Abstract
Xylella fastidiosa is a xylem-limited bacterium that causes various diseases, among them Pierce's disease of grapevine (PD) and almond leaf scorch (ALS). PD and ALS have long been considered to be caused by the same strain of this pathogen, but recent genetic studies have revealed differences among X. fastidiosa isolated from these host plants. We tested the hypothesis that ALS is caused by PD and ALS strains in the field and found that both groups of X. fastidiosa caused ALS and overwintered within almonds after mechanical inoculation. Under greenhouse conditions, all isolates caused ALS and all isolates from grapes caused PD. However, isolates belonging to almond genetic groupings did not cause PD in inoculated grapes but systemically infected grapes with lower frequency and populations than those belonging to grape strains. Isolates able to cause both PD and ALS developed 10-fold-higher concentrations of X. fastidiosa in grapes than in almonds. In the laboratory, isolates from grapes overwintered with higher efficiency in grapes than in almonds and isolates from almonds overwintered better in almonds than in grapes. We assigned strains from almonds into groups I and II on the basis of their genetic characteristics, growth on PD3 solid medium, and bacterial populations within inoculated grapevines. Our results show that genetically distinct strains from grapes and almonds differ in population behavior and pathogenicity in grapes and in the ability to grow on two different media.
The bacterium Xylella fastidiosa causes several plant diseases in California, such as Pierce's disease of grapevine (PD) (5), almond leaf scorch (ALS) (6), alfalfa dwarf (AD) (19), and oleander leaf scorch (29). Sharpshooter leafhoppers (Hemiptera: Cicadellidae) and spittlebugs (Hemiptera: Cercopidae) are vectors of X. fastidiosa for the natural spread of this pathogen (30, 31). Vector biology strongly influences the epidemiology of PD (see reference 17 for a review), but principal vectors for ALS have not been related to disease spread (24).
Extensive transmission experiments showed that X. fastidiosa isolates from grapes caused AD and isolates from alfalfa caused PD(12, 30). Davis et al. (6) found that three isolates from grapes and two isolates from almonds inoculated into almonds and grapes caused disease in both hosts. This suggested that PD, ALS, and AD are caused by the same strains. Strains of X. fastidiosa from oleander did not cause PD or ALS but were recovered from mechanically inoculated almond plants in the laboratory (28); neither the tested isolates from grapes nor those from almonds caused disease in or were recovered from oleander (29).
The major leafhopper vectors of X. fastidiosa for grapevines in the Central Valley (Xyphon [Carneocephala] fulgida and Draeculacephalaminerva) (26) are found in almond orchards and transmit the bacterium from almond and grape source plants to healthy almonds and grapes in all four combinations (24). But generally there is no ALS near PD-infected vineyards and vice versa (24). Apparently, another factor besides the presence of vectors may affect disease spread in such situations. An alternative hypothesis to explain the lack of ALS near PD hotspots is that most inoculations of PD strains of X. fastidiosa into almonds do not survive the winter; thus, the number of (symptoms) infections detected would be low in relation to the total number of infections. Data obtained by Davis et al. (6) suggested that this might be the case, since only 2 of 48 successful inoculations of grape strains into almonds in the field survived one winter. Unfortunately, those authors did not use isolates from natural ALS infections in the test because of concerns that the experimental area was located in a region free of ALS. Cold winters have been shown to cure PD in potted grapevines, and higher rates of recovery from infection occurred in vines exposed to winter conditions in areas in which temperatures were lower (25).
Adding to the complexity of the problem, Hendson et al. (10) found (using various genetic approaches) that most isolates from almonds are part of a distinct grouping whose members putatively form two clusters (groups I and II), are distinct from all isolates from grapes and include a minority of isolates from almonds. Thus, the questions about host specificity and whether grape and almond strains have overlapping host ranges are unresolved. We addressed questions raised by previous genetic analyses of X. fastidiosa strains (10) by determining whether various X. fastidiosa isolates from grapes and almonds in California could cause disease and attain similar population levels in both hosts. We also compared the abilities of grape and almond strains to overwinter in almonds and grapes.
(This research was conducted by Rodrigo P. P. Almeida in partial fulfillment of the requirements for a Ph.D. from the University of California, Berkeley.)
MATERIALS AND METHODS
Bacteria and plants.
X. fastidiosa isolates were obtained throughout California from grape, almond, and oleander (Table 1). Isolates were triply cloned and stored at −70°C. Genetic differences among some of the isolates have been previously described (10). Cells were grown and maintained on periwinkle wilt-modified (PWG) solid medium (13) unless specified otherwise. Grapevine (Vitis vinifera) seedlings of Cabernet sauvignon and Non-pareil almond (Prunus amygdalus) grafted on peach (Lovell variety) rootstock were used for pathogenicity tests. Plants were kept in a heated (∼25°C ± 1°C) greenhouse throughout the experiments unless noted otherwise.
