Abstract
Hepatitis C virus (HCV) RNA replication requires viral nonstructural proteins as well as cellular factors. Recently, a cellular protein, synaptotagmin-binding, cytoplasmic RNA-interacting protein (SYNCRIP), also known as NSAP1, was found to bind HCV RNA and enhance HCV IRES-dependent translation. We investigate whether this protein is also involved in the HCV RNA replication. We found that SYNCRIP was associated with detergent-resistant membrane fractions and colocalized with newly-synthesized HCV RNA. Knock-down of SYNCRIP by siRNA significantly decreased the amount of HCV RNA in the cells containing a subgenomic replicon or a full-length viral RNA. Lastly, an in vitro replication assay after immunodepletion of SYNCRIP showed that SYNCRIP was directly involved in HCV RNA replication. These findings indicate that SYNCRIP has dual functions, participating in both RNA replication and translation in HCV life cycle.
Keywords: HCV RNA replication, SYNCRIP, hnRNP Q, NSAP1, HCV RNA translation, siRNA knock-down, HCV replicon
Introduction
Hepatitis C virus (HCV) infection is a leading cause of chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma. HCV is an enveloped RNA virus with a positive-stranded RNA of 9.7 kb in length (Reed and Rice, 2000). It encodes a large polyprotein, which is then processed into structural proteins (C, E1, and E2) and nonstructural (NS) proteins, the latter of which participate in viral replication.
Besides the viral NS proteins, several host factors, including the human homologue of the 33-kDa vesicle-associated membrane protein-associated protein (hVAP-33) (Gao et al., 2004, Hamamoto et al., 2005), polypyrimidine-tract-binding protein (PTB) (Aizaki et al., 2006, Chang and Luo, 2006, Domitrovich et al., 2005), La antigen (Domitrovich et al., 2005) and host geranylgeranylated proteins and fatty acids (Kapadia and Chisari, 2005) have been shown to be involved in some steps of HCV replication cycle. Some of these host factors, such as PTB and La autoantigen, were initially found to regulate HCV protein translations (Ali and Siddiqui, 1997, Ito and Lai, 1999) by virtue of their binding to the 5′ and 3′-untranslated regions (UTR) of HCV RNA. Later studies showed that some of these host factors also directly regulate HCV RNA replication either by participating in the formation of the RNA replication complex (e.g., VAP-33) (Gao et al., 2004) or by binding to the viral RNA (e.g., La, PTB) (Ali and Siddiqui, 1995, Chang and Luo, 2006). A recent study showed that another host protein, synaptotagmin-binding, cytoplasmic RNA-interacting protein (SYNCRIP), also named NS-1-associated protein (NSAP1), binds to the N-terminal of the core protein-coding region of HCV RNA and enhances HCV Internal Ribosomal Entry Site (IRES)-dependent translation (Kim et al., 2004).
SYNCRIP is a member of cellular heterogeneous nuclear ribonucleoprotein (hnRNP) family, to which PTB also belongs. hnRNPs are well-known for their abilities to bind to cellular proteins and RNAs to facilitate many biological processes. Interestingly, SYNCRIP has previously been shown to be involved not only in cellular processes but also in mouse hepatitis virus (MHV) RNA replication (Choi et al., 2004b). Since SYNCRIP binds to HCV RNA at a site close to the 5′-end of the RNA, it is likely that SYNCRIP may also affect the RNA replication of HCV. If this is the case, SYNCRIP will have duel functions in both RNA replication and protein translation, similar to other duel-purpose hnRNPs, such as PTB. Our goal in this study is to investigate whether SYNCRIP is involved in HCV RNA replication in addition to its role in translation.
Results
SYNCRIP relocalized to detergent-resistant membrane fraction in HCV replicon cells
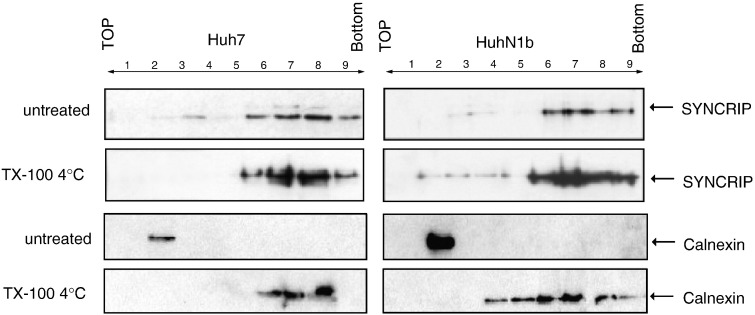
It has been shown that HCV RNA replication occurs in detergent-resistant membrane (DRM) fractions (Ali et al., 2002, El-Hage and Luo, 2003, Mizutani et al., 2000). The nonstructural proteins of HCV are associated with the DRM structures containing Caveolin-2, strongly suggesting that the viral replication complex has properties of lipid rafts (Gao et al., 2004, Mizutani et al., 2000). To determine whether SYNCRIP is in the RNA replication complex, we performed membrane flotation analysis of HCV replicon cells, followed by immunoblotting with anti-SYNCRIP antibody to examine the possible presence of SYNCRIP in the detergent-resistant membrane fractions, where the HCV replication complexes reside. We found that SYNCRIP was present mostly in the cytosolic fractions (fractions 6–9, Fig. 1 ) in both Huh7 and HuhN1b cells, but a small fraction was associated with the membrane (fractions 2–4, Fig. 1). After treatment with Triton X-100 at 4 °C, some SYNCRIP was still associated with the membrane in HuhN1b cells; in contrast, almost all of SYNCRIP was solubilized in Huh7 cells. Longer exposure of immunoblotting was performed, and SYNCRIP was still not found in the DRM fractions of TX-100 treated Huh7 cells. As a control, Calnexin, a marker protein of ER membrane, was concentrated exclusively in the membrane fractions (fraction 2–3) in the absence of detergent treatment in both Huh7 and HuhN1b cells (Fig. 1, panel 3). After the cells were treated with cold detergent, Calnexin was redistributed entirely to the soluble fractions, indicating that Calnexin was associated with detergent-soluble membrane, which is a known characteristic of unmodified ER. These data suggested that SYNCRIP is predominantly a cytoplasmic protein, but is relocalized to the DRM fractions (fraction 2–3 after TX-100 treatment) in the HuhN1b replicon cells. The relocalization of SYNCRIP protein, but not Calnexin, to the DRM fraction in the HuhN1b replicon cell indicated that SYNCRIP may be specifically recruited by HCV RNA to the replication complexes, since SYNCRIP binds to HCV RNA (Kim et al., 2004).
Fig. 1.
