides assurance of retention of near native structure. If the protein contains stretches of residues of intermediate hydrophobicity that cannot unambiguously be identified as membrane spanning, substitutions should be made approximately every 10 residues. The cysteine-less protein serves as the starting template for introducing single cysteine residues at desired positions as well as a negative labeling control to assure that residues such as lysine and histidine are not labeled by the reagents. Alternatively, templates containing natural cysteines can be utilized in this assay if they do not react with the thiol-specific reagents due to steric hindrance or membrane residency.
To obtain a minimal topological map a single cysteine replacement in each of the putative extramembrane loops should be expressed from a plasmid and analyzed in a host with wild type lipid composition. In practice, several cysteine replacements or complete cysteine scanning across extramembrane loops and into TM segments is required for a more precise mapping of topology. The host strain for plasmid expression should be deleted of the target protein gene if it contains native cysteines and is expressed at levels high enough to be detected in the assay. Since the target protein is expressed from a multicopy plasmid, it is often possible to analyze a protein in its normal host without deletion of the native protein.
3.2.2 Transformation of Phosphatidylethanolamine-Deficient E. coli Cells with Plasmids
Single cysteine replacements are expressed in the appropriate lipid host. The transformability of an engineered E. coli strain (AL95) lacking its major nonbilayer-prone and zwitterionic lipid phosphatidylethanolamine (PE, 75 mole% of total phospholipid) is compromised by a requirement for divalent cations (10–50 mM Mg+2) for viability, cell integrity, proper cell division, membrane impermeability and osmotic stress response (14–17). Most of the above phenotypes are corrected by introduction of the “foreign” non-bilayer-prone neutral glycolipids α-monoglucosyldiacylglycerol (MGlcDAG) or bilayer-prone diglucosyldiacylglycerol (GlcGlcDAG, but increased passive membrane permeability of the inner and outer membranes remains (17, 18). Therefore, variations of conventional transformation protocols are required to make different lipid hosts competent for plasmid DNA uptake. Electroporation, which requires suspension of cells in very low ionic strength media, cannot be used. In most cases cells can be made competent for transformation using a RbCl/CaCl2-containing solution, which is suitable for strains with normal as well as altered lipid compositions.
-
1
Dilute 0.1 ml of a fresh overnight culture of PE-lacking host cells grown in LB medium with 50 mM MgCl2 into 5 ml of 37°C LB media supplemented with 50 mM MgCl2 to support growth in the absence of PE (15 ml tube). Cells lacking PE must be kept in the presence of at least 20 mM MgCl2 or RbCl/CaCl2 during all manipulations (see note 2). MgCl2 can be eliminated in the above procedure for cells containing PE and more standard methods of transformation can be used. Cells containing plasmid pDD72 (encodes wild type gene required for PE synthesis) or its derivatives must be grown at 30 C because the plasmid is temperature sensitive for replication.
-
2
Grow cells with vigorous aeration for 3–4 h until cells reach mid-log phase (OD600 = 0.6).
-
3
Place 1 ml of culture into a sterile microfuge tube and incubate at 4°C for 30 min. Centrifuge in pre-chilled (+4°C) bench centrifuge 5417R (Eppendorf) at 14,000 rpm (20,800 x g) for 1 min to pellet cells and gently resuspend in 0.5 ml of ice-cold 1X RbCl/CaCl2 Transformation Salts.
-
5
After 30 min on ice, centrifuge to pellet cells and gently resuspend in 0.1 ml of ice-cold 1X RbCl/CaCl2 Transformation Salts. Keep cell suspension on ice.
-
6
Cells are now competent. Add plasmid DNA (100–200 ng) and incubate 30 min on ice.
-
7
Heat shock for 2 min at 42 C and place on ice for 2 min. Add 1 ml LB containing 50 mM MgCl2 and grow at 37°C for at least 2 hr.
-
8
Pellet the cells and resuspend in 50 μl LB containing 50 mM MgCl2 and plate on LB agar plates containing 50 mM MgCl2 and 100 μg/ml of ampicillin. Plates should be fresh or no more than a few days old for best results. Grow at least 2 days at 37 C. Individual colonies are streaked for single colonies on LB plates containing ampicillin and either with or without MgCl2. Single colonies are screened to verify MgCl2 dependence to eliminate potential contaiminants.
3.2.3 Cell growth and Regulated Expression of LacY and PssA Genes
-
1
Cells (see Fig 1a for strain description) carrying plasmids expressing single cysteine replacements in cysteine-less LacY under OPtac control are grown for at least two generations in LB medium supplemented with 50 mM MgCl2 to support growth in the absence of PE (see note 2), ampicillin (100 μg/ml) to maintain LacY plasmids, and IPTG (1 mM) to induce LacY expression.
-
2
To independently regulate expression of LacY and PE, strain AT2033 (PLtetO-1-pssA+ pss93::kanR lacY::Tn9 recA srl::Tn10) is used. In this strain the level of aTc in the growth medium regulates chromosomal pssA (encodes phosphatidylserine synthase for PE synthesis) expression and PE content of the cell. Expression of a plasmid copy of OPtac-lacY is regulated by IPTG. In order to determine the effect of a change in membrane lipid composition on the topological organization of LacY, cells are grown first in the presence of IPTG without aTc to allow synthesis and membrane insertion of LacY in the absence of PE. Then cells are switched to growth without IPTG in presence of aTc in order to permit biosynthesis of new PE in the absence of newly synthesized LacY.
