Abstract
The androgen receptor (AR) plays a key role in a regulating gene expression in a variety of tissues, including the prostate. In the latter role it is one of the primary targets in the development of new chemotherapeutics for treatment of prostate cancer, as well as being the target of the most widely prescribed current drug, bicalutamide (Bcu), for this disease. In view of it’s importance, and the absence of a crystal structure for any antagonist-AR complex, we have carried out a series of molecular dynamics based simulations of the AR-Bcu complex and quantum mechanical (QM) calculations of Bcu, to elucidate the structural basis for antagonism of this key target. The structures which emerge show that bicalutamide antagonizes AR by accessing an additional binding pocket (B-site) adjacent to the hormone binding site (HBS), induced by displacing helix 12. This distorts the coactivator binding site and results in the inactivation of transcription. An alternative equienergetic conformational state of bicalutamide was found to bind in an expanded hormone pocket without materially perturbing either helix 12 or the coactivator binding site. Thus both the structural basis of antagonism and the mechanism underlying agonist properties displayed by bicalutamide in different environments may be rationalized in terms of these structures. In addition the antagonist structure and especially the induced second site (B-site) provides a structural framework for the design of novel antiandrogens.
The Androgen Receptor (AR) is a nuclear receptor (NR) (1, 2) which responds to the hormones testosterone (T), and dihydrotestosterone (DHT), by modulating gene expression in a variety of cells including prostate and muscle tissue. The androgen-activated receptor enters the nucleus and anchors the assembly of a large complex of transcription factors at target promoters and enhancers. Some 40 years ago Huggins noted that prostate cancers (PC) require androgen for growth (3). Based on this observation, androgen ablation therapy and/or antiandrogens such as Casodex (bicalutamide) are the primary approaches to treating PC (4). Unfortunately, after 2–3 years of treatment, the cancer evolves to an “androgen independent” or hormone refractory state and current antiandrogens become ineffective (5).
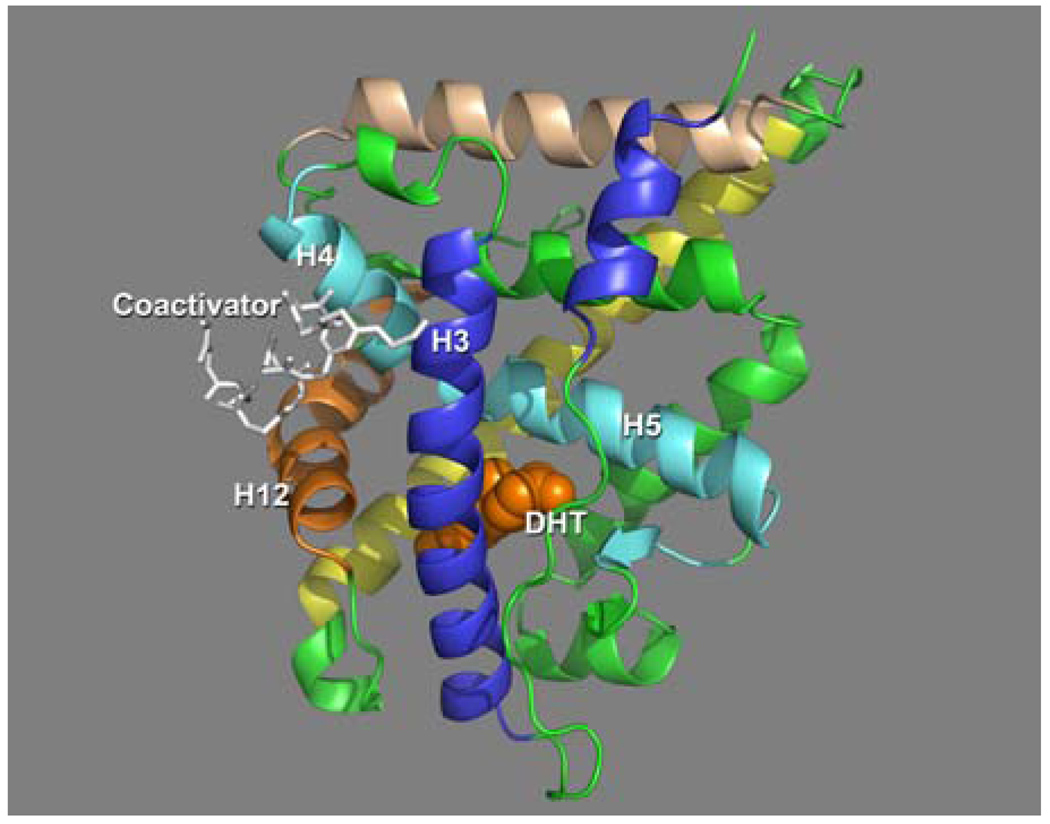
The ligand binding domain (LBD) is the heart of the receptor and has been the focus of AR based drug discovery. Crystal structures of the AR LBD containing numerous bound ligands and coactivator peptides have been solved (6–10). On binding of the hormone to the hormone binding site (HBS) an uncharacterized reorganization of the receptor occurs primarily assumed to be the registration of Helix 12 (H12) to bind coactivator. This reorganization results in the formation of an effective coactivator binding site (AF-2 site), a surface exposed HФ cleft involving helices H3, H4 and H12 (Fig 1).
Figure 1.
The AR LBD with DHT (orange spacefill) and coactivator peptide (white sticks) shown. Binding of DHT at the HBS organizes H12 which, along with H3 and H4 form the AF2 site allowing coactivators to bind (Estebanez-Perpina et al PDB 1T63).
The precise mechanism by which AR antagonists block the transcription of target genes remains elusive. The preponderance of evidence from X-ray structures of several NR-antagonist complexes suggests that the general mechanism involves perturbing Helix 12, displacing it from its hormone bound configuration, and distorting the AF-2 site (1, 2). This in turn interferes with coactivator binding, a requisite for transcription. Although indubitably this is the case in the AR as well, to date native AR in its antagonized state, i.e. a wild type (WT) AR-antagonist complex, has defied crystallization. Thus no structural information exists on the nature of the antagonist induced structural change. Perhaps even more of a mystery is how overexpression of AR or stimulation by the cytokine IL1-β, implicated in the transition to the androgen independent state, cause bicalutamide (Fig 2) and other antagonists to become agonists (11–13).
Figure 2.