TABLE 1.
Isolates of X. fastidiosa used in this study and their growth on PD3 solid medium; pathogenicity, bacterial populations, and overwinter survival in both host plants
| Growth in: | Host of origin/strain groupa | County of origin | Isolate on PD3b | No. of plants withc:
|
Log CFU/g (± SE) of indicated strain
|
Overwinter survival ofd:
|
|||
|---|---|---|---|---|---|---|---|---|---|
| PD | ALS | Grapes | Almonds | Grapes | Almonds | ||||
| ALS4 | Almond/A1 | San Joaquin | − | 0 | 5 | 5.2 | 7.2 ± 0.1 | 0/1 | 4/5 |
| ALS7 | Almond/A1 | San Joaquin | − | 0 | 5 | 4.5 | 7.4 ± 0.1 | 0/1 | 5/5 |
| Butte | Almond/A1 | Butte | − | 0 | 5 | 4.8 ± 0.2 | 7.5 ± 0.1 | 0/3 | 0/5 |
| Dixon | Almond/A1 | Solano | − | 0 | 5 | 3.8 ± 0.1 | 7.3 ± 0.1 | 1/2 | 5/5 |
| Glenn | Almond/A1 | Glenn | − | 0 | 5 | 5.6 | 7.5 ± 0.1 | 1/1 | 0/5 |
| Manteca | Almond/A1 | San Joaquin | − | 0 | 5 | 3.9 | 7.7 ± 0.1 | 0/1 | 4/5 |
| ALS6 | Almond/A2 | San Joaquin | + | 0 | 4 | 5.8 ± 0.2 | 7.3 ± 0.1 | 0/4 | 1/4 |
| Contra Costa | Almond/A2 | Contra Costa | + | 1 | 5 | 7.3 ± 0.2 | 6.8 ± 0.1 | 0/5 | 3/5 |
| Fresno-ALS | Almond/G | Fresno | + | 5 | 1 | 7.8 ± 0.2 | 7.5 ± 0.1 | 4/5 | 0/3 |
| Stanislaus | Almond/G | Stanislaus | + | 5 | 4 | 8.2 ± 0.1 | 6.4 ± 0.3 | 4/5 | 1/4 |
| Tulare | Almond/G | Tulare | + | 5 | 5 | 8.1 ± 0.1 | 7.6 ± 0.1 | 4/5 | 0/5 |
| Baja | Grape/G | Mexico | + | 4 | 4 | 7.9 ± 0.1 | 7.3 ± 0.1 | 4/5 | 2/5 |
| Bakersfield | Grape/G | Kern | + | 4 | 5 | 8.5 ± 0.1 | 7.5 ± 0.1 | 5/5 | 1/5 |
| Buena Vista | Grape/G | Kern | + | 5 | 3 | 7.1 ± 0.3 | 7.5 ± 0.1 | 4/5 | 1/3 |
| Conn Creek | Grape/G | Napa | + | 5 | 5 | 8.1 ± 0.1 | 7.3 ± 0.1 | 5/5 | 0/5 |
| Medeiros | Grape/G | Fresno | + | 5 | 5 | 8.4 ± 0.1 | 7.4 ± 0.1 | 5/5 | 0/5 |
| Pavich | Grape/G | Kern | + | 5 | 5 | 8.1 ± 0.1 | 7.4 ± 0.1 | 4/5 | 3/5 |
| STL | Grape/G | Napa | + | 5 | 5 | 8.3 ± 0.1 | 7.3 ± 0.1 | 5/5 | 0/5 |
| Temecula | Grape/G | Riverside | + | 5 | 5 | 8.4 ± 0.1 | 7.4 ± 0.1 | 5/5 | 0/5 |
| Traver | Grape/G | Tulare | + | 5 | 5 | 8.1 ± 0.1 | 7.5 ± 0.1 | 5/5 | 0/5 |
| UCLA | Grape/G | Los Angeles | + | 4 | 4 | 7.6 ± 0.1 | 7.4 ± 0.1 | 3/4 | 1/4 |
| Ann1 | Oleander/O | Riverside | − | 0 | 0 | None | 3.8 ± 0.1 | 0/0 | 0/3 |
A1, almond group I; A2, almond group II; G, grape; O, oleander (sensu reference 10 and this work).
+, single colonies present on PD3 solid medium; −, no growth.
Number exhibiting symptoms out of a total of 5 plants inoculated.
Number of plants from which X. fastidiosa was isolated at the end of the growing season after overwintering /number at 4 months after inoculation. The difference between the two numbers is the number that recovered after overwintering.
Growth on solid medium.
Cells were streaked on PWG and PD3 (7) solid media; both media were modified by the addition of 20 mg of phenol red per liter as a pH indicator. We maintained plates up to 4 weeks in an incubator at 28°C and observed growth of individual colonies with a 30× microscope. We replicated each test three times for each isolate on each medium. We assigned positive growth for plates in which single colonies were observed within 4 weeks after streaking.