-
2.1
Cells of strain AT2033 containing different plasmids expressing derivatives of LacY are first grown overnight at 37°C in LB medium supplemented with ampicillin (100 μg/ml, required for plasmid maintenance) and 50 mM MgCl2 to support growth at residual levels (ca. 2–3 mole%) of PE.
-
2.2
Then cells are diluted to OD600 of ca. 0.05 into 200 ml of medium supplemented with ampicillin (100 μg/ml) and 50 mM MgCl2, and the expression of the LacY derivatives are induced by growth in the presence of 1 mM IPTG for at least two generations (OD600 reading 0.20).
-
2.3
One-half of the cell culture (100 ml) is pelleted by centrifugation (3,000 x g) and stored on ice for LacY for SCAM TM analysis.
-
2.4
Remaining cells (100 ml) are pelleted by centrifugation and washed twice by centrifugation with pre-warmed sterile LB medium supplemented with 50 mM MgCl2 to remove IPTG and then re-suspended in 100 ml of pre-warmed medium supplemented with ampicillin (100 μg/ml) and 50 mM MgCl2. Expression of the pssA gene is induced by addition of aTc (1 μg/ml).
-
2.5
After 3 h of growth in the presence of inducer, cells are harvested and subjected to SCAMTM analysis.
3.2.4 General Protocol for SCAMTM
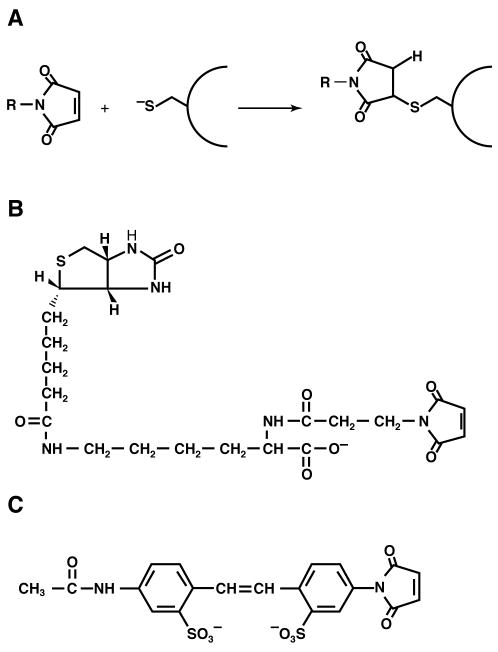
The chemical nature of the reactive portion of a labeling reagent should be highly reactive with and selective for thiol groups and form a stable non-exchangeable or non-hydrolysable derivative. Maleimide-based thiol reagents, which are available in a wide variety of forms, are particularly suited for SCAMTM (6). Maleimide reacts with the ionized form of a thiol group (Fig. 2a), and this reaction requires a water molecule as a proton acceptor. Maleimides are virtually unreactive until they encounter an available thiol group. For most water-exposed cysteine residues in proteins, pKa of the thiol of cysteine lies in the range of 8 to 9 and formation of cysteinyl thiolate anions is optimum in aqueous rather in a non-polar environment where the pKa of the thiol of cysteine is around 14. Therefore, the reaction rate of different sulfhydryls is controlled primarily by their water exposure making the residues that reside in regions of TM helices unfavorable for the generation of thiolate anions. Thus, the labeling characteristics of intramembrane (unreactive) and extramembrane (reactive) cysteines should be consistent with their localization either in a non-polar or polar environment, respectively (6). In most cases the unreactive cysteine residues are located within the membrane hydrophobic core or in a sterically hindered environment as described below.
Fig. 2. Structure of thiol-modifying reagents and their reaction with a thiol.
(a) Reaction of a maleimide with the thiolate of a protein bound cysteine to form a covalent adduct. (b) Structure of MPB. (c) Structure of AMS. Figure reproduced from (6) with permission.
SCAMTM using thiol-specific membrane–impermeable MPB (Fig 2b) has been extensively used to probe the topological organization of LacY (3, 5, 6, 8), high affinity phenylalanine permease (PheP) (9) and γ-aminobutyric acid permease (GabP) (19) as a function of membrane lipid composition. In this assay sonication of cells is used to disrupt cell membranes, making both periplasmic and cytoplasmic cysteines accessible to MPB, whereas cysteines located within a TM domain are still protected from labeling (6). Derivatization of cysteines in whole cells indicates periplasmic expose while derivatization only during sonication indicates cytoplasmic exposure, since desintegration allows thiol-specific reagent access to both sides of the membrane. It is important to note that the extent of biotinylation should be the same before and after sonication for an extracellular cysteine. If sonication results in an increase in biotinylation this may indicate a mixed topology.
Although single cysteine replacements could affect protein structure and expression, conclusions are based on comparison of the extent of labeling in whole cells and disrupted cells for the same protein in two different lipid environments. This approach simplifies interpretation of data obtained with a series of protein derivatives that may express at different levels since conclusions about topology are based on relative reactivity of cysteines in the same sample before and after cell disruption. Since sample pairs are analyzed on the same Western blot, no signal intensity normalization is required and the results are not dependent the expression level of the derivative. The level of expression of any given derivative will affect the absolute intensity of labeling but not the ratio of the labeling between the sample pairs.