Membrane flotation assay showed relocalization of SYNCRIP to DRM in HCV replicon cells. Cell lysates were prepared from HCV replicon (HuhN1b) or Huh7 cells by passing through a 25-gauge needle 20 times. Nuclei and unbroken cells were removed by centrifugation at 1000 ×g for 5 min in microcentrifuge at 4 °C. The supernatants treated with or without 1% TX-100 at 4 °C for 30 min were fractionated by discontinuous sucrose gradient centrifugation. Fractions were collected from the top, numbered from 1 to 9. Each fraction was concentrated by Centricon YM-30 (Millipore, MA) and immunoblotted by rabbit anti-SYNCRIP antibody or mouse anti-Calnexin antibody, respectively. SYNCRIP was found in both membrane and soluble fractions in the untreated Huh7 and HuhN1b cells, whereas in the HuhN1b cells, some SYNCRIP was localized to the DRM fractions. This phenomenon is not seen in the Calnexin profile.
SYNCRIP colocalized with de novo synthesized RNA in HCV replicon cells
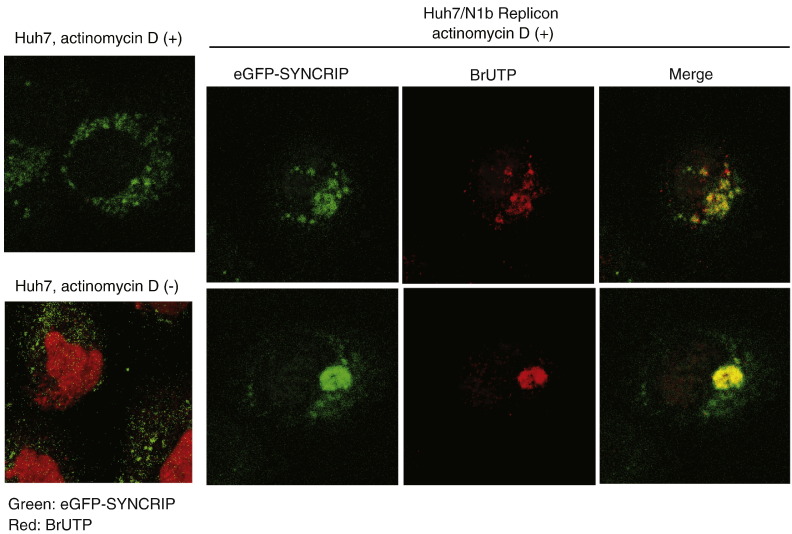
Previous studies on HCV replicon cells have shown that the newly synthesized HCV RNA and the viral nonstructural proteins colocalized with each other on the distinct speckle-like structure in the cytoplasm of the replicon cells (Gosert et al., 2003, Mizutani et al., 2000). To examine whether SYNCRIP is associated with HCV RNA synthesis in the speckle structures, BrUTP labeling was performed in HCV replicon cells transfected with peGFP-SYNCRIP (a gift from Dr. Mizutani, The University of Tokyo). Briefly, 2 days after transfection of peGFP-SYNCRIP, BrUTP was transfected into actinomycin D-pretreated cells (Kanestrom et al., 1998). Immunofluorescence staining with sheep anti-BrdU polyclonal antibody (Biodesign, ME) was then performed (Kanestrom et al., 1998). Under this condition, all of the BrU-label represents HCV RNA since the cellular transcription is inhibited by actinomycin D treatment. The BrU-labeled RNA was present either in distinct speckle-like structures or in large spherical particles in the cytoplasm of the replicon cell (Fig. 2 ), consistent with our previous report (Mizutani et al., 2000). These two patterns probably represent two different states of viral RNA synthesis. No BrU-labeled RNA was found in Huh7 cells without an HCV replicon. SYNCRIP was also localized in the cytoplasm in Huh7 cells without HCV replicon, but in a more diffuse pattern than that of BrU label in HCV replicon cells. It was found that eGFP-SYNCRIP was partially colocalized with BrU-labeled RNA in the replicon cells (Fig. 2), indicating that only a portion of SYNCRIP was recruited to the HCV RNA replication site. This finding is consistent with the fractionation profile, which showed that SYNCRIP is primarily a cytosolic protein and that only a portion of SYNCRIP is relocalized to the DRM fractions in the replicon cells (Fig. 1). This phenomenon was also observed with PTB in the replicon cell (Aizaki et al., 2006), in which only a small portion of PTB was relocalized to the cytoplasm, whereas the majority remained in the nucleus. These results suggested that a portion of SYNCRIP is localized to the HCV replication complex, implying that SYNCRIP is involved in HCV RNA replication.
Fig. 2.
SYNCRIP colocalization with de novo-synthesized HCV RNA in a HCV replicon cell. peGFP-SYNCRIP was transfected into Huh7 or HCV replicon (HuhN1b) cells by Fugene 6. Two days after transfection, Huh7 or HCV replicon cells were labeled with BrUTP for 15 min after one-hour treatment with Actinomycin D. Actinomycin D treatment inhibited BrUTP incorporation in Huh7 cells (left two panels), but not in HuhN1b replicon cells, where BrU label was detected in the cytoplasm (right 6 panels). Immunofluorescence staining was performed with sheep polyclonal antibody against BrdU (anti-BrdU) followed by Rhodamine-conjugated anti-sheep antibody (Jackson ImmunoResearch). Two different HuhN1b cells are shown, representing two different distribution patterns of BrUTP, as shown previously (Mizutani et al., 2000).