Cross-inoculation.
X. fastidiosa suspensions were prepared by suspending isolates in 1 ml of succinate-citrate buffer (18) after growth on PWG solid medium for 1 to 2 weeks, depending on the growth rate of each isolate. Suspensions were turbid, with estimates of cell concentrations of 108 to 109 CFU/ml. A 5-μl drop of the suspension was placed on a young stem of the test plant, and the tissue was pricked through the drop 5 times with a no. 0 entomological pin (14). Two sites per plant were inoculated; each isolate was inoculated into five almond and five grape plants. Mock inoculations were done with buffer only. After an incubation period of 2 to 4 months in the greenhouse, one symptomatic leaf from each plant with PD or ALS symptoms was collected and processed for X. fastidiosa identification and bacterial quantification by dilution plating (13). In cases in which plants had no symptoms, a mature leaf near one of the inoculation sites was collected. Samples testing negative by culture assays were tested twice. All plants were sampled up to 4 months after inoculation. After this period, plants which were nonsymptomatic and from which X. fastidiosa was not grown were considered not infected. We determined that a plant was symptomatic when it had two or more leaves with typical PD or ALS symptoms. After the 4-month period, all test plants were moved to an insect-proof screened cage kept outside the greenhouse and were watered as necessary (November 2002). After leaf abscission the plants were pruned and were taken in January 2003 to a cold box (4°C). In February 2003, plants were brought back into the greenhouse and kept for up to 6 months for symptom development. X. fastidiosa was again isolated from these plants by the culture method.
RAPD.
Random amplified polymorphic DNA (RAPD) assays were performed following the protocols used by Hendson et al. (10) with minor modifications. Because over half of the isolates used here have already been typed with various techniques (10), we tried to determine how the nontyped isolates would be grouped in relation to the typed isolates. Therefore, we only used 4 RAPD primers (OPAA-01, OPAA-02, OPAA-03, and OPAA-04; Operon Technologies, Inc., Alameda, Calif.) of the 10 previously determined to allow differentiation of X. fastidiosa strains (10). All isolates were tested at least twice with each primer. We did not score faint bands, and profiles were scored in a binary format. The FreeTree program (9) was used for genetic analysis, and TreeView (http://taxonomy.zoology.gla.ac.uk/rod/treeview.html) was used for construction of the diagram. We used the Jaccard similarity coefficient (32) to compare pairs of strains and the neighbor-joining tree-construction method to build the phenogram, with 250 bootstrap replicates to estimate branch support. Isolate Ann1 from oleander was used as an outgroup. All isolates were identified (using the specific RST31-33 primer set) (22) as X. fastidiosa; all isolates generated the expected ∼730-bp fragment (data not shown).
Overwinter survival of X. fastidiosa.
Experiments were conducted in the Armstrong orchard of the University of California at Davis and at the University of California Kearney Agricultural Center (Parlier, Calif.). The nine isolates used for this experiment are listed in Fig. 1. Cells were grown for 1 to 2 weeks on PWG solid medium before mechanical inoculation (14) and suspended on phosphate-buffered saline to slight turbidity (108 to 109 CFU/ml) immediately before inoculations were made. Suspensions were needle inoculated into new shoots of mature almonds (Non-pareil variety) in April 1997 by pin pricking through a 5-μl drop of the cell suspension. A total of 8 trees were inoculated in multiple branches at Davis, and 15 trees were inoculated at Parlier; 20 to 25 stems were inoculated per isolate in each location. Samples of inoculated plants were collected in the fall months of 1997, 1998, and 1999 in Davis but only in 1997 and 1998 in Parlier (the orchard was eliminated in 1999), and bacteria were quantified by dilution plating (13). Positive samples were tested only once a year, but stems with negative results were tested at least twice and usually three times every year. Negative controls were inoculated with phosphate-buffered saline only (n = 20). No mock inoculations caused ALS, and no X. fastidiosa was recovered at any period from these branches. Efficiency of mechanical inoculation was analyzed by the chi-square test using the Yates correction for continuity (1).
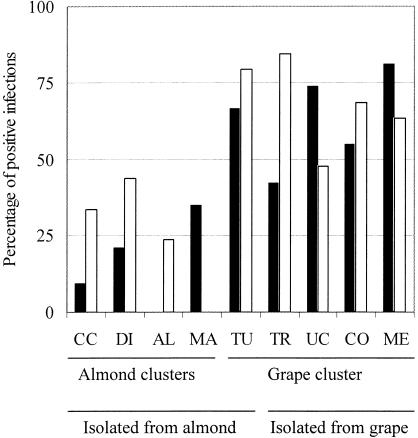
FIG. 1.