Harvest 100 ml mid-log phase (OD600 ~ 0.4–0.6) cells (see note 2) expressing a single cysteine derivative of LacY or protein of interest by centrifugation and suspend cell pellets in 1.5 ml of buffer A (adjusted to pH 7, 9 or 10.5 as indicated). Divide the sample into two equal aliquots (0.75 ml) in Microfuge polyallomer tubes (Beckman). To increase the reactivity of diagnostic cysteine residues (particularly those that might be in sterically hindered extramembrane domains), the reaction with MPB is carried out at pH 9 or 10.5 (see note 8).
Treat one set of samples with MPB at a final concentration of 100 μM (7.5 μl of 10 mM stock solution) (see notes 3 and 4) for 5 min at room temperature to label cysteines exposed to the periplasmic side of the inner membrane. Quench the reaction by the addition of β-mercaptoethanol to 20 mM (7.5 μl of 2 M stock solution). After labeling, cells are sonicated for 1 min using an amplitude of 15%.
To simultaneously label cysteines exposed to both sides of cell membrane, subject the remaining sample to sonication for 1 min in the presence of MPB at a final concentration of 100 μM. Incubate for 4 min at room temperature and quench the reaction by the addition of β-mercaptoethanol to 20 mM.
All sonicated samples are centrifuged at 4°C at 38,000 rpm (65,000 x g) (TLA-55 Beckman Coulter rotor) for 10 min followed by resuspension in 100 μl of Buffer A containing 20 mM β-mercaptoethanol by vortexing for 2 hr at room temperature using a Fisher Vortex Genie 2. The membranes are solubilizated by detergent, and the target protein is isolated by immunoprecipitation or affinity purification as described below.
Following modification with MPB and isolation by immunoprecipitation, the target protein is resolved by SDS PAGE, transferred to a solid support, and detected by Western blotting using avidin linked to horse radish peroxidase. SuperSignal West Pico chemiluminescent substrate (Thermo Scientific) is used to visualize biotinylated proteins. Biotinylation of exposed cysteine residues of whole cells (periplasmic exposure) or only during sonication (cytoplasmic exposure) is detected using a Fluor-STM MultiImager (Bio-Rad).
3.2.5 Sample Solubilization
After labeling, isolated membranes are solubilized with the appropriate detergent or detergent mixture such as SDS alone, Triton X-100 alone, SDS and Triton-X-100, CHAPS, octylglucoside, deoxycholate, cholate and Tween 20, β-D-dodecylmaltoside or nonidet P-40 and sodium deoxycholate (6). Conditions must be empirically determined that yield a non-aggregated soluble target protein throughout the remainder of the procedure. Many membrane proteins aggregate if boiled in SDS so that solubilization should be done between 37 C and 55 C. LacY forms irreversible polydisperse aggregates if solubilized by Triton X-100 alone. Therefore, a membrane-embedded LacY sample (100 μl) is solubilized by addition of an equal volume of solubilization buffer followed by vigorous vortexing for 15 min at room temperature, incubation at 37°C for 15 min, and an additional 15 min vortexing at room temperature. Prior to immunoprecipitation, the sample is diluted with buffer containing non-ionic detergent to neutralize the denaturing properties of SDS (as described below) and cleared by centrifugation in a pre-chilled (+4°C) bench centrifuge 5417R (Eppendorf) at 14,000 rpm (20, 800 x g) for 10 min.
3.2.6 Isolation of Derivatized Target Protein
The thiol reagents react with cysteine residues present in all other proteins in the membrane. Immunoprecipitation of the membrane protein of interest or a rapid purification step is necessary to eliminate other labeled proteins. A biotin-maleimide labeled protein can be recovered from cell lysates directly with streptavidin-agarose beads (20) and can then be detected by western blotting using a target-specific antibody. For immunoprecipitation of labeled protein from solubilized samples, polyclonal and monoclonal antibodies have been widely utilized. Antigen-antibody complexes can be isolated using precipitation with Pansorbin (Staphylococcus aureus cells) (3), protein A agarose (21) or protein A or G Sepharose beads (22, 23). If antibodies specific to the protein under study are not available, then epitope tags such as His6 can be incorporated at the C-terminus of the target protein for either immunoprecipitation or isolation by Ni2+ chelated affinity resin (3, 24, 25). Use of affinity methods with His-tagged proteins and small-scale batch purification procedures is becoming the method of choice since the labeled protein can be directly extracted from the resin with SDS-containing buffers followed by SDS PAGE (26). Of course protein function or topology should not be compromised by the presence of the tag. To allow appropriate antibody interaction with solubilized protein, SDS should be diluted with appropriate non-ionic detergent. In case of LacY, samples (200 μl from above) are diluted with 300 μl of cold Immunoprecipitation buffer followed by empirically established immunoprecipitation protocols (3).
Following modification and isolation, the target protein is resolved by SDS PAGE, transferred to a solid support, and detected by western blotting or one of the following techniques (6). Thiol reagents are available that contain a biotin group, a fluorescent group, or a radiolabel allowing detection of labeled proteins by avidin linked to horse radish peroxidase (avidin-HRP) and indirect chemilumiscence detection, fluorescence, or autoradiography, respectively. Signals can be quantified using available imaging systems and software.
3.2.6 SDS-PAGE
The final immunoprecipitates are solubilized in 30 μl of SDS sample buffer (10 mM Tris HCl (pH 6.8), 5.6% SDS, 200 mM DTT, 10% glycerol, 0.01% bromophenol blue) by vigorous vortexing for 15 min at room temperature, incubation at 37°C for 15 min, and an additional 15 min vortexing at room temperature using a Fisher Vortex Genie 2. Samples are subjected to SDS PAGE using standard protocols and then transferred from the gel to a solid support by Western blotting.