In vivo knock-down of SYNCRIP suppressed HCV replication
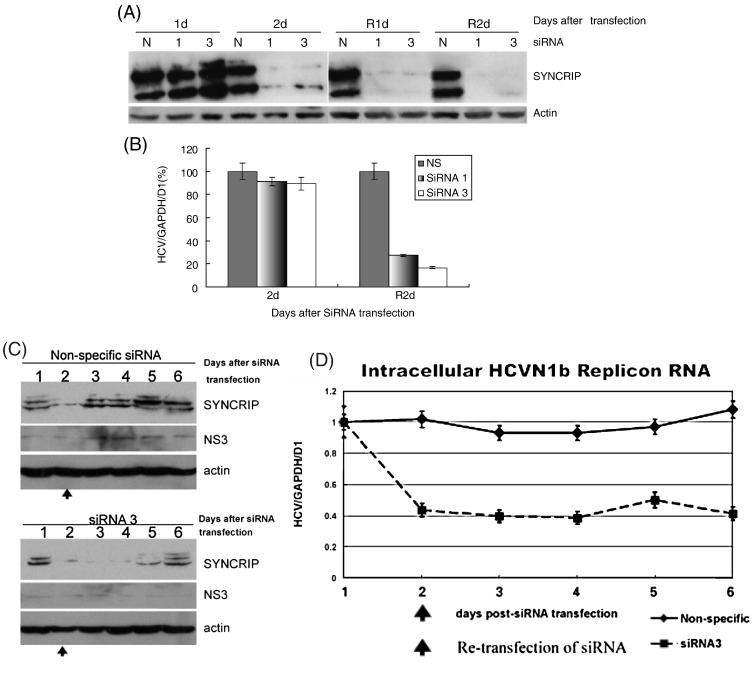
To determine the biological role of SYNCRIP in HCV RNA replication, we monitored HCV RNA levels in HCV replicon cells in which the endogenous SYNCRIP was knocked down with the RNA interference method (Aizaki et al., 2006, Wagner and Garcia-Blanco, 2002). HuhHyg replicon cells were transfected with either SYNCRIP-specific (siRNA 1 and 3) (Choi et al., 2004b) or nonspecific (NS) siRNA. Protein analysis by immunoblotting was performed with rabbit polyclonal anti-SYNCRIP antibody, and HCV RNA level was monitored by using Taqman quantitative realtime RT-PCR (Gao et al., 2004). The cells transfected with SYNCRIP siRNA showed a significant reduction of the endogenous SYNCRIP by day 2 post-transfection (Fig. 3A). One day after re-transfection with the same siRNAs respectively, almost no endogenous SYNCRIP could be detected (R1d, Fig. 3A). Correspondingly, SYNCRIP siRNA-transfected HCV replicon cells showed a 70–80% reduction of HCV RNA by day 2 after re-transfection with the siRNA as compared to that in the cells transfected with the nonspecific siRNA (R2d, Fig. 3B). The lag time of more than 1 day between the drop of SYNCRIP and that of HCV RNA was probably due to the relative stability of the HCV RNA.
Fig. 3.
SYNCRIP knock-down by siRNA in HCV replicon cells. The siRNAs against SYNCRIP (siRNA 1 and 3) (Choi et al., 2004b) or nonspecific siRNA were transfected into HCV replicon cells. Two days after the first transfection, each siRNA was re-transfected into the same cells to ensure complete knock-down of SYNCRIP. Endogenous SYNCRIP protein levels were monitored by immunoblotting (A), and HCV RNA levels were detected by realtime RT-PCR (B). (C), immunoblotting of SYNCRIP and NS3 expression in SYNCRIP siRNA knock-down replicon cells. (D), intracellular replicon RNA level was examined by realtime RT-PCR. R1d and R2d, one or 2 days after re-transfection of siRNA. siRNA 1 and 3, two different clones of siRNA; N, nonspecific siRNA.
HuhN1b replicon cells were also used to confirm the result obtained using HuhHyg replicon cells. SYNCRIP knock-down was achieved by siRNA transfection, and the viral protein and RNA levels were examined in a time-course study. While the nonspecific siRNA did not significantly affect SYNCRIP expression, the expression levels of SYNCRIP were dramatically decreased after the transfection of specific siRNA against SYNCRIP (siRNA 3) (Fig. 3C). Correspondingly, a 50% decrease in intracellular replicon RNA were detected from day 2 p.t. (Fig. 3D). The decrease in NS3 expression was also detected in siRNA 3-transfected cells; however, the viral protein was not decreased until 4 days after siRNA transfection (Fig. 3C). These results suggest that SYNCRIP affects both HCV RNA translation and RNA replication, but exerts these effects through different mechanisms.
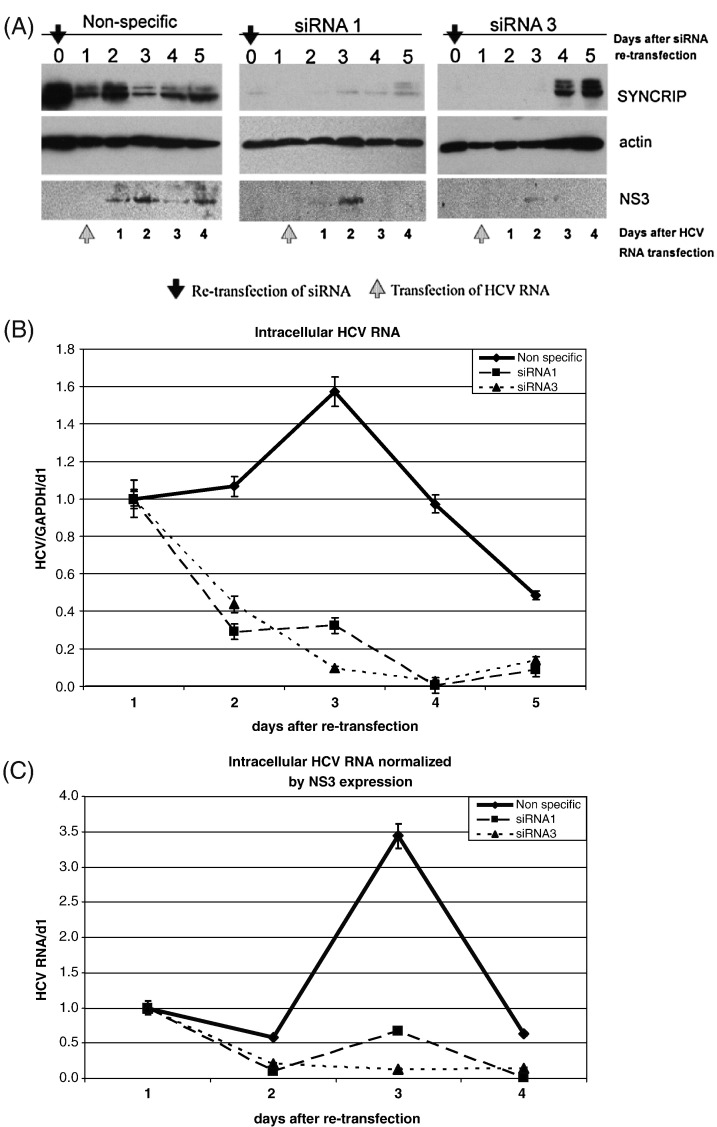
It has been reported that SYNCRIP interacts with HCV RNA fragment spanning nt 342 to 374, corresponding to the N-terminus of the core protein-coding region (Kim et al., 2004). Since this region is immediately downstream to the neomycin-phosphotransferase gene in the HCV replicon, it is possible that the observed involvement of SYNCRIP in the HuhN1b replicon cells was due to the possible effects of SYNCRIP on the expression of phosphotransferase. To rule out this possibility, we further examined the role of SYNCRIP in the replication of HCV full-length RNA (pHCV-1b-hyb) without the neomycin phosphotransferase gene. Huh7 cells were first transfected and re-transfected with SYNCRIP-specific siRNA to knock-down the endogenous SYNCRIP protein; 1 day after re-transfection of siRNA, the cells were transfected with the replication-competent full-length HCV RNA (HCV-1b). The SYNCRIP expressions and NS3 levels in siRNA-transfected cells were examined by immunoblotting. SYNCRIP expression was significantly decreased in cells transfected with specific siRNA (siRNA 1 or 3) (Fig. 4A). Correspondingly, NS3 expression was also affected by the specific SYNCRIP siRNA; in siRNA 1- and siRNA 3-transfected cells, NS3 was detected in the first 2 days post-transfection of HCV full-length RNA, but became undetectable thereafter, whereas in the nonspecific siRNA-transfected cells, NS3 was detected up to 4 days after HCV RNA transfection (Fig. 4A). Total intracellular HCV RNA was determined at various days