Rates of infection by needle inoculation for various X. fastidiosa isolates in almonds at Davis (solid bars) and Parlier (empty bars) in California. Percentages represent the proportion of leaves from which X. fastidiosa was recovered (August to October) after needle inoculation on stems earlier (April) in the same year. The almond strains used were Contra Costa (CC), Dixon (DI), ALS4 (AL), and Manteca (MA); the grape strains used were Tulare (TU), Traver (TR), UCLA (UC), Conn (CO), and Medeiros (ME). The strain grouping for the almond clusters follows the results shown in Fig. 2.
RESULTS
Growth on solid media.
All strains grew on PWG solid medium. Single colonies for grape strains could be seen within 1 week, but in general almond strains required at least 2 weeks for colonies to be seen. All strains from grapes grew on PD3 medium (Table 1). Almond strains from group II (isolates ALS6 and Contra Costa) grew on PD3 medium, but those from group I did not. Isolate Ann1 from oleanders also failed to grow on PD3 medium. Because we used phenol red as a pH indicator in both PWG solid medium and PD3 medium, we observed that PWG solid medium appeared to have a higher buffering capacity than PD3 medium; PWG solid medium rarely changed color, and this only happened a few weeks after inoculation. In contrast, PD3 medium turned pink within a few days in the areas of the medium in which X. fastidiosa was growing in high numbers. Colonies grew slightly slower on PD3 medium than on PWG solid medium, although we did not quantify that. Occasionally we observed scant growth of strains from almonds from group I on PD3 medium but only where cells were streaked in high concentrations, as indicated by change in color in medium.
Virulence in grape and almond.
Because we used the same bacterial suspension to inoculate grape and almond plants with each isolate, we assumed that similar numbers of cells were introduced into each plant. Thus, the differences in the frequency of infection by the same isolate on different plants probably reflect host specificity rather than variability in inoculation procedures (Table 1). Isolates from grapes infected all grapevines inoculated; only 3 (1 plant without symptoms and 2 plants with only initial foliar symptoms of reddening on leaf edges) of 50 plants did not show typical PD symptoms. As determined on the basis of pathogenicity results in grapes and growth on PD3 medium, three isolates (Fresno-ALS, Stanislaus, and Tulare) from almonds mimicked strains from grapes. Interestingly, Fresno-ALS caused little disease in almonds (only one plant showed typical ALS symptoms); in fact, this isolate was the only one (besides the almond-avirulent Ann1 isolate from oleanders) (29) with low virulence in almonds.
With the exception of isolates Fresno-ALS, Stanislaus, and Tulare, X. fastidiosa isolated from almonds did not cause PD (Fig. 1). Some of the isolates caused mild foliar symptoms (usually in only one of the five inoculated grapes). The Contra Costa isolate from almond caused mild PD symptoms. Because we kept the plants in a greenhouse with natural light, the experiment had to be completed within 4 months after inoculation due to a shortening photoperiod, which reduces plant growth and slows symptom development. All almond strains caused ALS, but isolates ALS4 and Contra Costa were notably more virulent to almond than any other tested almond or grape strains, causing advanced symptoms of marginal leaf necrosis 2 months after inoculation, at which time leaf scorch caused by all other strains were barely visible. We also observed that symptom development varied for both grape and almond strains in both host plants. For example, isolates UCLA and Baja consistently took 1 month longer to develop PD symptoms than other isolates from grapes.
Bacterial populations within grapes and almonds.
The numbers of live X. fastidiosa cells that we recovered from grapes were approximately 10- to 100-fold higher for the strains from grapes than for strains from almonds (Table 1). Similar population levels occurred in both hosts for isolates Fresno-ALS and Contra Costa, but Fresno-ALS caused PD in similarity to other strains from grapes, whereas Contra Costa was the sole strain from almonds that consistently caused mild PD symptoms of reddening and occasional necrosis limited to leaf margins under greenhouse conditions. We recovered X. fastidiosa from at least one of the five grapevines inoculated with each of the various almond strains but not from those inoculated with the oleander strain. From almonds, we recovered all strains with approximately identical population sizes and frequencies. The exception was the oleander strain, which we recovered only in low numbers (103 to 104 CFU/g of tissue) and which never caused ALS symptoms in three of the five isolate Ann1-inoculated almond plants from leaves that had grown after inoculation, indicating that systemic movement of the bacterium had occurred. Grape strains overwintered more successfully in grapes (57 of 65 plants) than in almonds (9 of 59 plants by chi-square test [P < 0.001; 1 df]), as did almond strains within almonds (22 of 39) compared to almond strains in grapes (2 of 18 plants by chi-square test [P < 0.001; 1 df]).
RAPD and enzyme restriction.