3.2.7 Western Blot Analysis
Protein samples are transferred from the SDS polyacrylamide gel to Protran BA 85 nitrocellulose membranes by electroblotting using a semi-dry electroblotting system.
After electrophoresis, place the SDS-PAGE gel in the cathode buffer to equilibrate for 10 min before blotting.
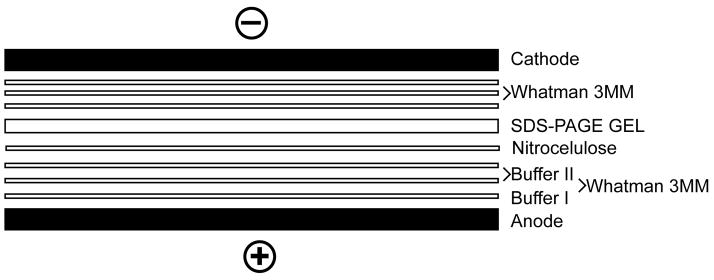
Cut out a piece of Protran BA 85 nitrocellulose transfer membrane and Whatman filter papers sized to fit the SDS polyacrylamide gel. See Fig. 3, which shows the procedure for building the blotting sandwich.
Soak the bottom layer of chromatography paper (Whatman 3 mm Cr) in Anode buffer No.1 and allow excess buffer to drain from the paper. Place a drop of this buffer in middle of positive electrode surface of the blotting apparatus, then lay down the soaked filter paper. Center and smooth out by rolling with a glass tube to establish good contact and eliminate air bubbles.
Place two layers of chromatography paper wetted with Anode buffer No. 2 on top of paper soaked in Anode buffer No.1. Roll a glass tube over the chromatography paper to smooth and remove trapped air bubbles.
Place a Protran BA 85 nitrocellulose transfer membrane wetted with distilled water on top of the three layers of chromatography paper and roll with a glass tube.
Now center the acrylamide gel onto the transfer membrane makng sure that air bubbles are not trapped in between the transfer membrane and acrylamide gel. Use a glass tube dipped in Cathode buffer to gently roll out any trapped air.
Place three layers of chromatography paper soaked in Cathode buffer on top of the acrylamide gel. Chromatography papers, gels and transfer membrane must be in the same dimensions to achieve a uniform transfer.
Place the lid (negative electrode) over the trans-unit sandwich making sure that the electrode rests on the sandwich.
Attach the power cables on Labconco Semi-Dry blotting system to a constant voltage power supply capable of supplying sufficient current for transfer. Adjust the power supply to provide initial current density of 2.5 mA/cm2 of gel area. The voltage reading should be approximately 10 volts with new electrodes.
Transfer time is somewhat dependent on the size of the proteins and percentage of gel used. In most cases complete transfer is achieved in 90 min.
Fig. 3.
Schematic illustration of the Western blotting sandwich used with a semi-dry electroblotting system.
3.2.8 Blocking Procedure
The nitrocellulose transfer membrane is then blocked overnight with blocking buffer. Note that 0.05% Nonidet P-40 is omitted during blocking prior to staining of biotinylated proteins with avidin-HRP.
3.2.9. Staining with Avidin-HRP
MPB is a thiol reagent containing a biotin group, which makes possible indirect chemilumiscence detection of labeled proteins after treatment with avidin linked to horseradish peroxidase (avidin-HRP).
The nitrocellulose transfer membranes are washed once with TBS buffer containing 0.3% BSA for 10 min.
Avidin–HRP is added at a final dilution of 1: 5000–10,000 of 2 mg/ml stock solution in TBS buffer containing 0.3% BSA and incubated for at least 1 h.
The sheets are washed two times with TBS buffer containing 0.3% BSA for 15 min each followed by another two washes with TBS/Nonidet P40 buffer and once with TBS buffer only.
To visualize biotinylated proteins, the sheets are incubated for 3 min with SuperSignal West Pico chemiluminescent substrates (Thermo Scientific) mixed immediately prior to use at a ratio of 1:1, and biotinylated proteins are visualized using a Fluor-S MaxTM MultiImager (Bio-Rad).
3.2.10 Image Acquisition and Processing
Western blots are imaged using a Fluor-S MaxTM MultiImager (Bio-Rad). Bio-Rad software Quantity OneTM versions 4.6.5.094 and 4.4.1 are used to collect and store the images as TIFF files, which can be imported later into Adobe Illustrator to construct figures. Images are expanded or reduced so that the horizontal strip containing the target protein images is sized appropriately and masked to only show the target protein results, which are then aligned with images from other gels and labeled. The only valid comparison in intensity is between whole cell and sonicated sets (images treated identically) run on the same gel.
3.2.9 Data Analysis and Interpretation
The criteria used for determining the location of an introduced cysteine are as follows. Labeling of a cysteine residue with a membrane-impermeable sulfhydryl reagent before disruption of whole cells is indicative of a periplasmic cysteine residue provided accessibility to a cytoplasmically localized control protein (see note 4) and a cysteine-less derivative of the target protein are negative (see note 8). Absence of labeling in whole cells but labeling during cell disruption indicates a cytoplasmic location for the cysteine-containing domain. No labeling with a sulfhydryl reagent before or during cell disruption implies localization to a hydrophobic membrane environment or unfavorable local orientation/positioning of introduced thiol groups which may prevent access by the reagent or result in an increase of the pKa of the thiol group as discussed below (see note 6).