by quantitative realtime RT-PCR. In the cells transfected with the nonspecific siRNA, HCV RNA titer gradually increased during the first 72 h post-transfection, in agreement with the published report (Choi et al., 2004a) (Fig. 4B). In contrast, in the cells transfected with the SYNCRIP-specific siRNA, HCV RNA titer decreased steadily over the same period of time (Fig. 4B). Although NS3 was detected in the first 2 days after HCV RNA transfection in SYNCRIP-knocked down cells (Fig. 4A), there was no sign of HCV replication (Fig. 4B). The intracellular HCV RNA level normalized by NS3 expression level was shown in Fig. 4C. Regardless the NS3 expression detected in SYNCRIP knock-down cells, HCV RNA titer constantly decreased after full-length HCV RNA transfection. There was a slight increase in intracellular HCV RNA level at day 5 post-transfection of HCV full-length RNA, probably due to the increase in SYNCRIP protein level. This result suggested that endogenous SYNCRIP is directly involved in HCV replication, but not through the suppression of the expression of neomycin phosphotransferase gene. These results combined indicate that SYNCRIP is involved in HCV replication by affecting either HCV RNA replication or translation, or both.
Fig. 4.
Deficiency of full-length HCV replication in SYNCRIP knock-down Huh7 cells. The siRNAs against SYNCRIP (siRNA 1 and 3) or nonspecific siRNA were transfected into Huh7 cells twice to knock-down endogenous SYNCRIP level as described in Fig. 3. One day after re-transfection of siRNA, in vitro synthesized full-length HCV-1b-hyb RNA was transfected to siRNA-transfected Huh7 cells respectively. (A), immunoblotting of endogenous SYNCRIP after siRNA re-transfection, and NS3 expression levels at various days after HCV RNA transfection. (B), intracellular HCV RNA levels determined by quantitative RT-PCR, and (C), intracellular HCV RNA levels normalized by NS3 expression levels. Huh7 cells were transfected with SYNCRIP-specific or nonspecific siRNA as in (A). One day after siRNA transfection, the replication-competent full-length HCV RNA was transfected into the cells, and HCV RNA levels were detected by realtime RT-PCR on different days after the HCV RNA transfection. The relative amounts of HCV RNA are expressed as in Fig. 3B.
SYNCRIP inhibited HCV RNA replication in vitro
The siRNA knock-down approaches showed that once SYNCRIP protein level was decreased, the HCV RNA titer would be correspondingly decreased. Since SYNCRIP has been shown to be directly involved in HCV translation (Kim et al., 2004), the inhibition of HCV RNA replication in SYNCRIP-knock-down cells may have resulted from the indirect effect of inhibition of translation; namely, the viral NS protein synthesis was inhibited, and thereby viral RNA synthesis was decreased.
To distinguish the effect of SYNCRIP on RNA replication from that on translation, we designed experiments to separate viral RNA replication from viral translation. We employed an in vitro replication assay using crude membrane fractions of the HCV subgenomic replicon cells (Ali et al., 2002, Gao et al., 2004), after the endogenous SYNCRIP had been knocked down by the siRNA approach. We also performed in vitro RNA replication assay after SYNCRIP was depleted with the anti-SYNCRIP antibody from the cell lysates.
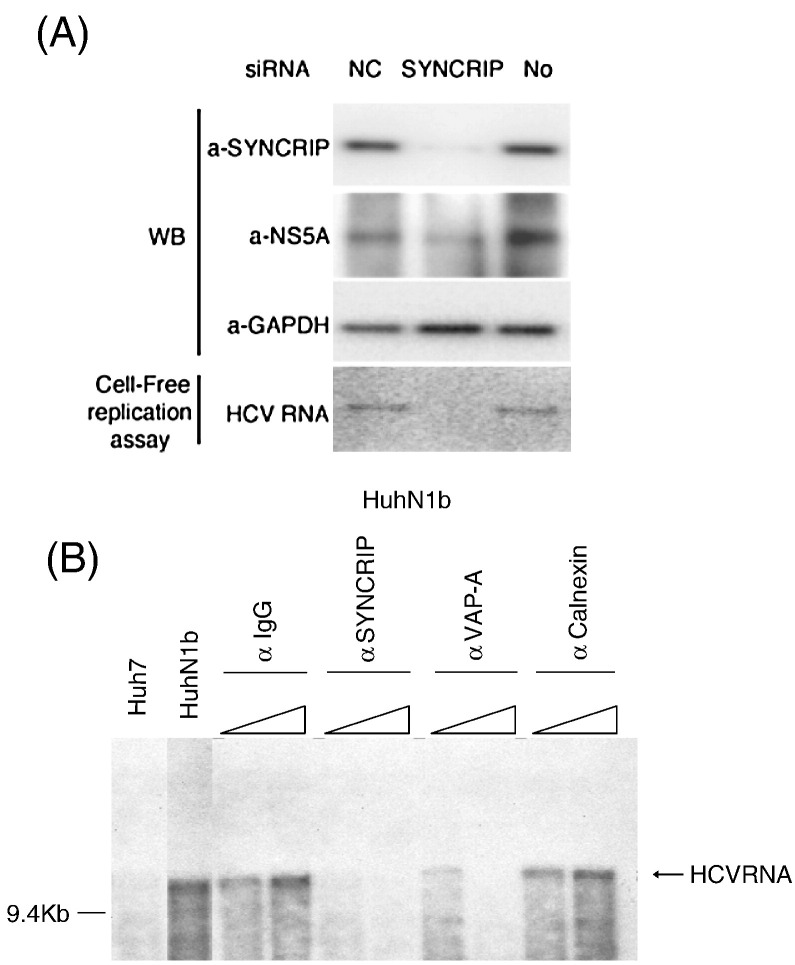
Immunoblotting showed that the amount of SYNCRIP in the HuhN1b replicon cells was substantially reduced by the specific siRNA treatment for 2 days (Fig. 5A). At this time, the amount of NS5A was only partially reduced. Cell lysates from siRNA-transfected replicon cells were treated with TX-100 at 4 °C for 30 min and fractionated by sucrose gradient centrifugation to isolate DRM fraction (Aizaki et al., 2006, Ali et al., 2002). The DRM fractions from these cell lysates were then used for in vitro replication assay. The HCV RNA synthesis was detected as single band of 32P-labeled RNA. The result showed that there was no detectable RNA replication activity at all in the DRM fractions from the SYNCRIP siRNA-transfected replicon cells when compared with those from the non-transfected or nonspecific siRNA-transfected replicon cells (Fig. 5A). Since there was still a significant amount of NS5A remaining in the siRNA-transfected cells, the total lack of the in vitro replication activity in SYNCRIP knocked-down replicon cells suggested a direct role of SYNCRIP in HCV RNA replication.