Schematic of Bicalutamide (Bcu) and S1 (S1)
In order to understand the molecular interactions underlying AR antagonism, we have exploited in silico simulations including molecular dynamics (MD), Quantum mechanics (QM), and a “slow growth” technique employing free energy perturbation theory (FEP) of the AR-Bcu complex to explore the binding modes accessible to Bcu in the AR-LBD. MD has recently been used in an analogous way to determine the effects of ligand binding to the coactivator affinity in the nuclear receptor LRH-1 (14). Similarly Zhou et al have carried out computational studies of hydroxyflutamide binding to wild type and the mutant T877A AR (15), and these studies suggest that the transition from antagonist to an agonist in the latter system corresponds to H12 adopting an agonist conformation, consistent with the importance of conformational changes in H12 to the mechanism of antagonism in wild type AR. Other MD studies of AR have looked at the effect of pathogenic mutations to AR that lead to Androgen Insensitivity Syndrome (16), where mutations in the receptor were shown computationally to cause changes in the ligand binding site which lead to an inactive AR (17, 18). We seek to understand the structural basis for the pharmacological activity of this drug and, if possible, to gain insight into the molecular mechanisms underlying conversion of antagonists into agonists. In addition, although the use of structure-activity relationships (SAR) have proven very successful in developing new clinical candidates of antiandrogens (19–23) not all patients respond to these 2nd generation antagonists and resistance still emerges (20) and thus 3rd generation antiandrogens are required. The elucidation of the antagonist bound structure should provide a better basis for us and others to exploit structure-based design in the development of more potent and effective therapeutics – especially compounds that overcome “Androgen Independent” or refractory PC (5).
MATERIALS AND METHODS
The first step in the MD simulations involves building the system. The 1T63 structure of AR bound to DHT and a coactivator peptide, Figure 1, from the Fletterick lab was used as the basis for all simulations (9). Missing atoms and hydrogens were built in. Two different initial structures of the Bcu – AR complex were constructed, exploiting the AR W741L- Bcu and AR-S1 complexes as a basis for initial ligand placement. A third, independent trajectory was carried out by the FEP-Slow Growth method, also described below. For comparison and as a test of the simulation method the experimentally determined AR complexes with DHT and the mutant W741L AR with Bcu were also simulated.
Initial Structure of WT AR-Bcu complexes
As noted above no structure of Bcu bound to the wild-type receptor has been solved. However a mutant AR, W741L, arose in the LNCaP prostate cancer cell line on androgen depletion and treatment with bicalutamide. At first these cells did not grow, but after 6–13 weeks Bcu stimulated cell growth (rather than antagonizing it). Transactivation assays confirmed that mutation of W741 to Leu converts Bcu into an agonist (24), and this complex has been crystallized and solved (25). Although this AR mutation, in complex with Bcu, results in an agonist conformation of the receptor (presumably the reason it could be crystallized), we exploited the Bcu configuration in this complex by superposing it onto the 1T63 structure from the Fletterick lab (9) and used the resulting Bcu coordinates in the HBS (hormone binding site) of 1T63. Not surprisingly Bcu clashes sterically with W741 in this construct. To address this both the B-ring of Bcu (Fig 2) and W741 were shifted slightly to relieve the overlap prior to equilibration.
In order to overcome bias or artifacts introduced by a single set of initial conditions and a single run we also simulated a system starting from Bcu in the structure of AR with agonist S1 (26). S1 differs from Bcu only in the replacement of the sulfonyl group by an ether linkage and an NO2 replacing the cyano group in the para position of the A-ring (Fig 2), yet it is an agonist and stabilizes the agonist conformation of AR (26). To build this construct, key side chains in the 1T63 HBS, especially W741, M745, and M895 were positioned in their configurations in the S1-AR structure and Bcu was inserted into the resulting S1 configuration.
Force Field, Ligand Charges and Torsional Barriers
Simulations were carried out with the OPLS all-atom potential for AR, as implemented in GROMACS (27). Although potentials for proteins have been well characterized for some years now (28, 29), this is not the case for arbitrary heterocyclic ligands such as Bcu. Fortunately OPLS had bond stretching and angle bending parameters for the functional groups found in DHT and S1. QM calculations of the more critical rotational barriers of each ligand were carried out and torsion parameters generated to fit these barriers were used. Partial atomic charges on the ligand were determined by QM. These calculations were performed using DFT at the 6–31G** level with the Jaguar package (Schrodinger).
System Setup and Equilibration
The AR-ligand complexes were solvated with ~10,000 water molecules in a dodecahedral box extending 6Å around the receptor, using genbox (Gromacs). In addition five chloride counterions were added to neutralize the system (using Genion). The solvated system was then minimized, with heavy atoms restrained to reorient hydrogen atoms. Following minimization we carried out 10ps of MD, at 0.1°K followed by 10ps at 306°K with restraints of 1000 kJ /nm2 on the protein heavy atoms to allow the solvent structure to accommodate to the protein complex. Further equilibration of the system for 200 ps at 306°K, using weaker restraints (10 kJ nm−1) of the protein heavy atoms to their crystallographic positions, allowed distorted geometry and steric clashes in less well-determined regions of the protein to be relaxed.
MD, and FEP simulations
Following equilibration we used a variety of MD based simulations to explore the configurational landscape available to Bcu and the residues comprising the HBS. MD simulations were performed in an NVT ensemble with a cutoff of 12.0Å for coulombic interactions and 9.0Å for Van der Waals interactions. The pragmatic approach to the intractability of carrying out simulations with infinite cutoffs is problematic and has led to differing approaches and recommendations. Cheatham et. al. (30) recommend the use of particle mesh Ewald (PME) as superior to a cutoff. However, Norberg, J., and Nilsson, L. point out these conclusions were drawn for a 9 Å cutoff and have shown results similar to PME (31) are obtained with conditions used here without the introduction of the artifactual infinite lattice coulombic interactions of PME in a system designed to emulate solution conditions. The studies of ligand binding to AR by Bisson et. al. (32) were performed using similar conditions to ours but with the somewhat smaller cutoff of 9 Å. Nevertheless this remains an open issue and is one of the reasons we have used multiple simulations and techniques. A timestep of 2fs was employed with a Nose-Hoover thermostat with a coupling parameter of 0.1 (27). After equilibration, the simulations were continued out to 5.3ns.
Slow Growth – Free Energy Perturbation
Although FEP theory has been used for calculation of the free energy of ligand binding, for this purpose it is only valid for small perturbations of ligand structure (e.g. a methyl group mutated to a chloride) (33, 34). Instead we have exploited it here to reduce the bias in ligand placement. In this protocol the ligand is “grown slowly” in situ (by gradually turning on the nonbond potential). Our objective, rather that determining binding free energy, is to allow time for the protein structure to adapt to the new ligand gently rather than creating a bias by “forcing” the insertion of the ligand into the 1T63 structure. This is accomplished according to the equation
| (1) |
The FEP stage was performed with a timestep of 1fs and the nonbonded interactions between Bcu and AR were turned on continuously over a 1ns interval by scaling lambda linearly, from 0.0 to 1.0, while the temperature was linearly annealed from 0.1 K to 306 K for the first 0.5 ns, then remaining constant at 306 K for the final 0.5ns of the FEP stage. Following this the simulation was continued another 5ns.