3.2.10 Topological Assignment of Problematic Regions
No definitive conclusion can be made for the location of a domain based on lack of reaction of the cysteine (see notes 5 and 6). An unreactive cysteine can be due to its location within a TM or proximal environmental effects, which affect the thiol pKa or sterically restrict access (6). Cysteine scanning in the neighborhood of the unreactive cysteine may be required to differentiate between a TM or an unreactive cysteine within a larger exposed domain. Unfavorable orientation of a thiol group due to local secondary structure or properties of neighboring amino acids may restrict or prevent access by large thiol reagents. Cysteine residues closer to the membrane interface generally react slower than those near the center of extramembrane domains. Since derivatization is favored by formation of the cysteinyl thiolate anion (Fig. 2a), increasing the reaction buffer pH would favor alkylation of an extramembrane cysteine as well as disrupt local restrictive secondary structure while truly membrane imbedded cysteines would not be expected react. Moreover diagnostic cysteines can be hidden within membrane domains that partially penetrate the membrane, which have been termed membrane-dipping, shallow-penetrating, re-entry, mini-, or U-shaped loops. Such unusual topological arrangements are present in aquaporins, potassium chloride channels and protein conducting channels (27). Under strongly alkaline conditions (pH >11), soluble and peripheral membrane proteins (i.e, those that do not contain any domains that span the membrane bilayer) are released in soluble forms. Integral membrane proteins remain embedded in the lipid bilayer and can be isolated by centrifugation. Extramembrane domains that are sterically hindered or exhibit elevated pKa’s can be derivatized with increasing pH up to 10.5 without compromising membrane integrity. However, appropriate controls such as inaccessibility of known cytoplasmically exposed domains and lack of label of a cysteine-less target protein should be done (sse note 8). The same may apply for TMs that form aqueous channels across or part way across the membrane. Cysteines in mini-loop domains are exposed by NaOH with loss of membrane integrity, but a true TM remains unreactive
To expose hindered cysteines the pH of the reaction mix is raised either step-wise to pH 10.5 in a series of reactions (see note 8). Cell integrity is maintained at pH 10.5 as evidenced by labeling of cytosolic domains only during sonication. Cysteine-less protein and protein with an intramembrane cysteine are not labeled at pH 10.5 (5). To detect cysteines in a mini-loop domain, cells are treated with NaOH prior to reaction with a thiol reagent.
Cell aliquots in buffer A are mixed with an equal volume of cold 0.2 N NaOH, incubated for 5 min on ice, and separated into a pellet and supernatant fraction by centrifugation at 40,000 rpm (70,000 x g) (TLA-55 Beckman Coulter rotor) for 10 min.
The pellets are washed three times by suspension in Buffer A using sonication for 1 min at an amplitude of 15% followed by centrifugation as above. The final pellet is resuspended in Buffer A and subjected to SCAMTM at pH 7.5.
3.2.10.11 Indirect and Direct Labeling and Identification of Mixed Topologies
Central to the method is the use of detectable thiol-specific reagents to differentiate intracellular from extracellular domains of membrane protein which in some cases may adopt mixed or dual TM topologies within the same membrane (28) (see note 9). Confirmation of labeling of external water-exposed cysteines by MPB can be achieved by first blocking putative external cysteines in intact E. coli cells with a thiol-specific reagent that is transparent in the detection phase of the procedure. Such a pre-blocking step also allows selective labeling of luminal (exposed to cytoplasm) cysteines after cell permeabilization or disruption and therefore detection of mixed topologies co-existing within the same cell membrane (3, 6). A set of impermeable blocking reagents that effectively react with thiols exposed to solvent but are transparent in the detection phase of the procedure is available for SCAMTM (6). One such reagent is 4-acetamido-4′-maleimidylstilbene-2,2′-disulfonic acid (AMS, Fig. 3c) which is membrane-impermeable due to its size, two charged sulfonate groups and high solubility in water.
The degree of mixed topology can be assessed with a two-step protocol as follows. Intact cells are treated either with or without AMS followed by labeling with MPB of intact cells and cells during disruption. Excess AMS is removed prior to any subsequent treatments by centrifuging the reaction mixture through a small column of gel filtration resin to avoid lysis of fragile cell preparations (6). In the case of mixed topology, biotinylation of the target cysteine by MPB will occur with whole cells not pre-treated with AMS and to a greater extent after cell disruption. AMS treatment will prevent any biotinylation with whole cells and reduce the amount of biotinylation observed in disrupted cells. The protective effect of AMS is almost complete and has been used to quantify the degree of mixed topology (29).
To confirm labeling of external (periplasmic) water-exposed cysteines by MPB and detect possible mixed topology of LacY or protein of interest cell pellets derived from 200 ml of mid-log phase (OD600 ~ 0.4–0.6) cells expressing a single cysteine derivative are resuspended in 3.5 ml of buffer A and divided into four equal aliquots (0.75 ml) in Microfuge polyallomer tubes (Beckman).
Two cell aliquots are incubated for 30 min at 25°C with AMS at a final concentration of 5 mM (37.5 μl of 100 mM aqueous stock solution) to block periplasmic water-accessible cysteine residues from the outside of cells. Then AMS is removed either by centrifugation through a small gel filtration column or by two cycles of centrifugation and resuspension in 0.75 ml of buffer A.