Fig. 5.
siRNA-knock-down or immunodepletion of SYNCRIP inhibited HCV replication activity in in vitro replication assay. (A) SYNCRIP was knocked down by transfection two times with specific siRNA in HCV replicon cells as in Fig. 3. SYNCRIP, NS5A and GAPDH protein expression were determined by immunoblotting. Cell-free RNA replication assays using the DRM fraction isolated from the cell lysates, as described in Material and methods, were performed. 32P-CTP-labeled HCV RNA product was detected by autoradiography after separation by agarose gel electrophoresis. NC, nonspecific siRNA control; SYNCRIP, SYNCRIP-specific siRNA; No, no siRNA transfection. (B) Partially purified lysates of HCV replicon cells were incubated with anti-IgG, anti-SYNCRIP, anti-VAP-A (VAP-33), or anti-Calnexin antibodies. Then samples were incubated with α-32P-CTP in a cell-free RNA-dependent RNA polymerase assay. The RNA product was separated by formaldehyde agarose gels and identified by autoradiography.
We further performed an immunodepletion experiment to remove SYNCRIP form the DRM fraction and assessed the effects on HCV RNA replication in vitro. For immunodepletion, the DRM fractions from replicon cell lysates were incubated with a rabbit anti-SYNCRIP polyclonal antibody to deplete the endogenous SYNCRIP from the lysate. After incubation, samples were used for cell-free synthesis of HCV RNA. The results showed that the treatment with anti-SYNCRIP antibody inhibited the replication activity in an antibody concentration-dependent manner, whereas a control anti-Ig antibody did not inhibit any activity at the same or an even higher antibody concentration (Fig. 5B and data not shown). As a control, anti-VAP33 (VAP-A) antibody also inhibited HCV RNA replication, similar to the previous result (Hamamoto et al., 2005), whereas anti-Calnexin antibody did not.
These results combined suggested that SYNCRIP is directly involved in HCV RNA replication, in addition to its role in regulating the translation of HCV RNA. Since SYNCRIP is colocalized with the newly synthesized HCV RNA, it is likely that SYNCRIP is a part of the HCV RNA replication complex and participates in viral RNA synthesis.
Discussion
Our results show that SYNCRIP can modulate HCV RNA replication. It has been previously reported that SYNCRIP can also enhance HCV IRES-dependent translation (Kim et al., 2004). Thus, similar to PTB and La autoantigen, SYNCRIP has dual functions in HCV life cycle. This may be a common characteristic of HCV RNA-binding proteins.
In our study, the relocalization of SYNCRIP to the DRM fractions in HCV replicon cells indicates that SYNCRIP is associated with the RNA replication complex, which is localized in this membrane fraction. It is interesting to note that the distribution of Calnexin was slightly different between the control and the replicon cells; there was some shift of Calnexin toward the lighter sucrose gradient fractions, probably cased by the alteration of cellular membrane structures and the associations of HCV NS proteins to ER membrane structure in HCV replicon cells (Egger et al., 2000, El-Hage and Luo, 2003, Gosert et al., 2003, Mottola et al., 2002). Nevertheless, SYNCRIP was clearly localized in the DRM fraction, whereas Calnexin was not.
The immunodepletion experiments in the current and previous studies have shown that antibody against PTB, hVAP-A, hVAP-B, and SYNCRIP can inhibit HCV RNA replication activities specifically. Previous studies have suggested that the HCV RNA replication complexes are protein complexes with the newly-synthesized RNA being contained within (Yang et al., 2004), and that subtilisin protease treatment could disrupt the replication complexes. However, it was also reported that the HCV replication complexes were resistant to proteinase K treatment at room temperature (Aizaki et al., 2004, Quinkert et al., 2005). The latter study suggested that the HCV replication complexes were very compact, and therefore the accessibility of immunoglobulin to the specific protein target in the replication complex may be limited. However, the ability of these antibodies to inhibit HCV RNA replication suggested that those complexes may not be so compact and are accessible by immunoglobulin molecules under these conditions.
The genome of positive-stranded RNA viruses, such as HCV, poliovirus, and coronavirus, serve as a template for both translation and the synthesis of negative-strand RNA, the latter of which is, in turn, the template for synthesizing more positive-strand RNA. The positive-strand RNA can also be packaged to form new viral particles. Since the same positive-strand RNA can participate in different steps of the viral life cycle, the temporal or spatial regulation is very important. It is likely that the regulation is through RNA–protein interactions. When in complex with specific RNPs, the RNA can be utilized specifically in different steps. With limited numbers of genes in the viral genome, the regulation likely requires the participation of various host factors interacting with the viral RNA or viral proteins.