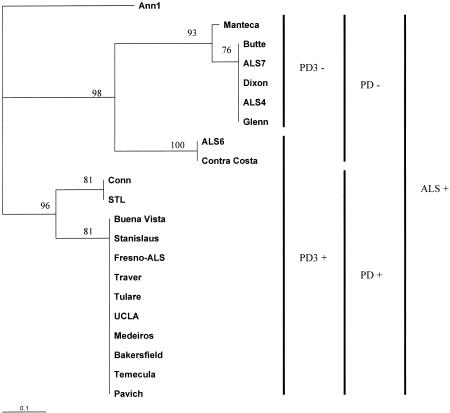
We confirmed the trends observed by Hendson et al. (10): all isolates from grapes were grouped together genetically, but most almond isolates were distinctive (Fig. 2). Three isolates from almonds, Tulare, Stanislaus, and Fresno-ALS, were grouped with grape strains and behaved on media and within plants similarly to grape strains. Even though we used only four RAPD primers, the two coastal California isolates from grapes (Conn Creek and STL) were genetically distinct from Central Valley and southern California isolates from grapes. The two almond groups identified previously (10) were also separated in our analysis, with the exception of the Manteca isolate, which behaved as a strain from almonds in biological assays and in our RAPD assay but was previously grouped with strains from grapes; therefore, we believe that the same isolate was not used in both studies. When the RST31-33 PCR-amplified product was digested with RsaI, all almond strains generated two fragments (∼550 and ∼200 bp) and no grape strain was cut (data not shown), as previously reported (10).
FIG. 2.
Tree determined on the basis of distance values obtained by RAPD-PCR analysis using the Jaccard similarity coefficient. Bootstrap values (percent of 250 repetitions) are shown for each branch point. Vertical lines and groupings indicate whether associated strains in the diagram (i) grew on PD3 solid medium (PD3+) or not (PD3−), (ii) caused PD (PD+) or not (PD−), or (iii) caused ALS (ALS+).
Overwinter survival of X. fastidiosa in orchards.
Needle inoculation of eight X. fastidiosa isolates in mature almonds in the field resulted in successful infection, with no differences observed between the Davis and Parlier locations for groupings of grape or almond strains (chi-square test [P > 0.05 for both combinations; 1 df]). The rate of infection that we observed in both locations was significantly lower for isolates within the almond groupings (I and II) than for those in the grape group (Fig. 1). Chi-square test result values representing the proportion of successfully inoculated (infected) plants in the same year of inoculation (for both locations) were significantly higher (P < 0.001 for both combinations; 1 df) for grape strains than for almond strains. X. fastidiosa was recovered from inoculations made in 1997, with strains from the grape group infecting 64 and 75% of branches in the same year of inoculation (1997) for the Davis and Parlier sites, respectively. We recovered almond stains from 21 and 33% of inoculated branches from the respective areas during the year of inoculation.
At Parlier, a total of 22 of 23 infections for all almond group strains combined survived the 1997-to-1998 winter. A total of 45 of 51 plants infected with strains from the grape cluster survived the winter. For the Davis location we had data for 1997, 1998, and 1999; therefore, overwinter survival was estimated according to the levels of X. fastidiosa being recovered from inoculated branches in 1998 or 1999. We also used only those branches that had tested positive in 1997 and that we had evaluated at least once again in one of the successive years. At Davis, 5 of 6 infections with X. fastidiosa almond strains survived and 24 of 57 infections by grape cluster strains tested positive after one or two winters (P > 0.05 [comparing survival of pooled grape and almond strains and using chi-square testing with a Yates correction]). We did not observe differences of symptom severity among strains or localities, although symptoms (consisting of the presence of 2 to 7 leaves with marginal leaf scorch) were mild during the first year (1997). Such first-year symptoms were so light that it is likely that few of these would have been noticed in orchard surveys. Symptomatic leaves tended to more readily fall from the stem, increasing the difficulty of seeing disease symptoms.
DISCUSSION
X. fastidiosa isolates from grapes and almonds have been considered to be biologically indistinguishable (see, e.g., references 6 and 24), but recent genetic data (10) showed that there are at least two and probably three distinctive genetic groupings among these isolates. We found that isolates of almond group I did not grow on PD3 medium, whereas almond group II and all isolates from grapes (10) grew on PD3 medium. Grape strains caused both PD and ALS. All almond strains caused ALS and multiplied within grapes but did not cause typical PD symptoms. As determined on the basis of our results, X. fastidiosa grape and almond strains are apparently biologically distinct, with characteristics associated with genetic groupings of the strains (10).
We examined the overwintering survival rates of X. fastidiosa in almonds to determine whether grape strains survived winters more poorly than almond strains. In a previous study that examined only grape strains inoculated into almond trees in two southern Central Valley locations, overwintering survival was very poor (4.2%) (6). The hypothesis that strains from almonds survived winters better than grape strains might explain the lack of ALS in orchards adjacent to PD hotspots in the southern Central Valley. Our data from field-grown trees did not support this hypothesis, but our data on overwintering potted plants did. Both strain groups survived winter dormancy for 2 years at about the same rates in central California. Differences in winter severity may have contributed to the higher rates of survival of X. fastidiosa recorded in our study compared to those determined in the earlier study (6). In addition, our use of mechanical inoculation may have resulted in faster or wider spread of X. fastidiosa within plants than that caused by vector inoculation. It was surprising to find that some infections of almonds that survived the first winter (1997 to 1998) failed to survive the second winter. Our laboratory experiments showed results for overwintering survival of strains in grapes and almonds which were especially surprising because of the milder temperatures at Berkeley compared to those in the Central Valley. Future research with more field sites distributed throughout California is needed to further clarify this epidemiological aspect of both strains.