Two cell aliquots, one pre-treated with AMS from step 2 and one not treated with AMS, are biotinylated by adding MPB at a final concentration of 100 μM (7.5 μl of 10 mM stock solution) followed by incubation for 5 min at 25°C and quenching of the reaction by the addition of β-mercaptoethanol to 20 mM. Cells are then disrupted by sonication.
The two remaining samples, one pre-treated with AMS from step 2 and one not treated with AMS, are biotinylated by adding MPB at a final concentration of 100 μM (7.5 μl of 10 mM stock solution) during sonication for 1 min followed by incubation for another 4 min at room temperature before quenching the reaction by the addition of β-mercaptoethanol to 20 mM.
All samples are processed for detection of biotinylation as described above.
Acknowledgments
This work was supported by NIH grant GM20478 and funds from the John S. Dunn Foundation awarded to W. D.
Footnotes
Pre-chilled (+4°C) 50 mM Tris-HCl (pH 8.1) should be used to prepare IP buffer with SDS/non-ionic detergent ratio 1/5.
Cells lacking PE must not be exposed to solutions with MgCl2 concentrations less than 10 mM. However, when comparing cells with different lipid compositions to wild type cells, the MgCl2 concentration should be the same for all samples and in the range of 10–50 mM with the higher concentration being optimal for growth of most PE-lacking strains.
Final concentration of DMSO used to dissolve MPB should never exceed 0.5%.
SCAMTM is based on the controlled membrane permeability of sulfhydryl reagents. The results of SCAMTM are valid only if the modifying reagent is thiol-specific and membrane impermeable, cells are intact and cell disruption does not expose sterically hindered or water inaccessible cysteine residues (3, 5, 6). Various reagents including MPB will cross the membrane in a concentration, time and temperature dependent manner and permeability varies with genetic background of the host cells. Therefore, conditions must be empirically determined to minimize derivatization of intracellular cysteines. Cell lysis during a labeling experiment or during manipulation of cells will also result in labeling of intracellular cysteines. Pre-blocking external cysteines with AMS before labeling intact cells with MPB provides a means of estimating the degree of labeling due to low permeability under different experimental conditions.
The membrane permeability of a thiol-specific labeling reagent can be tested and labeling conditions (concentration, time and temperature) established by quantification of the degree of labeling of an abundant cytoplasmic protein that is rich in surface exposed cysteine residues. In E. coli β-galactosidase is ideal for this purpose due to its content of cysteine, mobility in a region on SDS PAGE devoid of other major proteins, availability of mutants lacking the enzyme as a control, and availability of antibody against β-galactosidase (Sigma ImmunoChemicals). Other cytosolic bacterial markers such a glutathione or elongation factor Tu have been used to access membrane permeability. Significant differences in the permeability of different host strains emphasize the need to screen host strains for reagent permeability prior to initiating experiments (6–8).
Whole and disrupted cells are treated with various concentrations of reagent from 10 μM to 1 mM, at temperatures from 0 C to 25°C, and for various lengths of time from 5 min to 1 h. In most cases low concentration of MPB (100 μM) and relatively short incubation period (5 min) at room temperature favor biotinylation of extramembrane thiol groups. In this case a labeling experiment with both intact cells and disrupted cells is carried out except that soluble proteins rather than the membrane fraction are retained for immunoprecipitation with antibody against β-galactosidase followed Western blotting analysis.
A major problem is the variability between samples due to mechanical loss during the work up or due to differences in expression level of individual replacements. Since conclusions are only made by comparing sample pairs (labeling without or during cell disruption), differences in expression or labeling efficiency between individual replacements are not important. Corrections for mechanical loss can be made by quantitative Western blot analysis using a target specific antibody. However, if no labeling occurs with the thiol reagent, it is important to verify that the protein was expressed and is present on the Western blot. To verify the presence of the target protein or to ensure the presence of equal amounts of target protein in each paired sample, blots can be stripped using RestoreTM Western Blot Stripping Buffer (Thermo Scientific) and reprobed with appropriated antibody.
Caution must be used in assigning an intramembrane location to a cysteine residue because it is unreactive to hydrophilic thiol reagent in both intact and disrupted cells. Lack of or low levels of labeling may result from any of the following reasons: (1) steric hindrance due to local secondary structure; (2) internalization into the compact fold of the protein; (3) lack of ionization of the thiol group due to a hydrophobic environment; (4) local environment with the same charge as the thiol reagent; (5) increased pKa of the thiol due to the high negative charge density of neighboring residues or anionic lipids; (6) facing other helices. Periplasmic extramembrane domains tend to be shorter (sometimes only three amino acids in length) than cytoplasmic domains. Therefore, there may be little or no protrusion of these loops into the extracellular space, thus preventing reaction of the cysteine residues in these locations with relatively bulky reagents. Alkylating reagents appear to react better with cysteines towards the middle of extended hydrophilic loops than near the TM interfacial domain. Cysteine scanning across a domain is an effective means for identifying useful replacement sites and differentiating between local effects and true TMs. Scanning can be coupled with alkaline treatment as discussed above. Finally, reagents of different size and charge can be tested (6).