There are many known host and viral RNA-binding proteins that can facilitate positive-strand RNA replication, such as Tat protein binding to the TAR structure in HIV1 RNA (Dingwall et al., 1989, Wagner and Garcia-Blanco, 2002) and poly(rC)-binding protein binding to the cloverleaf structure of poliovirus RNA (Blyn et al., 1996, Gamarnik and Andino, 2000). However, not so many RNA-binding proteins have been reported to have dual functions in viral RNA replication and translation. Recently, La and PTB, are found to regulate both RNA replication and translation of HCV, probably as a result of their ability to bind to HCV RNA (Aizaki et al., 2006, Domitrovich et al., 2005). Host factors with dual-regulatory functions may play important roles in switching the RNA from translation to replication or replication to translation. For example, PCBP regulates translation–replication switch in poliovirus life cycle (Back et al., 2002, Gamarnik and Andino, 1998). It was reported previously that stem–loop I and II are critical for HCV RNA replication, and stem–loop II, III, and IV are important for HCV RNA translation (El-Hage and Luo, 2003, Fukushi et al., 2001, Qi et al., 2003). La autoantigen was shown to bind to loop IV of HCV 5′NTR (Ali and Siddiqui, 1997). Although there is no evidence that La binds to stem–loop I or II, La can, nevertheless, regulate HCV replication (Domitrovich et al., 2005). Similarly, SYNCRIP was reported to bind at nt 342 to 374 (Kim et al., 2004), a region essential for HCV IRES-driven translation but not HCV replication. Yet we found significant positive regulatory effect of SYNCRIP in HCV RNA replication. It is possible that the binding of SYNCRIP to HCV RNA alters the secondary structure of the RNA or recruits other required factors to facilitate the assembly of the replication complex.
The mechanism of switching between translation and replication of HCV RNA is still unclear; conceivably, it may be regulated by these dual-function proteins which are involved in both replication and translation. It will be interesting to determine in the future whether the relative ratio of these proteins may trigger the switch.
Materials and methods
Cells
Huh7 cells were grown at 37 °C in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and nonessential amino acids. Huh7N1b and HuhHyg replicon cells harboring an HCV subgenomic replicon RNA derived from the HCV-N strain (Guo et al., 2001) were grown in the same medium containing 0.5 mg/ml of G418 or 100 μg/ml of Hygromycin (Mizutani et al., 2000).
Antibodies and drugs
The primary antibodies used for the analyses in this study were sheep anti-BrdU polyclonal antibody (BioDesign, ME), mouse anti-BrdU monoclonal antibody (Caltag, CA), anti-Calnexin monoclonal antibody (Abcam, MA), anti-GS27 monoclonal antibody (Abcam, MA). Brefeldin A and Nocodazole were purchased from Sigma, and Actinomycin D was from Fisher. The polyclonal anti-SYNCRIP antibody was generated in rabbits by peptide (amino acid 140 to 152) injection (Mizutani et al., 2000).
Labeling and immunofluorescence staining of de novo-synthesized viral RNA
Labeling of de novo-synthesized viral RNA, immunofluorescence staining and confocal microscopy were modified from the previously described procedures (Kanestrom et al., 1998). Briefly, Huh7 or replicon cells were plated on 8-well chamber slides at a density of 1 × 104 cells per well. Two days after seeding, cells were incubated with actinomycin D (10 μg/ml) for 1 h to inhibit cellular RNA synthesis. Subsequently, 2 mM of bromouridine triphosphate (BrUTP) was transfected into cells at 4 °C for 15 min using FuGENE 6 transfection reagent according to the manufacturer's instructions (Roche Molecular Biochemicals, IN). The cells were washed with phosphate-buffered saline (PBS) twice and cultured at 37 °C for different incubation durations with DMEM supplemented with 10% FBS. After incubation, cells were washed twice with PBS and subsequently fixed by 4% formaldehyde for 1 h at 4 °C. For permeabilization, the cells were treated with 0.1% Triton X-100 (TX-100) (Sigma-Aldrich, St. Louis, MO) in PBS supplemented with 1% FBS for 30 min at room temperature. Primary antibodies were diluted in PBS containing 1% bovine serum albumin (BSA) and incubated with cells for 1 h at room temperature. After three washes in PBS, the cells were incubated with fluorescein isothiocyanate (FITC)-conjugated or Rhodamine-conjugated secondary antibodies diluted at a 1:100 with PBS containing 5% BSA for 1 h at room temperature. The cells were then washed three times in PBS and mounted in Vectashield (Vector Laboratories, Burlingame, CA).
Membrane flotation, detergent solubilization assay
The membrane flotation assay was performed as previously described (Mizutani et al., 2000). Briefly, cells were first lysed in 1 ml of hypotonic buffer [10 mM Tris–HCV (pH 7.5), 10 mM KCl, 5 mM MgCl2] and passed through a 25-gauge needle 20 times. Nuclei and unbroken cells were removed by centrifugation at 1000 g for 5 min in microcentrifuge at 4 °C. Cell lysates were then mixed with 3 ml of 72% sucrose in low-salt buffer [LSB, comprising 50 mM Tris–HCl (pH 7.5), 25 mM KCl, and 5 mM MgCl2] and overlaid with 4 ml of 55% sucrose in LSB, followed by 1.5 ml of 10% sucrose in LSB. The sucrose gradient was centrifuged at 38,000 rpm in a Beckman SW41 Ti rotor for 14 h for 4 °C. After centrifugation, 1-ml fractions were taken from the top of the gradient, and each was added 1.7 ml of LSB to dilute sucrose and concentrated by being passed through a Centricon YM-30 filter unit (Millipore, Bedford, MA). One half of each sucrose gradient fraction was separated by 12% SDS-PAGE and transferred to nitrocellulose membrane. After blocking, the membrane was incubated with the primary antibody for 1 h at 37 °C, followed by the appropriate species-specific horseradish peroxidase conjugate, for an additional 1 h at 37 °C. Bound antibody was detected by the ECL-plus system (Amersham, Piscataway, NJ).
Transfection of siRNAs and HCV full-length RNA
The siRNAs against SYCRIP are 19-nt sequences located at nt 189–107 and nt140–1438, respectively, of SYNCRIP open reading frame (ORF) and were synthesized by Integrated DNA Technologies, Inc. (Coralville, IW). siRNAs were designed to target two different sites of the human SYNCRIP gene (5′-CUAUCGUGGUGGAUAUGAAGATT-3′, and 5′-AGACAGUGAUCUCUCUCAUGUTT-3′) chosen with the siRNA target finder software from Ambion (http://www.ambion.com/techlib/misc/siRNA_finder.html) (Choi, Mizutani, and Lai, 2004b). Replicon or Huh7 cells were grown in 10% FBS-DMEM without antibiotics. For transfection, cells were plated to a density of 105 cells per well in a 24-well plate on day 1. Three microliters of a 20-μM stock of siRNA duplex was mixed with 47 μl of Opti-MEM (Invitrogen, CA) on day 2. In a separate tube, 3 μl of Lipofectamine 2000 (Invitrogen, CA) was resuspended in 12 μl of Opti-MEM, followed by incubation at RT for 7 min. The two mixtures were combined and allowed to sit at RT for 25 min. After the incubation, 35 μl of Opti-MEM was added and the 100 μl mixture was directly added to the well containing 500 μl of growth medium. On day 3, cells were trypsinized and split into a well of the 12-well plate. On day 4, cells were re-transfected using 6 μl of siRNA with 6 μl of Lipofectamine 2000. On day 5, cells were harvested either for Western blot analysis or for RNA isolation.