The limitations in cultivating X. fastidiosa in vitro were only overcome in 1978 (5), and in the following years other media were developed to grow this bacterium. In general, little difference has been found among different media in the ability to support growth of X. fastidiosa from grapes or almonds (3, 4) but the number of isolates and media combinations tested for media specificity has been limited. We believe that Davis et al. (6) did not test isolates from almonds avirulent to grapes because, at that time, only PD3-like media were available (Davis et al. used PD2 medium for their work). PW (the base medium for PWG solid medium) was developed a few years later for the growth of more fastidious strains of X. fastidiosa, such as those found in peaches (4). Therefore, although the conclusions of the studies are different, our findings corroborate their results.
Hopkins (16) first quantified X. fastidiosa populations within plants, associating bacterial numbers with isolate virulence. The technique was later used to determine differences in resistance and susceptibility to PD among grape varieties (8). X. fastidiosa populations within plants were also correlated with vector acquisition efficiency (15) and used to identify natural reservoirs of this bacterium (14, 27). Recently, independent results of quantification of X. fastidiosa populations from citrus by culture (2) and quantitative PCR (23) techniques suggest that the number of live cells recovered from culture assays is very similar to the total number of cells estimated by PCR. We found that for grape strains pathogenic to both grapes and almonds, the number of live X. fastidiosa cells within host plants is dependent on the host and not the strain type.
The reason for the lack of virulence of almond strains in grapes has not been addressed, but we suggest that limited colonization (103 to 105 CFU/g) and movement precludes the virulence of almond strains in grapes. Because X. fastidiosa multiplies and causes symptoms significantly faster in the greenhouse than in the field (A. H. Purcell, personal observations), we expect that the almond strains from group II, which caused mild PD symptoms in our experiment, would be virtually avirulent to grapes under field conditions. The recently published complete genome sequence for a grape strain (Temecula) (33) included in our study and a draft sequence for the Dixon isolate (http://www.jgi.doe.gov/JGI_microbial/html/xylella_almond/xyle_almnd_homepage.html) in group I of the almond strains should facilitate searches for the genetic basis of resemblances between these strains that cause differentiated pathology in grapes.
Our results suggest that no PD is found near ALS orchards because ALS strains likely do not cause PD under field conditions. In addition, all ALS isolates that we and Hendson et al. (10) typed were obtained from northern counties in the Central Valley (areas with little evidence of PD). All isolates from almonds identified as grape strains came from southern areas of central California. Because grape strains cause ALS, we hypothesize that the rare cases of infected almond plants in these areas are due to accidental infections by infective vectors. This still leaves unanswered the issue of why ALS has been rare or not detected at all in almond orchards adjacent to vineyards with a high incidence of PD. The principal insect vectors for PD can be found only rarely on grapes (11, 26), as appears to be the case for these insects on almonds (24; A. H. Purcell, unpublished data). It is possible that the seasonality determining when the insects are most likely to feed on almonds differs substantially from that determining when the insects are most likely to feed on grapes, which could lead to lower rates of disease development in almonds. Since RsaI differentially digests the PCR product of the X. fastidiosa primer set RST31-33, which is widely used for detection of X. fastidiosa in plants, it may provide an easy way to type isolates collected from vineyards or almond orchards for their pathogenicity to almonds or grapes. The future impact of the invasive vector Homalodisca coagulata on the spread of these different X. fastidiosa types is unknown, but it may modify the epidemiology of both diseases, because it feeds on grapes and almonds (28).
Despite the biological differences we found between almond strains and those from grapes, we suggest that the pathovar classification approach is not useful for studies involving X. fastidiosa. For example, citrus and coffee strains from Brazil are genetically distant from grape strains and are more distant and nonsympatric than almond strains but cause PD in grapes (20, 21). The relevance of the number of isolates tested, the location of collection, the isolation media, the host specificity, and other unknown factors might be larger for X. fastidiosa diversity studies than previously expected on the basis of comparisons with other plant pathogenic bacteria.
Acknowledgments
We thank Stuart Saunders for technical assistance with field experiments. We acknowledge assistance from Tina Wistrom, Edward Norberg, Meg Tung, Renee Mann, Dawn Chiniquy, and Karyn Newman. We thank members of our lab and Steven Lindow for helpful suggestions on the manuscript. Bruce C. Kirkpatrick, University of California, Davis, and Beth L. Teviotdale, University of California's Kearney Agricultural Center, provided access to experimental almond orchards.