RESULTS AND DISCUSSION
Trajectories
In order to explore the structural details of the binding modes of bicalutamide in the AR-LBD we carried out a variety of MD based simulations of the hydrated AR-Bcu complexes described in Methods. For comparison we also simulated the AR complex with its endogenous ligand, Dihydrotestosterone (DHT). The latter along with a simulation of the experimentally determined AR W741L-Bcu construct in which Bcu is an agonist, not only provide a basis for understanding the perturbations induced by Bcu in the wild type receptor, but also, by comparing with the experimental structures, provide a benchmark for assessing the validity of the simulations and the significance of the antagonist induced structural changes. The systems simulated, along with the nomenclature used to describe them, are given in Table 1.
Table 1.
Simulation Trajectories of AR-Ligand Complexes
| Name | System | Init. Ligand Orientation | Runs |
|---|---|---|---|
| Experimental Structures | |||
| sDHT | AR-DHT | AR-DHT | 3 |
| sBcuExp | AR W741L-Bcu | ARW741L-Bcu | 3 |
| Antagonist Complexes | |||
| sBcu | AR-Bcu | ARW741L-Bcu | 3 |
| sBcuS1 | AR-Bcu | AR-S1 | 3 |
| sBcuFEP | AR-Bcu | ARW741L-Bcu | 3 |
The overall evolution of the AR structure in different complexes as well as an overall comparison with the experimentally determined structures may be obtained by plotting the RMS deviation from the initial state for each complex. These trajectories are given in Fig 3. As can be seen the averaged trajectories of both experimentally determined complexes, WT AR-DHT (sDHT: green) and W741L AR-Bcu (sBcuExp: red) plateau at about the same RMS deviations of ~1Å, showing good agreement with the experimental structures by this measure and as shown by the superpositions of the structures in Fig 4. On the other hand the two trajectories of the WT AR complexes with Bcu, starting from the configurations of Bcu in the W741L and S1 complexes, both reflect significant distortions, exhibiting deviations from the agonist structures of ~1.7Å (cyan and magenta ligands respectively). Of note is the behavior of the free energy perturbation simulations of the antagonist complex (sBcuFEP, yellow curve in Fig. 3). Although initially the structure diverges dramatically from the agonist structure it returns after a ns. to follow the antagonist trajectories until 4ns at which time the deviation decreases and approaches that of the agonist structures.
Figure 3.
Plot of RMS deviations of C-alpha atoms in helical scaffold of AR versus time for simulations of experimental and antagonist structures. The experimental agonist complexes are given in green (sDHT) and in red (sBcuExp). The Trajectories for the antagonist structures are given in cyan (sBcu): the “slow growth” (FEP) simulation (sBcuFEP) in yellow, and the structure in which the sidechain conformations of key LBS residues were taken from the S1-AR (sBcuS1) in magenta. Each curve is an average of 3 separate simulations with different initial velocities. The error bars indicate the standard deviation at that time of the 3 simulations.
Figure 4.
Superposition of the experimental (colored as in figure 1) and simulated structures (colored magenta) of the AR-DHT (a) and W741L AR-Bcu (b) complexes. The helix scaffold Calpha atom RMSD of these structures is a little over 1Å and as can be seen from these figures, the overall AR structure including the helical scaffold is accurately recapitulated at this level. The ligands, DHT in (a) and Bcu in (b), are shown in sticks (the experimental ligand structure is colored orange and the simulated configuration cyan).
A closer look at the three individual trajectories comprising the average of the FEP trajectories (not shown) reveals an intriguing behaviour. Unlike the other trajectories the individual FEP trajectories behave very differently. One of the trajectories essentially follows the path of the agonist trajectories sDHT and sBcuExp, following a plateau a little over 1Å RMS. The other two initially display large divergence from the initial structure (seen as the peak in sBcuFEP at ~1ns in Fig. 3) and then after roughly 4ns begin to converge back toward the agonist structure. The behavior of the trajectory following the agonist structures is unexpected and we will return to the underlying structure of the complex below.
Mechanism of AR Antagonism by Bicalutamide. Structure of an AR Antagonist Complex
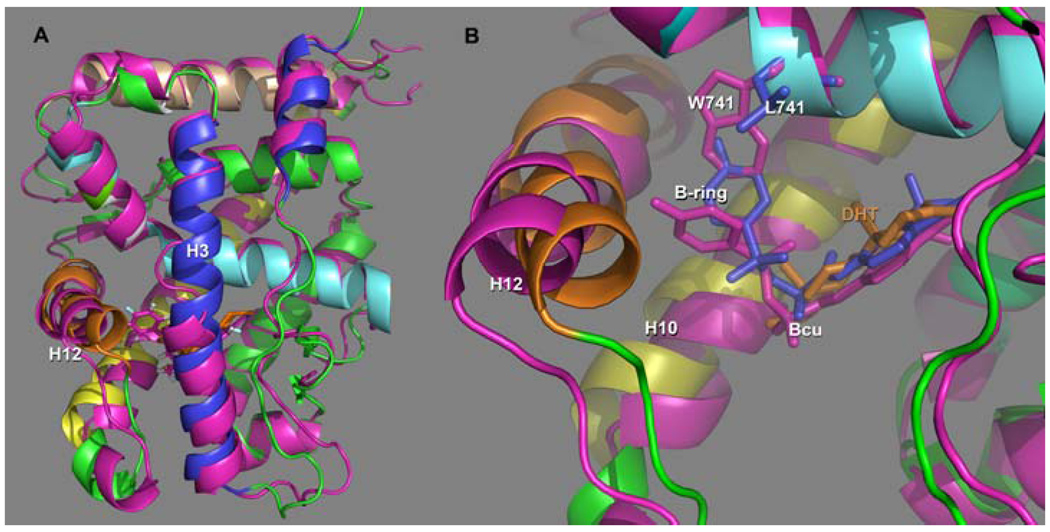
The distortions underlying the deviations in the 5ns trajectories of bicalutamide may be seen by superposing the 5ns structure of the AR-Bcu (sBcu) complex on the AR-DHT complex (Figure 5). As can be seen in these figures the displacements in AR induced by Bcu binding are not solely restricted to the region of the HBS but are also distributed throughout the receptor. Perhaps the most noteworthy distortion occurs in the helices comprising the AF2 site. Here, there are distortions in all three helices forming the AF2 site, H3, H4 and H12, with the largest displacement in H12 (Fig 5b). This is consistent with the theme of structural displacements underlying antagonism observed in other NRs (1, 2). A window into this mechanism has been provided by the naturally occurring mutant W741L (24). The crystal structure of this complex demonstrated that indeed W741L-AR adopted an agonist conformation on binding to Bcu (RMS from AR-DHT of 0.33) (25). The structure revealed that the B-ring of Bcu occupied the cavity created by the absence of the Trp indole in the mutated complex (25), while the A-ring is virtually in the same environment as the A-ring of DHT (Fig 5b). Thus, although a structural description of the AR antagonist mechanism still defied elucidation, this structure gave insight into possible interactions in the binding of Bcu to AR, and along with MD allows for the description of one of the mechanisms by which Bcu antagonizes AR dependent transcription. The MD studies of the WT AR-Bcu complex show that, as expected, W741 precludes the B-ring of Bcu from accessing that region of the HBS (Fig 5b). Instead it is displaced “downward” toward H12, as seen from comparison of B-rings in the W741L mutant (blue) and the WT MD simulation (magenta). In the agonist conformation this repositioning of the B-ring (magenta) would introduce several severe steric conflicts with M895 and I899 in helix 12 (e.g. I899Cδ - C17 2.02Å; M895Cα- F19 2.24Å), which, in turn, cause H12 to shift outward to accommodate the B-ring. We note that the A-ring pocket is basically conserved in the simulation, residing in essentially the same site that is occupied by the A-ring in the W741L complex and the DHT steroid hormone A-ring.
