Abstract
The examination of 137 non-O1/O139 Vibrio cholerae isolates from Newport Bay, California, indicated the presence of diverse genotypes and a temporal succession. Unexpectedly, the cholera toxin gene (ctxA) was found in 17% of the strains, of which one-third were also positive for the zot gene. This suggests that ctxA is prevalent in the region of nonepidemicity and is likely to have an environmental origin.
Vibrio cholerae, a gram-negative bacterium belonging to the genus Vibrio, is the causative agent of the severe dehydrating diarrheal disease cholera. Contrary to the traditional belief that V. cholerae is a solely clinical bacterium that only survives in the aquatic environment for a short time, V. cholerae is now known to be indigenous to brackish waters (5). While it has not caused a major outbreak in the United States for nearly half a century, V. cholerae has been isolated from water bodies in many coastal regions (13-15, 18, 21, 22, 25). However, it is a common belief (based on the results of immunological and biological tests) (6, 9, 20, 28) that most of the environmental strains do not produce cholera toxin (CT) and therefore are of negligible importance in epidemic potential. CT is encoded by a transferable filamentous phage, CTXΦ (30), and previous reports have implied the acquisition of these CT genes under conditions similar to those of the aquatic environment (9, 10).
The presence of virulence genes among environmental strains of V. cholerae was examined among isolates from freshwater lakes and ponds in the eastern part of Calcutta, India (3). Virulence genes, including ctxAB, were found among environmental strains. However, since the India subcontinent is an area in which cholera is endemic, possibly these virulence genes are contributed by human waste from a diseased population. When a limited number of isolates was used, the occurrence of ctxA was also found among 10% of non-O1/O139 environmental isolates from coastal Brazil (24). Here we report on the diversity and prevalence of virulence genes ctxA and zot among non-O1/O139 V. cholerae isolates from the Newport Bay, California, watershed, a region of nonepidemicity.
A total of 137 V. cholerae isolates from San Diego Creek and Newport Bay, California, obtained over a year were examined (15). Each isolate was given a 7-digit code. The first two characters indicate the isolation site; the following four digits indicate the month and year of isolation. The final character indicates the identity of individual strains. For example, C10899a indicates strain a isolated from site C1 in August 1999. Water samples for isolation were taken monthly from seven sites. Sites C1 to C3 were located in San Diego Creek, a major tributary of the Newport Bay, with an additional three sites (UC, UNB, and BC) spread throughout upper and low Newport Bay. The seventh site was at the Pacific Ocean front at the Balboa Pier (BP). There is no known source of direct sewage influence to any of the sampling sites. However, all locations are influenced by urban runoff during the winter rainy season. Since there has not been a case of cholera in the area for over 50 years, the level of occurrence of toxigenic V. cholerae in sewage and urban runoff is expected to be low or nonexistent. Details on sampling locations and isolation and confirmation methods can be found in the report of Jiang and Fu (15).
Isolates were cultured in Luria-Bertani broth (Difco Inc.), and genomic DNA was extracted using a Wizard genomic DNA purification kit and protocol (Promega). DNA purity and quantity were determined using UV spectrophotometry (Beckman). DNA extracts were stored at 4°C for less than 2 days before fingerprint analysis and PCR analysis of virulence genes were conducted.
Enterobacterial repetitive intergenic consensus sequence-PCR (ERIC-PCR) genomic fingerprinting was performed essentially as described by Rivera et al. (23), with minor modifications. Briefly, bacterial genomic DNA was heated to 65°C for 10 min before conducting PCR. Two primers (ATGTAAGCTCCTGGGGATTCAC and AAGTAAGTGACTGGGGTGAGCG) for PCR were used in the program and under the conditions described by Rivera et al. (23). Each 50-μl reaction mixture contained 1× PCR buffer, deoxynucleoside triphosphates (50 nM each), 50 pmol of each primer, and 1.25 U of Taq DNA polymerase (Perkin Elmer). Two V. cholerae reference strains, O1 El Tor nontoxigenic (isolated from Mexico) and classical (ATCC 11623), were used as internal controls to ensure run-to-run reproducibility. A 1-kb DNA stepladder and a 200-bp DNA stepladder (Promega) were run at multiple gel locations for correction of distortion and normalization between gels. Digitized fingerprints were analyzed using GelCompar II (Applied Maths, Sint-Martens-Latem, Belgium) software, following the manufacturer's instructions. In brief, fingerprint types were defined, target lanes were searched, and manually refined, gel-to-gel variations were normalized on the basis of external references (molecular-weight markers run at multiple locations on the gels) and within-gel common bands were aligned using internal references. The bands were selected using the autosearch function and then eye refined. A band-based comparison (employing the similarity coefficient defined by Jaccard) was used to create a similarity matrix. The clustering method of Ward was used to create the dendrogram. The Ward method is intended for interval-scaled measurements and makes use of Euclidean distances (19). A band position tolerance of 2% was allowed in the comparison.