This work was supported by the Almond Board of California and USDA/CSREES Viticultural Consortium grant 2001-34360-10328. R.P.P.A. had a scholarship from CNPq-Brazil.
REFERENCES
- 1.Agresti, A., and B. Finlay. 1997. Statistical methods for the social sciences. Prentice-Hall Inc., Upper Saddle River, N.J.
- 2.Almeida, R. P. P., E. F. Pereira, A. H. Purcell, and J. R. S. Lopes. 2001. Multiplication and movement of a citrus strain of Xylella fastidiosa within sweet orange. Plant Dis. 85:382-386. [DOI] [PubMed] [Google Scholar]
- 3.Chang, C. J., and J. T. Walker. 1988. Bacterial leaf scorch of northern red oak: isolation, cultivation, and pathogenicity of a xylem-limited bacterium. Plant Dis. 72:730-733. [Google Scholar]
- 4.Davis, M. J., W. J. French, and N. W. Schaad. 1981. Axenic culture of the bacteria associated with phony disease of peach and plum leaf scald. Curr. Microbiol. 6:309-314. [Google Scholar]
- 5.Davis, M. J., A. H. Purcell, and S. V. Thomson. 1978. Pierce's disease of grapevines: isolation of the causal bacterium. Science 199:75-77. [DOI] [PubMed] [Google Scholar]
- 6.Davis, M. J., S. V. Thomson, and A. H. Purcell. 1980. Etiological role of a xylem-limited bacterium causing Pierce's disease in almond leaf scorch. Phytopathology 70:472-475. [Google Scholar]
- 7.Davis, M. J., R. F. Whitcomb, and A. G. Gillaspie, Jr. 1981. Fastidious bacteria of plant vascular tissue and invertebrates (including so called rickettsia-like bacteria), p. 2172-2188. In M. P. Starr, H. Stolp, H. G. Truper, A. Balows, and H. G. Schlegel (ed.), The prokaryotes: a handbook on habits, isolation, and identification of bacteria. Springer-Verlag, Heidelberg, Germany.
- 8.Fry, S. M., and R. D. Milholland. 1990. Response of resistant, tolerant, and susceptible grapevine tissues to invasion by the Pierce's disease bacterium, Xylella fastidiosa. Phytopathology 80:66-69. [Google Scholar]
- 9.Hampl, V., A. Pavlicek, and J. Flegr. 2001. Construction and bootstrap analysis of DNA fingerprinting-based phylogenetic trees with the freeware program FreeTree: application to trichomonad parasites. Int. J. Syst. E vol. Microbiol. 51:731-735. [DOI] [PubMed] [Google Scholar]
- 10.Hendson, M., A. H. Purcell, D. Chen, C. Smart, M. Guilhabert, and B. Kirkpatrick. 2001. Genetic diversity of Pierce's disease strains and other pathotypes of Xylella fastidiosa. Appl. Environ. Microbiol. 67:895-903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hewitt, W. B., N. W. Frazier, J. H. Freitag, and A. J. Winkler. 1949. Pierce's disease investigations. Hilgardia 19:207-264. [Google Scholar]
- 12.Hewitt, W. B., B. R. Houston, N. W. Frazier, and J. H. Freitag. 1946. Leafhopper transmission of the virus causing Pierce's disease of grape and dwarf of alfalfa. Phytopathology 36:117-128. [PubMed] [Google Scholar]
- 13.Hill, B. L., and A. H. Purcell. 1995. Acquisition and retention of Xylella fastidiosa by an efficient vector, Graphocephala atropunctata. Phytopathology 85:209-212. [Google Scholar]
- 14.Hill, B. L., and A. H. Purcell. 1995. Multiplication and movement of Xylella fastidiosa within grapevine and four other plants. Phytopathology 85:1368-1372. [Google Scholar]
- 15.Hill, B. L., and A. H. Purcell. 1997. Populations of Xylella fastidiosa in plants required for transmission by an efficient vector. Phytopathology 87:1197-1201. [DOI] [PubMed] [Google Scholar]
- 16.Hopkins, D. L. 1981. Seasonal concentration of the Pierce's disease bacterium in grapevine stems, petioles, and leaf veins. Phytopathology 71:415-418. [Google Scholar]
- 17.Hopkins, D. L., and A. H. Purcell. 2002. Xylella fastidiosa: cause of Pierce's disease of grapevine and other emergent diseases. Plant Dis. 86:1056-1066. [DOI] [PubMed] [Google Scholar]
- 18.Hopkins, D. L., and C. M. Thompson. 1984. Seasonal concentration of the Pierce's disease bacterium in “Carlos” and “Welder” muscadine grapes compared with “Schuyler” bunch grape. Hortscience 19:419-420. [Google Scholar]