Figure 5.
a) Superposition of the 5ns structure of Bcu bound AR, (magenta) with the native AR-DHT complex (Helices colored as in Fig1). DHT and Bcu are rendered as gold and magenta sticks respectively. As can be seen Bcu induces global deformations in the AR structure including some unraveling in the center of H3, and significant displacement of H12 both comprising components of the AF2 site. b) In this view we remove H3 to better focus in on the HBS. We have also depicted Bcu and L741 in the W741L mutant (both colored blue). The shift in the position of the B-ring of Bcu (magenta) by Trp 741 is readily apparent when compared to its position when Leu is present at the 741 position (blue). Again we can see the concomitant displacement of the key AF2 Helix H12 impairing the ability of coactivator to bind at this site and consistent with the mechanism of antagonism in other NRs.
Some similar interactions were found by Bisson et. al. (32) in their simulations of Bcu, although notable differences were observed as well. For example, the interactions between M895 and Bcu are observed in both results, but on the other hand interactions we observe with M745 are not mentioned in that study. The most significant difference however is the lack of perturbation of the position of H12 in their results. This may be due to the fact that they analyzed only a single simulation carried out over a shorter time frame (2.15ns) with a shorter cutoff. It is possible that a combination of these factors prevented the propagation of perturbations induced by the ligand from reaching H12, though this is of course, speculation.
Plasticity of the Hormone Binding Pocket – Determining Ligand Functionality
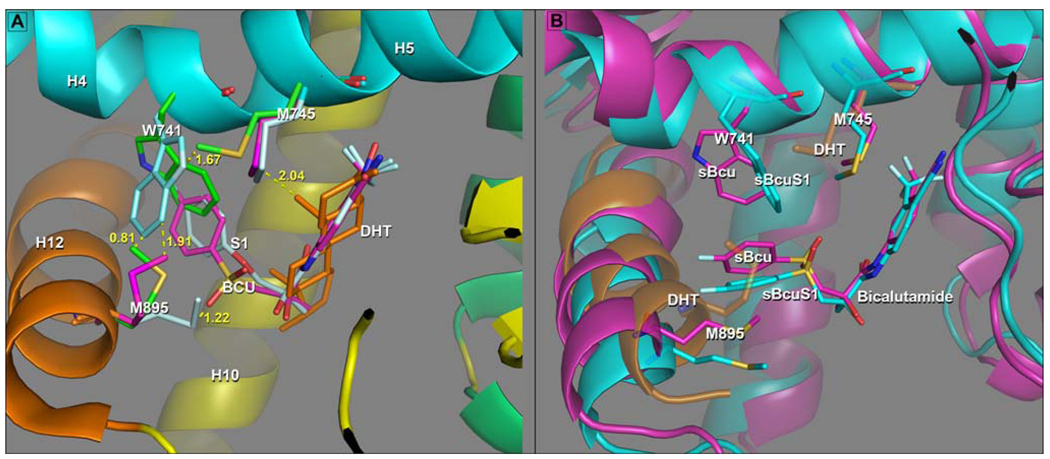
In figure 6 we focus on the hormone binding site, to explore the detailed mechanism by which Bcu antagonizes the androgen response. Although the agonist activity of Bcu in the W741L mutant is fairly definitive in identifying W741 as playing a key role in the determination of a ligand's functional consequences (and therefore in future design efforts), it is not the sole determinant. This is demonstrated compellingly by the activity of the selective agonist S1 in WT AR (Fig. 2b) (25). The substitution of an–NO2 at the para position on the A-ring is unlikely to be related to the agonist activity of S1 as many, if not most, antiandrogens have this functional group (35). In addition, the B-ring is retained in S1. The result of structural studies, shown in Fig 6a, reveal how this is accomplished in the S1 complex but precluded with Bcu (26).
Figure 6.
Adaptation of AR LBP to the B-ring of Bcu and S1 in experimental and simulated complexes. In both panels A and B Helix 3 is not displayed to provide an unobstructed view of binding pocket interactions. A) When the sidechains can adapt to ligand in an agonist structure. Superposition of S1 (cyan), Bcu (magenta) and DHT (ligand gold: protein sidechains green, H12 gold, H4–H5 cyan, H10 yellow) experimental AR structures. This figure shows the shift of M895 from its position in the DHT complex (green carbons), away from H12 towards the ether oxygen of S1 (cyan) in order to accommodate the repositioned W741. This S1 configuration doesn’t accommodate Bcu in the agonist AR structure as when Bcu is bound, M895 in this position clashes with the sulfonyl group of Bcu. The orientation adopted by W741 in S1 to accommodate the B-ring brings it too close to the M745 side chain in DHT (green) and the configuration of M745 found in the S1 and Bcu complexes, which accommodates the rotated Trp741, is very close to the A-ring axial methyl group in DHT thus being disallowed in the DHT complex. B) When the sidechains are unable to adapt to ligand in agonist structure. Superposition of the 5ns structures for the sBcu simulation, colored magenta and the sBcuS1 simulation colored cyan. The semitransparent structure shows H12 and positions of M895 and M745 position in the reference AR-DHT complex (gold). This shows the re-positioning of the B-ring of Bcu “downward” and the concomitant displacement of H12 away from the LBS position, to create an enlarged pocket to accommodate the shifted B-ring. Repositioning of the B-ring is accomplished primarily by a rotation about the C11–C13 bond from ~ −60° to +60°, while the A-ring occupies the identical position it occupies in the W741L X-ray structure (Fig 4b). The displacement of H12 allows M895 to adopt an extended conformation back into the LBS without clashing with the sulfonyl group of Bcu or with the B-ring of Bcu as seen in the co-location of M895 in the DHT complex (gold) and the B-rings of Bcu. This shift of H12 occurs in both trajectories as does the positioning of M745 into a space similar to that it occupies in the S1 and W741L AR crystal structures.