pHCV-1bhyb, which contains a full-length HCV RNA hybrid sequence of genotype 1a and 1b under the control of T7 polymerase promoter, has been described previously (Choi et al., 2004a). Full-length HCV RNA was in vitro transcribed through T7 promoter to obtain a positive-sense HCV RNA of about 9.6 kb. HCV full-length RNA was then transfected into cells 1 day after siRNA re-transfection with Mirus Trans-IT mRNA transfection reagents (Mirus Bio, WI). Briefly, 1 μl of Booster reagent and 1 μl of Trans-IT reagent were added into 100 μl OPTI-MEM sequentially, followed by 1.5 μg of in vitro transcribed HCV RNA. The mixture was incubated for 3 min at RT and added into each well of a 12-well plate containing 1 ml of fresh DMEM supplemented with 10% FBS. Cellular RNA was isolated from each well at 0 to 4 days after HCV RNA transfection.
Cell-free replication assay and immunodepletion experiment
Cell lysate of replicon or control Huh7 cells were prepared by a modified protocol (Ali, Tardif, and Siddiqui, 2002). The cells grown in 100-mm-diameter dishes were washed with cold washing buffer (150 mM sucrose, 30 mM HEPES [pH 7.4], 33 mM ammonium chloride, 7 mM KCl, 4.5 mM magnesium acetate), followed by treatment with lysolecithin buffer (250 μg/ml of washing buffer) for 2 min. Three milliliters of washing buffer were added to each culture plate. The buffer was removed by aspiration. The cells were collected by scraping in 120 μl of incomplete replication buffer (100 mM HEPES [pH 7.4]; 50 mM ammonium chloride; 7 mM potassium chloride; 1 mM spermidine; 1 mM [each] ATP, GTP, and UTP; 10 μM CTP), transferred to a new tube, and lysed gently by pipetting 15 times. The cell suspension was centrifuged at 1600 rpm in a microcentrifuge for 5 min at 4 °C.
For immunodepletion experiment, 40 μl of cytoplasmic fraction (supernatant) obtained as above was treated with 1% Nonidet P-40 (NP-40) (Boehringer Mannheim, Quebec, Canada) at 4 °C for 1 h and incubated with 0.1 μg or 1.0 μg of the indicated antibody with an adjusted amount of PBS at 4 °C for 4 h with rotation. After incubation, sample was incubated with 32P-CTP (30 μCi; 800 Ci/mmol), 10 μg of actinomycin D per ml, and 800 U of RNase inhibitor per ml (Promega Corporation, Wis.) for 3 h at 30 °C. Extraction of RNA from the total mixture was performed with the TRI Regent (Molecular Research Center, Inc., Cincinnati, OH). The RNA were precipitated and eluted in 10 μl of RNase-free water. The replication products were analyzed by gel electrophoresis on 1% formaldehyde agarose gel.
References
- Aizaki H., Lee K.J., Sung V.M., Ishiko H., Lai M.M. Characterization of the hepatitis C virus RNA replication complex associated with lipid rafts. Virology. 2004;324(2):450–461. doi: 10.1016/j.virol.2004.03.034. [DOI] [PubMed] [Google Scholar]
- Aizaki H., Choi K.S., Liu M., Li Y.J., Lai M.M. Polypyrimidine-tract-binding protein is a component of the HCV RNA replication complex and necessary for RNA synthesis. J. Biomed. Sci. 2006;13(4):469–480. doi: 10.1007/s11373-006-9088-4. [DOI] [PubMed] [Google Scholar]
- Ali N., Siddiqui A. Interaction of polypyrimidine tract-binding protein with the 5′ noncoding region of the hepatitis C virus RNA genome and its functional requirement in internal initiation of translation. J. Virol. 1995;69(10):6367–6375. doi: 10.1128/jvi.69.10.6367-6375.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali N., Siddiqui A. The La antigen binds 5′ noncoding region of the hepatitis C virus RNA in the context of the initiator AUG codon and stimulates internal ribosome entry site-mediated translation. Proc. Natl. Acad. Sci. U. S. A. 1997;94(6):2249–2254. doi: 10.1073/pnas.94.6.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali N., Tardif K.D., Siddiqui A. Cell-free replication of the hepatitis C virus subgenomic replicon. J. Virol. 2002;76(23):12001–12007. doi: 10.1128/JVI.76.23.12001-12007.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Back S.H., Kim Y.K., Kim W.J., Cho S., Oh H.R., Kim J.E., Jang S.K. Translation of polioviral mRNA is inhibited by cleavage of polypyrimidine tract-binding proteins executed by polioviral 3C(pro) J. Virol. 2002;76(5):2529–2542. doi: 10.1128/jvi.76.5.2529-2542.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blyn L.B., Swiderek K.M., Richards O., Stahl D.C., Semler B.L., Ehrenfeld E. Poly(rC) binding protein 2 binds to stem–loop IV of the poliovirus RNA 5′ noncoding region: identification by automated liquid chromatography–tandem mass spectrometry. Proc. Natl. Acad. Sci. U. S. A. 1996;93(20):11115–11120. doi: 10.1073/pnas.93.20.11115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang K.S., Luo G. The polypyrimidine tract-binding protein (PTB) is required for efficient replication of hepatitis C virus (HCV) RNA. Virus Res. 2006;115(1):1–8. doi: 10.1016/j.virusres.2005.06.012. [DOI] [PubMed] [Google Scholar]
- Choi J., Lee K.J., Zheng Y., Yamaga A.K., Lai M.M., Ou J.H. Reactive oxygen species suppress hepatitis C virus RNA replication in human hepatoma cells. Hepatology. 2004;39(1):81–89. doi: 10.1002/hep.20001. [DOI] [PubMed] [Google Scholar]
- Choi K.S., Mizutani A., Lai M.M. SYNCRIP, a member of the heterogeneous nuclear ribonucleoprotein family, is involved in mouse hepatitis virus RNA synthesis. J. Virol. 2004;78(23):13153–13162. doi: 10.1128/JVI.78.23.13153-13162.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingwall C., Ernberg I., Gait M.J., Green S.M., Heaphy S., Karn J., Lowe A.D., Singh M., Skinner M.A., Valerio R. Human immunodeficiency virus 1 tat protein binds trans-activation-responsive region (TAR) RNA in vitro. Proc. Natl. Acad. Sci. U. S. A. 1989;86(18):6925–6929. doi: 10.1073/pnas.86.18.6925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domitrovich A.M., Diebel K.W., Ali N., Sarker S., Siddiqui A. Role of La autoantigen and polypyrimidine tract-binding protein in HCV replication. Virology. 2005;335(1):72–86. doi: 10.1016/j.virol.2005.02.009. [DOI] [PubMed] [Google Scholar]