Conclusions based on full reactivity of diagnostic residues should be made with caution, since cysteine residues facing a hydrophilic pore or near a substrate-binding site maybe within a TM segment but chemically reactive due to water channels or pockets. Therefore an extramembrane domain and an aqueous pore are not easily distinguished by SCAMTM. Substitution at positions crucial to overall protein structure and stability cannot be used, but such substitutions often result in low levels of the protein or loss of activity and are informative.
Since the formation of cysteinyl thiolate anions is favored by increasing the solution pH (optimum pH 8.0–8.5), increasing the pH during labeling should favor the reaction. However, maleimides are known to react with primary amines at pH values above 7.5. Therefore, attempts to increase efficiency of labeling by raising the pH of the assay should be thoroughly controlled in order to ensure that the modification is confined to cysteine. An effective control to rule out non-thiol modifications is to use a cysteine-less target protein.
TM topology studies assume that all copies of the target protein have the same orientation. The labeling patterns are the result of end-point titrations and assume a relatively fixed conformation for extramembrane loops, ignoring the presence of regions with heightened mobility and flexibility or the possibility of topological inversions on the time scale of the labeling. Topological models derived from accessibility patterns depict a static TM topology whereas the actual structure in a membrane is more likely to be dynamic. Through the application of SCAMTM some examples exist of dynamic changes in topological organization induced post-assembly of membrane proteins as well as some proteins that appear to exist with multiple topological organizations (6). Even with optimal assays and reagents, membrane proteins that assume multiple conformations either within the same or between different membranes may yield confusing and conflicting results. Therefore, the static nature of methods that measure topological organization may have missed more dynamic properties of membrane proteins. In particular low yield of modification due to slow rate of reaction may be due to dynamic movement of a domain into and out of an accessible region. There are examples of cryptic intramembrane regions that become exposed to the aqueous phase and extramembrane domains that translocate to the opposite side of the membrane which limit topology studies in intact cells where the protein is turning over or metabolic conditions influence organization (6).
References
- 1.Drews J. What’s in a number? Nat Rev Drug Discov. 2006;5:975. doi: 10.1038/nrd2205. [DOI] [PubMed] [Google Scholar]
- 2.Goder V, Junne T, Spiess M. Sec61p contributes to signal sequence orientation according to the positive-inside rule. Mol Biol Cell. 2004;15:1470–1478. doi: 10.1091/mbc.E03-08-0599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bogdanov M, Heacock PN, Dowhan W. A polytopic membrane protein displays a reversible topology dependent on membrane lipid composition. EMBO J. 2002;21:2107–2116. doi: 10.1093/emboj/21.9.2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bogdanov M, Mileykovskaya E, Dowhan W. Lipids in the Assembly of Membrane Proteins and Organization of Protein Supercomplexes: Implications for Lipid-linked Disorders. Subcell Biochem. 2008;49:197–239. doi: 10.1007/978-1-4020-8831-5_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bogdanov M, Xie J, Heacock P, Dowhan W. To flip or not to flip: lipid-protein charge interactions are a determinant of final membrane protein topology. J Cell Biol. 2008;182:925–935. doi: 10.1083/jcb.200803097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bogdanov M, Zhang W, Xie J, Dowhan W. Transmembrane protein topology mapping by the substituted cysteine accessibility method (SCAMTM): application to lipid-specific membrane protein topogenesis. Methods. 2005;36:148–171. doi: 10.1016/j.ymeth.2004.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang X, Bogdanov M, Dowhan W. Topology of polytopic membrane protein subdomains is dictated by membrane phospholipid composition. EMBO J. 2002;21:5673–5681. doi: 10.1093/emboj/cdf571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xie J, Bogdanov M, Heacock P, Dowhan W. Phosphatidylethanolamine and monoglucosyldiacylglycerol are interchangeable in supporting topogenesis and function of the polytopic membrane protein lactose permease. J Biol Chem. 2006;281:19172– 19178. doi: 10.1074/jbc.M602565200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang W, Bogdanov M, Pi J, Pittard AJ, Dowhan W. Reversible topological organization within a polytopic membrane protein is governed by a change in membrane phospholipid composition. J Biol Chem. 2003;278:50128–50135. doi: 10.1074/jbc.M309840200. [DOI] [PubMed] [Google Scholar]
- 10.Dowhan W. Molecular Genetic Approaches to Defining Lipid Function. J Lipid Res. 2009;50 doi: 10.1194/jlr.R800041-JLR200. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Iwaarden PR, Pastore JC, Konings WN, Kaback HR. Construction of a functional lactose permease devoid of cysteine residues. Biochemistry. 1991;30:9595–9600. doi: 10.1021/bi00104a005. [DOI] [PubMed] [Google Scholar]