The indole ring of W741 rotates away from H4 and toward H12. This creates a pocket into which the B-ring of S1 inserts. Perhaps not surprisingly, as seen in Fig 6a, it occupies essentially the same pocket inhabited by the B-ring of Bcu in the W741L construct, created by the Trp to Leu mutation (25). The question then arises as to why W741 can’t adopt this configuration in the WT receptor to accommodate Bcu? What emerges is a network of residues (M745- W741-M895) surrounding the B-ring pocket and adjacent to H12, which interact with the ligand and themselves. The degree to which this network can adapt in a concerted fashion to a ligand without distorting the overall conformation of the receptor, and especially the position of H12, plays a major role in determining the functional consequences of a ligand occupying this pocket. Thus, the repositioning of Trp to accommodate the B-ring of S1 requires a concerted shift by M895 away from H12 and into the vicinity of the ether oxygen of S1 (cyan sidechains in Fig 6a). This reorientation of M895 would be precluded by the sulfonyl group of Bcu in this configuration (d(SO2-Sδ) ~1.22 Fig 6a). We also note that the orientation adopted by W741 in the S1 complex is incompatible with the position of the M745 (gold) in the DHT complex (d(Nε-Cε) ~1.67Å). Thus WT-AR must find another way to accommodate Bcu, which leads to an antagonistic response.
We also note the configuration of M745 found in the AR S1 (cyan) and W741L Bcu (magenta) complexes, which accommodates the rotation of Trp741 is incompatible with the A-ring axial methyl group in DHT (d(Sδ-C19 ~2.04). Thus we would surmise for example that a substitution on the A-ring of S1 occupying the C19 position of DHT might reposition M745 in turn forcing Trp back into the B-ring pocket and converting the S1 analog into an antagonist much as the sulfonyl linkage in bicalutamide does acting through M895.
Conformational Response of AR to Casodex – What happens when the network can’t accommodate the B-ring in the agonist AR structure?
In Figure 5 we showed the overall distortion of AR engendered by binding of Bcu and compared it to the AR-DHT and AR W741L-Bcu complexes. Here we further focus our attention on the response to Bcu of the HBS residues and H12. To this end the 5ns structures starting with W741, M745 and M895 in their orientations in the DHT and S1 complexes, sBcu and sBcuS1 respectively, are given in Figure 6b, and compared to their configurations in the experimental structures. As seen from this figure the different trajectories, result in the same overall deformation of the AR structure but differ somewhat in the details after 5ns. This is to be expected of MD trajectories, and is in fact why we carry out multiple trajectories starting from different states and use different methodologies. The overall distortions results in expansion of the ligand binding pocket mainly by displacement of H12 away from the ligand binding site (LBS), as the B-ring of Bcu is repositioned “downward” into the pocket created to accommodate it. As noted above the repositioning of the B-ring brings it into the space occupied by M895, and I899 (not displayed) in the W741L-Bcu construct. The displacement of H12 and the concomitant reorientation and translation of M895 alleviate these clashes, and allows M895 to adopt an extended conformation back into the LBS without clashing with the sulfonyl group of Bcu. This reorganization occurs in both trajectories, as does the positioning of M745 into a space similar to that which it occupies in the S1 and W741L AR structures. Of these three key residues only the configuration of the Trp741 side chain differs somewhat in the two 5ns structures.
The A-ring and Conserved Water Molecule
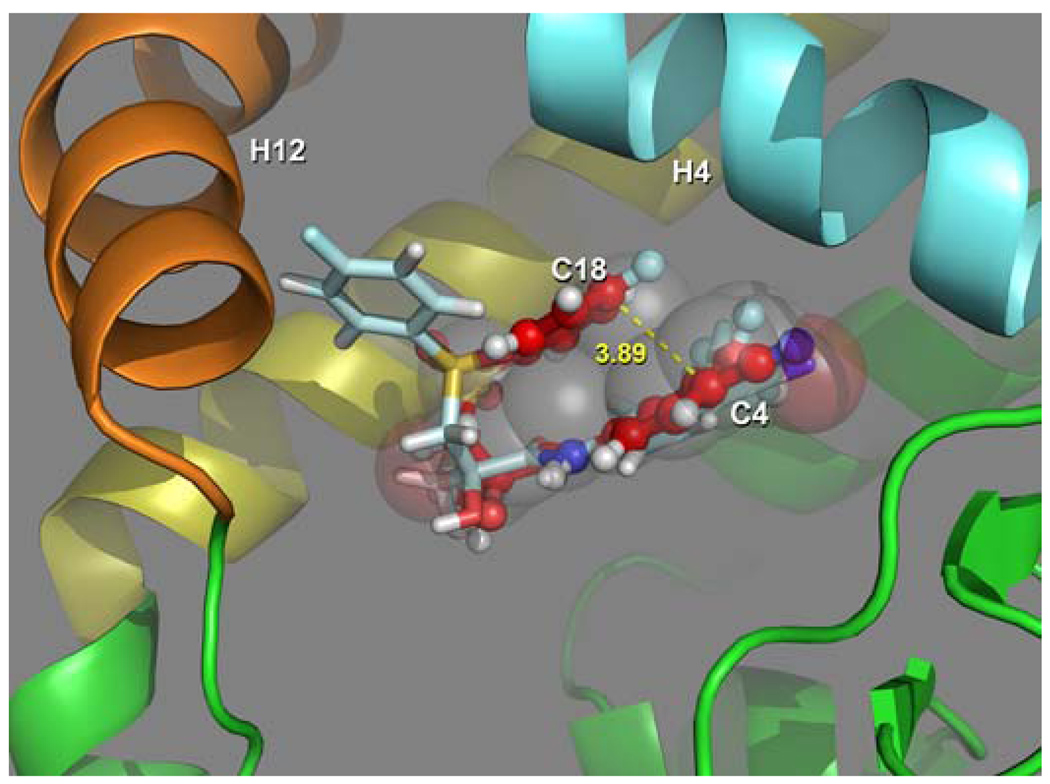
In contrast to the B-ring, the A-ring of Bcu occupies virtually the identical position in the HBS of WT AR in both simulations, sBcuS1, and sBcu, as it does in the W741L mutant. The A-ring sits in a hydrophobic pocket bounded by helices H3 (L704, L707), H5 (M745, V746), H7 (M787), H10 (L873), and strand β3 (F764) where it participates in the same interactions as in the W741L structure. The nature of the binding site and a subset of these interactions in the X-ray (W741L) and MD structure sBcuS1 are compared in Figure 7. As can be seen the interactions are highly conserved even though side chain conformations, such as M745, may reorient in some cases. In general the corresponding interatomic distances in the two structures differ only by a few tenths of an angstrom. This binding mode of the A-ring appears to be highly favored, as in addition to the antagonist structures reported here X-ray structures indicate that the corresponding A-rings of essentially every ligand solved, including DHT (Fig 5b and ref (7)), S-1 (26), R-3 an agonist which is an analog of the antagonist hydroxyflutamide (26) all lie in the same orientation in this pocket. In addition the conserved water molecule found in the LBS of AR structures is also found in the Bcu antagonist structures at the same location. It should be pointed out that it was not placed there initially, but rather emigrated from the initial, arbitrary position it was placed in by the solvation procedure to this location (see methods).