- Egger D., Teterina N., Ehrenfeld E., Bienz K. Formation of the poliovirus replication complex requires coupled viral translation, vesicle production, and viral RNA synthesis. J. Virol. 2000;74(14):6570–6580. doi: 10.1128/jvi.74.14.6570-6580.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Hage N., Luo G. Replication of hepatitis C virus RNA occurs in a membrane-bound replication complex containing nonstructural viral proteins and RNA. J. Gen. Virol. 2003;84(Pt. 10):2761–2769. doi: 10.1099/vir.0.19305-0. [DOI] [PubMed] [Google Scholar]
- Fukushi S., Okada M., Kageyama T., Hoshino F.B., Nagai K., Katayama K. Interaction of poly(rC)-binding protein 2 with the 5′-terminal stem loop of the hepatitis C-virus genome. Virus Res. 2001;73(1):67–79. doi: 10.1016/s0168-1702(00)00228-8. [DOI] [PubMed] [Google Scholar]
- Gamarnik A.V., Andino R. Switch from translation to RNA replication in a positive-stranded RNA virus. Genes Dev. 1998;12(15):2293–2304. doi: 10.1101/gad.12.15.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamarnik A.V., Andino R. Interactions of viral protein 3CD and poly(rC) binding protein with the 5′ untranslated region of the poliovirus genome. J. Virol. 2000;74(5):2219–2226. doi: 10.1128/jvi.74.5.2219-2226.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L., Aizaki H., He J.W., Lai M.M. Interactions between viral nonstructural proteins and host protein hVAP-33 mediate the formation of hepatitis C virus RNA replication complex on lipid raft. J. Virol. 2004;78(7):3480–3488. doi: 10.1128/JVI.78.7.3480-3488.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosert R., Egger D., Lohmann V., Bartenschlager R., Blum H.E., Bienz K., Moradpour D. Identification of the hepatitis C virus RNA replication complex in Huh-7 cells harboring subgenomic replicons. J. Virol. 2003;77(9):5487–5492. doi: 10.1128/JVI.77.9.5487-5492.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J.T., Bichko V.V., Seeger C. Effect of alpha interferon on the hepatitis C virus replicon. J. Virol. 2001;75(18):8516–8523. doi: 10.1128/JVI.75.18.8516-8523.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamamoto I., Nishimura Y., Okamoto T., Aizaki H., Liu M., Mori Y., Abe T., Suzuki T., Lai M.M., Miyamura T., Moriishi K., Matsuura Y. Human VAP-B is involved in hepatitis C virus replication through interaction with NS5A and NS5B. J. Virol. 2005;79(21):13473–13482. doi: 10.1128/JVI.79.21.13473-13482.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T., Lai M.M. An internal polypyrimidine-tract-binding protein-binding site in the hepatitis C virus RNA attenuates translation, which is relieved by the 3′-untranslated sequence. Virology. 1999;254(2):288–296. doi: 10.1006/viro.1998.9541. [DOI] [PubMed] [Google Scholar]
- Kanestrom A., Andresen V., Szilvay A.M., Kalland K.H., Haukenes G. Histographic recording of human immunodeficiency virus type 1 (HIV-1) regulatory protein Rev and nuclear factors. Arch. Virol. 1998;143(2):279–294. doi: 10.1007/s007050050286. [DOI] [PubMed] [Google Scholar]
- Kapadia S.B., Chisari F.V. Hepatitis C virus RNA replication is regulated by host geranylgeranylation and fatty acids. Proc. Natl. Acad. Sci. U. S. A. 2005;102(7):2561–2566. doi: 10.1073/pnas.0409834102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.H., Paek K.Y., Ha S.H., Cho S., Choi K., Kim C.S., Ryu S.H., Jang S.K. A cellular RNA-binding protein enhances internal ribosomal entry site-dependent translation through an interaction downstream of the hepatitis C virus polyprotein initiation codon. Mol. Cell. Biol. 2004;24(18):7878–7890. doi: 10.1128/MCB.24.18.7878-7890.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani A., Fukuda M., Ibata K., Shiraishi Y., Mikoshiba K. SYNCRIP, a cytoplasmic counterpart of heterogeneous nuclear ribonucleoprotein R, interacts with ubiquitous synaptotagmin isoforms. J. Biol. Chem. 2000;275(13):9823–9831. doi: 10.1074/jbc.275.13.9823. [DOI] [PubMed] [Google Scholar]
- Mottola G., Cardinali G., Ceccacci A., Trozzi C., Bartholomew L., Torrisi M.R., Pedrazzini E., Bonatti S., Migliaccio G. Hepatitis C virus nonstructural proteins are localized in a modified endoplasmic reticulum of cells expressing viral subgenomic replicons. Virology. 2002;293(1):31–43. doi: 10.1006/viro.2001.1229. [DOI] [PubMed] [Google Scholar]
- Qi Z.T., Kalkeri G., Hanible J., Prabhu R., Bastian F., Garry R.F., Dash S. Stem–loop structures II–IV of the 5′ untranslated sequences are required for the expression of the full-length hepatitis C virus genome. Arch. Virol. 2003;148(3):449–467. doi: 10.1007/s00705-002-0933-0. [DOI] [PubMed] [Google Scholar]
- Quinkert D., Bartenschlager R., Lohmann V. Quantitative analysis of the hepatitis C virus replication complex. J. Virol. 2005;79(21):13594–13605. doi: 10.1128/JVI.79.21.13594-13605.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed K.E., Rice C.M. Overview of hepatitis C virus genome structure, polyprotein processing, and protein properties. Curr. Top. Microbiol. Immunol. 2000;242:55–84. doi: 10.1007/978-3-642-59605-6_4. [DOI] [PubMed] [Google Scholar]
- Wagner E.J., Garcia-Blanco M.A. RNAi-mediated PTB depletion leads to enhanced exon definition. Mol. Cell. 2002;10(4):943–949. doi: 10.1016/s1097-2765(02)00645-7. [DOI] [PubMed] [Google Scholar]
- Yang G., Pevear D.C., Collett M.S., Chunduru S., Young D.C., Benetatos C., Jordan R. Newly synthesized hepatitis C virus replicon RNA is protected from nuclease activity by a protease-sensitive factor(s) J. Virol. 2004;78(18):10202–10205. doi: 10.1128/JVI.78.18.10202-10205.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]