- 19.Houston, B. R., K. Esau, and W. B. Hewitt. 1947. The mode of vector feeding and the tissues involved in the transmission of Pierce's disease virus in grape and alfalfa. Phytopathology 37:247-253. [Google Scholar]
- 20.Li, W. B., C. H. Zhou, W. D. Pria, D. C. Teixeira, V. S. Miranda, E. O. Pereira, A. J. Ayres, and J. S. Hartung. 2002. Citrus and coffee strains of Xylella fastidiosa induce Pierce's disease in grapevine. Plant Dis. 86:1206-1210. [DOI] [PubMed] [Google Scholar]
- 21.Mehta, A., and Y. B. Rosato. 2001. Phylogenetic relationships of Xylella fastidiosa strains from different hosts, based on 16S rDNA and 16S-23S intergenic spacer sequences. Int. J. Syst. E vol. Microbiol. 51:311-318. [DOI] [PubMed] [Google Scholar]
- 22.Minsavage, G. V., C. M. Thompson, D. L. Hopkins, R. M. V. B. C. Leite, and R. E. Stall. 1994. Development of a polymerase chain reaction protocol for detection of Xylella fastidiosa in plant tissue. Phytopathology 84:456-461. [Google Scholar]
- 23.Oliveira, A. C., M. A. Vallim, C. P. Semighini, W. L. Araujo, G. H. Goldman, and M. A. Machado. 2002. Quantification of Xylella fastidiosa from citrus trees by real-time polymerase chain reaction assay. Phytopathology 92:1048-1054. [DOI] [PubMed] [Google Scholar]
- 24.Purcell, A. H. 1980. Almond leaf scorch: leafhopper and spittlebug vectors. J. Econ. Entomol. 73:834-838. [Google Scholar]
- 25.Purcell, A. H. 1980. Environmental therapy for Pierce's disease of grapevines. Plant Dis. 64:388-390. [Google Scholar]
- 26.Purcell, A. H., and N. W. Frazier. 1985. Habitats and dispersal of the principal leafhopper vectors of Pierce's disease bacterium in the San Joaquin Valley. Hilgardia 53:1-32. [Google Scholar]
- 27.Purcell, A. H., and S. R. Saunders. 1999. Fate of Pierce's disease strains of Xylella fastidiosa in common riparian plants in California. Plant Dis. 83:825-830. [DOI] [PubMed] [Google Scholar]
- 28.Purcell, A. H., and S. R. Saunders. 1999. Glassy-winged sharpshooters expected to increase plant disease. California Agriculture. 53:26-27. [Google Scholar]
- 29.Purcell, A. H., S. R. Saunders, M. Hendson, M. E. Grebus, and M. J. Henry. 1999. Causal role of Xylella fastidiosa in oleander leaf scorch disease. Phytopathology 89:53-58. [DOI] [PubMed] [Google Scholar]
- 30.Severin, H. H. P. 1949. Transmission of the virus of Pierce's disease of grapevines by leafhoppers. Hilgardia 19:190-206. [Google Scholar]
- 31.Severin, H. H. P. 1950. Spittle-insect vectors of Pierce's disease virus II. Life history and virus transmission. Hilgardia 19:357-381. [Google Scholar]
- 32.Sneath, P. H. A., and R. R. Sokal. 1973. Numerical taxonomy. W. H. Freeman and Co., San Francisco, Calif.
- 33.Van Sluys, M. A., M. C. de Oliveira, C. B. Monteiro-Vitorello, C. Y. Miyaki, L. R. Furlan, L. E. A. Camargo, A. C. R. da Silva, D. H. Moon, M. A. Takita, E. G. M. Lemos, M. A. Machado, M. I. T. Ferro, F. R. da Silva, M. H. S. Goldman, G. H. Goldman, M. V. F. Lemos, H. El-Dorry, S. M. Tsai, H. Carrer, D. M. Carraro, R. C. de Oliveira, L. R. Nunes, W. J. Siqueira, L. L. Coutinho, E. T. Kimura, E. S. Ferro, R. Harakava, E. E. Kuramae, C. L. Marino, E. Giglioti, I. L. Abreu, L. M. C. Alves, A. M. do Amaral, G. S. Baia, S. R. Blanco, M. S. Brito, F. S. Cannavan, A. V. Celestino, A. F. da Cunha, R. C. Fenille, J. A. Ferro, E. F. Formighieri, L. T. Kishi, S. G. Leoni, A. R. Oliveira, J. Rosa, V. E., F. T. Sassaki, J. A. D. Sena, A. A. de Souza, D. Truffi, F. Tsukumo, Y. G. M., L. G. Zaros, E. L. Civerolo, A. J. G. Simpson, N. F. Almeida, Jr., J. C. Setubal, and J. P. Kitajima. 2003. Comparative analyses of the complete genome sequences of Pierce's disease and citrus variegated chlorosis strains of Xylella fastidiosa. J. Bacteriol. 185:1018-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]