Figure 7.
Superposition of the W741L experimental structure and WT AR Bcu simulated structure showing the high conservation of the A-ring pocket. (Bcu is clipped at the amide attached to the A ring). The A-ring in the experimental structure is shown with green carbons and the simulated structure with cyan. The conserved water position in the experimentally determined DHT and Bcu-W741L complexes are shown as a blue and red spheres, while the stick model depicting the position from sBcuS1 shows this interaction to be well accounted for in the simulated results. (the water position is similarly conserved in all simulated structures)
Free Energy Perturbation Results in an Intriguing Folded Conformation of Bcu
As noted above the FEP trajectory of the Bcu-AR complex resembled those of the agonists more than those of the “standard” trajectories of Bcu. Inspection of the structure of the complex, given in Fig 8, shows why. As seen in this figure, Bcu adopts an entirely different conformation, with the B-ring rotated and stacked in a compact folded structure over the A-ring (ball and stick red carbons). The B-ring no longer extends into H12 and the AF-2 region as it does in structures obtained in the standard MD simulations (stick; cyan carbons) described above. As can be seen in the figure the A-ring position is still conserved resulting in the folded structure occupying an expanded pocket enveloping the DHT site (Shown in transparent spacefill). The elastic response of the receptor enables it to adapt to this Bcu conformation in an agonist-like conformation, with little distortion of the H12 helix seen in the antagonist conformation. As can be seen from Table 2 the displacements of the helices bordering the AF-2 site are significantly larger in the extended Bcu complexes as compared to the DHT or Bcu folded structure. This is especially true for H12 where the displacement is 3-fold greater in the former.
Figure 8.
Free Energy Perturbation docking results in an unanticipated configuration and binding mode of bicalutamide. The folded configuration resulting from this slow growth procedure is shown in ball and stick rendering with red carbons. The extended configuration of Bcu is shown in stick rendering with cyan carbons. The DHT molecule is shown in transparent spacefill. The FEP slow growth MD results in bicalutamide folding over into the pocket occupied by DHT, which expands somewhat to accommodate it. In this conformation Bcu no longer sterically impacts H12 or the AF2 site.
Table 2.
Substructural Deviations from DHTexp (Backbone)
| Substructure | DHT5ns | BCU-FEP | BCUS1 | BCU5ns |
|---|---|---|---|---|
| H3 | 1.19 | 1.28 | 1.63 | 1.64 |
| H4 | 1.39 | 1.1 | 1.44 | 1.16 |
| H5 | 0.87 | 0.67 | 0.92 | 1.23 |
| H12 | 0.58 | 0.72 | 2.69 | 1.94 |
| AF2 site | 1.15 | 1.11 | 1.81 | 1.52 |
Structures superimposed fitting all backbone heavy atoms; RMS (in Å) is computed for backbone heavy atoms of the substructures without further superimposition.
Is the folded ligand conformation plausible? Energetic analysis
In order to confirm that the conformational states of bicalutamide found in the MD simulations are energetically reasonable and not the result of a fortuitous rare statistical fluctuation, we have carried out high-level Density Functional QM calculations, (DFT), of the conformational energetics of Bcu. The results are given in the first column of Table 3. As seen from this Table there are two equi-energetic minima on the Bcu energy surface: an extended structure similar to that found in the W741L construct (and the two structures resulting from the standard MD simulations); and a stacked structure similar to that found bound to AR in the FEP simulation (QMmin-Ext and QMmin-Folded respectively). To assess the strain energies on binding we minimized the energy of Bcu restrained to the bound configurations. As can be seen, the induced strain energy is ~ 9 Kcal for both conformational states, with the extended structures having slightly higher strain. Based on these results the novel folded conformation of Bcu, stabilizing an agonist-like configuration of the AR-Bcu complex is readily accessible.
Table 3.
Binding and Strain Energy of Agonist and Antagonist Like Bcu Conformations
| AR_BCU Complex |
ESTRAIN (QM) | <Eprot-lig> | Eprot-lig (Min) |
|---|---|---|---|
| QMmin-Folded | 0.00 | ||
| QMmin-Ext | 0.04 | ||
| 5ns | 9.92 | −45.76 | −49.37 |
| 5ns S1 | 9.89 | −50.08 | −49.26 |
| 5ns FEP | 8.53 | −45.21 | −48.28 |
Energy in Kcal/mol
Is the Binding Energy of the folded conformation of bicalutamide comparable to the extended structure?
The question then arises as to what the protein-ligand intermolecular energies are for the different poses of Bcu. Does the folded conformation induce significantly unfavorable interactions in the binding site, resulting in weaker binding? To address this question we have evaluated both the time-averaged, and minimized binding energies of the three Bcu-AR complexes. These are given in the 2nd and 3rd columns of Table 3. Again, as can be seen from these results, both the minimized and time-averaged binding energies are comparable, with the extended structures being slightly more favorable. The structure resulting from the initial S1 configuration is ~5Kcal more favorable in the dynamically averaged calculation and ~1Kcal in the minimized structures. Thus both the strain and binding energy suggest that two distinct AR-Bcu complexes are accessible, an extended Bcu complex which significantly distorts the AF2 pocket and the orientation of H12, accounting for its antagonist properties, and intriguingly, a folded conformation which could be the species responsible for its agonist activity under increased AR expression levels or other agonist producing environments.