- 12.Frillingos S, Sahin-Toth M, Wu J, Kaback HR. Cys-scanning mutagenesis: a novel approach to structure function relationships in polytopic membrane proteins. Faseb J. 1998;12:1281–1299. doi: 10.1096/fasebj.12.13.1281. [DOI] [PubMed] [Google Scholar]
- 13.Elofsson A, von Heijne G. Membrane Protein Structure: Prediction vs Reality. Annu Rev Biochem. 2007;76:125–140. doi: 10.1146/annurev.biochem.76.052705.163539. [DOI] [PubMed] [Google Scholar]
- 14.DeChavigny A, Heacock PN, Dowhan W. Sequence and inactivation of the pss gene of Escherichia coli. Phosphatidylethanolamine may not be essential for cell viability. J Biol Chem. 1991;266:5323–5332. [PubMed] [Google Scholar]
- 15.Mileykovskaya E, Dowhan W. Role of membrane lipids in bacterial division site selection. Curr Opin Microbiol. 2005;8:135–142. doi: 10.1016/j.mib.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 16.Rietveld AG, Chupin VV, Koorengevel MC, Wienk HL, Dowhan W, de Kruijff B. Regulation of lipid polymorphism is essential for the viability of phosphatidylethanolamine-deficient Escherichia coli cells. J Biol Chem. 1994;269:28670– 28675. [PubMed] [Google Scholar]
- 17.Wikström M, Xie J, Bogdanov M, Mileykovskaya E, Heacock P, Wieslander Å, Dowhan W. Monoglucosyldiacylglycerol, a foreign lipid, can substitute for phosphatidylethanolamine in essential membrane-associated functions in Escherichia coli. J Biol Chem. 2004;279:10484–10493. doi: 10.1074/jbc.M310183200. [DOI] [PubMed] [Google Scholar]
- 18.Wikström M, Kelly A, Georgiev A, Eriksson H, Rosen-Klement M, Bogdanov M, Dowhan W, Wieslander Å. Lipid-engineered Escherichia coli membranes reveal critical lipid head-group size for protein function. J Biol Chem. 2009:284. doi: 10.1074/jbc.M804482200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang W, Campbell HA, King SC, Dowhan W. Phospholipids as determinants of membrane protein topology. Phosphatidylethanolamine is required for the proper topological organization of the gamma-aminobutyric acid permease (GabP) of Escherichia coli. J Biol Chem. 2005;280:26032–26038. doi: 10.1074/jbc.M504929200. [DOI] [PubMed] [Google Scholar]
- 20.Sato Y, Zhang YW, AAT, Rudnick G. Analysis of transmembrane domain 2 of rat serotonin transporter by cysteine scanning mutagenesis. J Biol Chem. 2004;279:22926–22933. doi: 10.1074/jbc.M312194200. [DOI] [PubMed] [Google Scholar]
- 21.Wada T, Long JC, Zhang D, Vik SB. A novel labeling approach supports the five-transmembrane model of subunit a of the Escherichia coli ATP synthase. J Biol Chem. 1999;274:17353–17357. doi: 10.1074/jbc.274.24.17353. [DOI] [PubMed] [Google Scholar]
- 22.Cao W, Matherly LH. Characterization of a cysteine-less human reduced folate carrier: localization of a substrate-binding domain by cysteine-scanning mutagenesis and cysteine accessibility methods. Biochem J. 2003;374:27–36. doi: 10.1042/BJ20030301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu Q, Lee DW, Casey JR. Novel topology in C-terminal region of the human plasma membrane anion exchanger, AE1. J Biol Chem. 2003;278:3112–3120. doi: 10.1074/jbc.M207797200. [DOI] [PubMed] [Google Scholar]
- 24.Fujihira E, Tamura N, Yamaguchi A. Membrane topology of a multidrug efflux transporter, AcrB, in Escherichia coli. J Biochem. 2002;131:145–151. doi: 10.1093/oxfordjournals.jbchem.a003069. [DOI] [PubMed] [Google Scholar]
- 25.Long JC, DeLeon-Rangel J, Vik SB. Characterization of the first cytoplasmic loop of subunit a of the Escherichia coli ATP synthase by surface labeling, cross-linking, and mutagenesis. J Biol Chem. 2002;277:27288–27293. doi: 10.1074/jbc.M202118200. [DOI] [PubMed] [Google Scholar]
- 26.Valiyaveetil FI, Fillingame RH. Transmembrane topography of subunit a in the Escherichia coli F1F0 ATP synthase. J Biol Chem. 1998;273:16241–16247. doi: 10.1074/jbc.273.26.16241. [DOI] [PubMed] [Google Scholar]
- 27.Lasso G, Antoniw JF, Mullins GL. A combinatorial pattern discovery approach for the prediction of membrane dipping (re-entrant) loops. Bioinformatics. 2006;22:290–297. doi: 10.1093/bioinformatics/btl209. [DOI] [PubMed] [Google Scholar]
- 28.Gafvelin G, von Heijne G. Topological “frustration” in multispanning E. coli inner membrane proteins. Cell. 1994;77:401–412. doi: 10.1016/0092-8674(94)90155-4. [DOI] [PubMed] [Google Scholar]
- 29.Kimura T, Ohnuma M, Sawai T, Yamaguchi A. Membrane topology of the transposon 10-encoded metal-tetracycline/H+ antiporter as studied by site-directed chemical labeling. J Biol Chem. 1997;272:580–585. doi: 10.1074/jbc.272.1.580. [DOI] [PubMed] [Google Scholar]
- 30.Martinez-Morales F, Schobert M, Lopez-Lara IM, Geiger O. Pathways for phosphatidylcholine biosynthesis in bacteria. Microbiology. 2003;149:3461–3471. doi: 10.1099/mic.0.26522-0. [DOI] [PubMed] [Google Scholar]
- 31.Shiba Y, Yokoyama Y, Aono Y, Kiuchi T, Kusaka J, Matsumoto K, Hara H. Activation of the Rcs signal transduction system is responsible for the thermosensitive growth defect of an Escherichia coli mutant lacking phosphatidylglycerol and cardiolipin. J Bacteriol. 2004;186:6526–6535. doi: 10.1128/JB.186.19.6526-6535.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]