Summary
We have derived a structural model for an antagonist bound conformation of AR and the possible binding modes of bicalutamide, using MD and QM simulations, This provides a structural rationale for the antagonist activity of Bcu and a model for further design of AR modulators. Through an analysis of experimental and simulated structures, we have shown how a network of interacting sidechains in the hormone binding site play a key role in the functional consequences of agonist or antagonist activity and reorganize on the binding of Bcu. As might be expected, a considerable shift in the position of H12 results from these interactions resulting in distortion of the AF2 coactivator binding site.
A “Slow Growth” procedure based on FEP methodology led to a novel docking pose for Bcu, a “folded” conformation with the A and B-rings stacked. QM calculations of this folded configuration reveal that Bcu has two equienergetic minima; one similar to the “extended” conformation observed in the W741L structure and the two standard MD runs, and the second analogous to the folded configuration resulting from the FEP simulation, suggesting the latter’s accessibility. The docked “folded” conformation of Bcu shows similar effects on the AR protein structure to the physiological agonist DHT and therefore could be responsible for the agonist activity of Bcu. In addition it may provide a model for the design of a new class of AR modulators, exploiting an expanded ligand binding cavity around the DHT site.
Acknowledgements
We thank Dr Robert Fletterick and Dr. Sherin Abdul-Meguid for helpful comments on the manuscript.
This work was supported by the National Institutes of Health Grant R43 CA132538, and the National Science Foundation Grant CNS 0551500.
Glossary
- AR
Androgen Receptor
- NR
Nuclear Receptor
- Bcu
Bicalutamide
- T
Testosterone
- DHT
Dihydrotestosterone
- LBD
Ligand Binding Site
- WT
Wild Type
- MD
Molecular Dynamics
- QM
Quantum Mechanical
- FEP
Free Energy Perturbation
- DFT
Density Functional Theory
- PC
Prostate Cancer
- HBS
Hormone Binding Site
- sDHT
DHT Simulation
- sBcuExp
Bcu Experimental Simulation
- sBcu
Bcu Simulation
- sBcuS1
Bcu S1 Simulation
- sBcuFEP
Bcu FEP Simulation
- RMS
root mean square
References
- 1.Gronemeyer H, Gustafsson JA, Laudet V. Principles for modulation of the nuclear receptor superfamily. Nat Rev Drug Discov. 2004;3:950–964. doi: 10.1038/nrd1551. [DOI] [PubMed] [Google Scholar]
- 2.Moore JT, Collins JL, Pearce KH. The nuclear receptor superfamily and drug discovery. ChemMedChem. 2006;1:504–523. doi: 10.1002/cmdc.200600006. [DOI] [PubMed] [Google Scholar]
- 3.Huggins C. Endocrine-induced regression of cancers. Science. 1967;156:1050–1054. doi: 10.1126/science.156.3778.1050. [DOI] [PubMed] [Google Scholar]
- 4.Wirth MP, Hakenberg OW, Froehner M. Antiandrogens in the treatment of prostate cancer. European urology. 2007;51:306–314. doi: 10.1016/j.eururo.2006.08.043. [DOI] [PubMed] [Google Scholar]
- 5.Feldman BJ, Feldman D. The development of androgen-independent prostate cancer. Nature reviews. 2001;1:34–45. doi: 10.1038/35094009. [DOI] [PubMed] [Google Scholar]
- 6.Matias PM, Donner P, Coelho R, Thomaz M, Peixoto C, Macedo S, Otto N, Joschko S, Scholz P, Wegg A, Basler S, Schafer M, Egner U, Carrondo MA. Structural evidence for ligand specificity in the binding domain of the human androgen receptor. Implications for pathogenic gene mutations. J Biol Chem. 2000;275:26164–26171. doi: 10.1074/jbc.M004571200. [DOI] [PubMed] [Google Scholar]
- 7.Sack JS, Kish KF, Wang C, Attar RM, Kiefer SE, An Y, Wu GY, Scheffler JE, Salvati ME, Krystek SR, Jr, Weinmann R, Einspahr HM. Crystallographic structures of the ligand-binding domains of the androgen receptor and its T877A mutant complexed with the natural agonist dihydrotestosterone. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:4904–4909. doi: 10.1073/pnas.081565498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hur E, Pfaff SJ, Payne ES, Gron H, Buehrer BM, Fletterick RJ. Recognition and accommodation at the androgen receptor coactivator binding interface. PLoS biology. 2004;2:E274. doi: 10.1371/journal.pbio.0020274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Estebanez-Perpina E, Moore JM, Mar E, Delgado-Rodrigues E, Nguyen P, Baxter JD, Buehrer BM, Webb P, Fletterick RJ, Guy RK. The molecular mechanisms of coactivator utilization in ligand-dependent transactivation by the androgen receptor. J Biol Chem. 2005;280:8060–8068. doi: 10.1074/jbc.M407046200. [DOI] [PubMed] [Google Scholar]
- 10.Ostrowski J, Kuhns JE, Lupisella JA, Manfredi MC, Beehler BC, Krystek SR, Jr, Bi Y, Sun C, Seethala R, Golla R, Sleph PG, Fura A, An Y, Kish KF, Sack JS, Mookhtiar KA, Grover GJ, Hamann LG. Pharmacological and x-ray structural characterization of a novel selective androgen receptor modulator: potent hyperanabolic stimulation of skeletal muscle with hypostimulation of prostate in rats. Endocrinology. 2007;148:4–12. doi: 10.1210/en.2006-0843. [DOI] [PubMed] [Google Scholar]
- 11.Baek SH, Ohgi KA, Nelson CA, Welsbie D, Chen C, Sawyers CL, Rose DW, Rosenfeld MG. Ligand-specific allosteric regulation of coactivator functions of androgen receptor in prostate cancer cells. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:3100–3105. doi: 10.1073/pnas.0510842103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen CD, Welsbie DS, Tran C, Baek SH, Chen R, Vessella R, Rosenfeld MG, Sawyers CL. Molecular determinants of resistance to antiandrogen therapy. Nature medicine. 2004;10:33–39. doi: 10.1038/nm972. [DOI] [PubMed] [Google Scholar]
- 13.Shen MM, Abate-Shen C. Molecular genetics of prostate cancer: new prospects for old challenges. Genes Dev. 2010;24:1967–2000. doi: 10.1101/gad.1965810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burendahl S, Treuter E, Nilsson L. Molecular dynamics simulations of human LRH-1: the impact of ligand binding in a constitutively active nuclear receptor. Biochemistry. 2008;47:5205–5215. doi: 10.1021/bi7025084. [DOI] [PubMed] [Google Scholar]
- 15.Zhou J, Liu B, Geng G, Wu JH. Study of the impact of the T877A mutation on ligand-induced helix-12 positioning of the androgen receptor resulted in design and synthesis of novel antiandrogens. Proteins. 2010;78:623–637. doi: 10.1002/prot.22592. [DOI] [PubMed] [Google Scholar]
- 16.Quigley CA, De Bellis A, Marschke KB, el-Awady MK, Wilson EM, French FS. Androgen receptor defects: historical, clinical, and molecular perspectives. Endocr Rev. 1995;16:271–321. doi: 10.1210/edrv-16-3-271. [DOI] [PubMed] [Google Scholar]
- 17.Wu JH, Gottlieb B, Batist G, Sulea T, Purisima EO, Beitel LK, Trifiro M. Bridging structural biology and genetics by computational methods: an investigation into how the R774C mutation in the AR gene can result in complete androgen insensitivity syndrome. Hum Mutat. 2003;22:465–475. doi: 10.1002/humu.10279. [DOI] [PubMed] [Google Scholar]
- 18.Elhaji YA, Stoica I, Dennis S, Purisima EO, Lumbroso R, Beitel LK, Trifiro MA. Impaired helix 12 dynamics due to proline 892 substitutions in the androgen receptor are associated with complete androgen insensitivity. Human molecular genetics. 2006;15:921–931. doi: 10.1093/hmg/ddl009. [DOI] [PubMed] [Google Scholar]
- 19.Scher HI, Beer TM, Higano CS, Anand A, Taplin ME, Efstathiou E, Rathkopf D, Shelkey J, Yu EY, Alumkal J, Hung D, Hirmand M, Seely L, Morris MJ, Danila DC, Humm J, Larson S, Fleisher M, Sawyers CL. Antitumour activity of MDV3100 in castration-resistant prostate cancer: a phase 1–2 study. Lancet. 2010;375:1437–1446. doi: 10.1016/S0140-6736(10)60172-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen Y, Clegg NJ, Scher HI. Anti-androgens and androgen-depleting therapies in prostate cancer: new agents for an established target. Lancet Oncol. 2009;10:981–991. doi: 10.1016/S1470-2045(09)70229-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tran C, Ouk S, Clegg NJ, Chen Y, Watson PA, Arora V, Wongvipat J, Smith-Jones PM, Yoo D, Kwon A, Wasielewska T, Welsbie D, Chen CD, Higano CS, Beer TM, Hung DT, Scher HI, Jung ME, Sawyers CL. Development of a second-generation antiandrogen for treatment of advanced prostate cancer. Science. 2009;324:787–790. doi: 10.1126/science.1168175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xiao HY, Balog A, Attar RM, Fairfax D, Fleming LB, Holst CL, Martin GS, Rossiter LM, Chen J, Cvjic ME, Dell-John J, Geng J, Gottardis MM, Han WC, Nation A, Obermeier M, Rizzo CA, Schweizer L, Spires T, Jr, Shan W. Gavai A, Salvati ME, Vite G. Design and synthesis of 4-[3,5-dioxo-11-oxa-4,9-diazatricyclo[5.3.1.0(2,6)]undec-4-yl]-2-trifluoro methyl-benzonitriles as androgen receptor antagonists. Bioorganic & medicinal chemistry letters. 2010;20:4491–4495. doi: 10.1016/j.bmcl.2010.06.034. [DOI] [PubMed] [Google Scholar]
- 23.Attar RM, Jure-Kunkel M, Balog A, Cvijic ME, Dell-John J, Rizzo CA, Schweizer L, Spires TE, Platero JS, Obermeier M, Shan W, Salvati ME, Foster WR, Dinchuk J, Chen SJ, Vite G, Kramer R, Gottardis MM. Discovery of BMS-641988, a novel and potent inhibitor of androgen receptor signaling for the treatment of prostate cancer. Cancer research. 2009;69:6522–6530. doi: 10.1158/0008-5472.CAN-09-1111. [DOI] [PubMed] [Google Scholar]
- 24.Hara T, Miyazaki J, Araki H, Yamaoka M, Kanzaki N, Kusaka M, Miyamoto M. Novel mutations of androgen receptor: a possible mechanism of bicalutamide withdrawal syndrome. Cancer research. 2003;63:149–153. [PubMed] [Google Scholar]
- 25.Bohl CE, Gao W, Miller DD, Bell CE, Dalton JT. Structural basis for antagonism and resistance of bicalutamide in prostate cancer. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:6201–6206. doi: 10.1073/pnas.0500381102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bohl CE, Miller DD, Chen J, Bell CE, Dalton JT. Structural basis for accommodation of nonsteroidal ligands in the androgen receptor. J Biol Chem. 2005;280:37747–37754. doi: 10.1074/jbc.M507464200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJ. GROMACS: fast, flexible, and free. Journal of computational chemistry. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 28.Kitson DH, Avbelj F, Moult J, Nguyen DT, Mertz JE, Hadzi D, Hagler AT. On achieving better than 1-A accuracy in a simulation of a large protein: Streptomyces griseus protease A. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:8920–8924. doi: 10.1073/pnas.90.19.8920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaminski GA, Friesner RA, Tirado-Rives J, Jorgensen WL. Evaluation and Reparametrization of the OPLS-AA Force Field for Proteins via Comparison with Accurate Quantum Chemical Calculations on Peptides. The Journal of Physical Chemistry B. 2001;105:6474–6487. [Google Scholar]
- 30.Cheatham TE, III, Miller JL, Fox T, Darden TA, Kollman PA. Molecular Dynamics Simulations on Solvated Biomolecular Systems: The Particle Mesh Ewald Method Leads to Stable Trajectories of DNA, RNA, and Proteins. Journal of the American Chemical Society. 1995;117:4193–4194. [Google Scholar]
- 31.Norberg J, Nilsson L. On the Truncation of Long-Range Electrostatic Interactions in DNA. Biophysical journal. 2000;79:1537–1553. doi: 10.1016/S0006-3495(00)76405-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bisson WH, Abagyan R, Cavasotto CN. Molecular basis of agonicity and antagonicity in the androgen receptor studied by molecular dynamics simulations. Journal of Molecular Graphics and Modelling. 2008;27:452–458. doi: 10.1016/j.jmgm.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 33.Simonson T, Archontis G, Karplus M. Continuum Treatment of Long-Range Interactions in Free Energy Calculations. Application to Protein-Ligand Binding. The Journal of Physical Chemistry B. 1997;101:8349–8362. [Google Scholar]
- 34.Kollman P. Free energy calculations: Applications to chemical and biochemical phenomena. Chemical reviews. 1993;93:2395–2417. [Google Scholar]
- 35.Gao W, Bohl CE, Dalton JT. Chemistry and structural biology of androgen receptor. Chemical reviews. 2005;105:3352–3370. doi: 10.1021/cr020456u. [DOI] [PMC free article] [PubMed] [Google Scholar]