Amplification of ctxA and zot genes was performed as previously described (12, 24). The primers for ctxA were 94F (CGG GCA GATTCT AGA CCT CCT G) and 614R (CGA TGA TCT TGG AGC ATT CCC AC). The primers for zot were 225F (TCG CTT AAC GAT GGC GCG TTT T) and 1129R (AAC CCC GTT TCA CTT CTA CCC A). Each 25-μl reaction mixture contained 1× PCR buffer, deoxynucleoside triphosphates (50 nM each), 0.8 μM of each primer, and 0.75 U of Taq DNA polymerase. Amplifications were performed at 94°C for 2 min followed by 30 cycles of 94°C for 1 min, 60°C for 1 min, and 72°C for 1 min, with a final extension at 72°C for 10 min.
For ctxA confirmation, PCR products were Southern transferred to nylon membranes and hybridized with a γ-P32-end-labeled internal probe (ACGGGATTTGTTAGGCACG). Hybridizations were carried out at 45°C overnight and stringently washed twice with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 1% sodium dodecyl sulfate, and 0.05% sodium pyrophosphate at 55°C for 1 h each time. Positive hybridizations were determined by autoradiography.
For sequence analysis, PCR amplicons were gel excised and purified using a QIAquick gel purification kit (Qiagen Inc., Valencia, Calif.), cloned into pGEM-T, and transformed into Escherichia coli DH5α-competent cells, following the instructions of the manufacturer (Promega). Positive transformants (white colonies) were picked and reamplified to confirm the presence of an insert. Plasmid DNA was purified using a QIAquick mini prep kit (Qiagen Inc.), and inserts were sequenced bidirectionally using an ABI BigDye Terminator Cycle Sequencing Ready Reaction kit (Applied Biosystems, Foster City, Calif.) and following the manufacturer's recommendations. Multiple alignments were performed using ClustalX (27) with default settings.
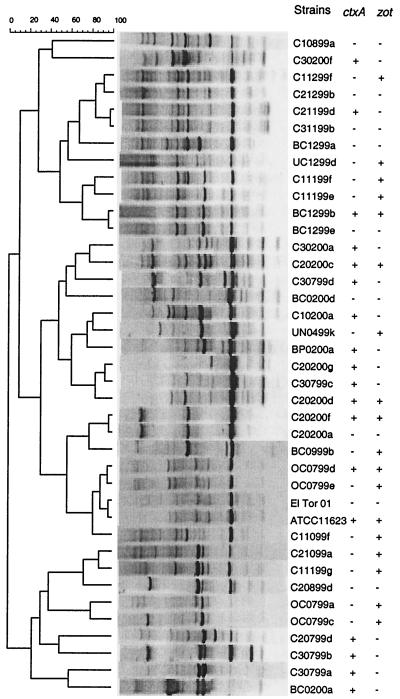
Examination (using ERIC-PCR) (23) of 137 environmental isolates yielded 104 unique fingerprint patterns. Figure 1 shows 37 of 104 unique fingerprints that represent the grouping pattern. The figure was first produced to include all strains and duplicates; selective toxin gene-negative strains were subsequently removed to reduce the size of the figure without dramatically impacting the groupings. All strains positive for at least one toxin gene, as well as toxin-negative strains that had very similar fingerprints to those of toxin-positive strains, were left in the figure. No environmental isolate had a pattern identical to those of either O1 classical or nontoxigenic O1 El Tor V. cholerae strains. Similarity analysis grouped all isolates into three major clusters. There was no clear separation of isolates from different locations. However, the groupings related to the date of isolation. For example, most isolates obtained during the winter of 1999 (November and December) were grouped together and were more closely related to the group primarily containing isolates from February and March 2000 than to the group of isolates from the summer and fall of 1999 (July, August, and October). This result is in agreement with the results of a previous study of genetic diversity of V. cholerae isolates from the Chesapeake Bay (16). The seasonal succession of genotypes of environmental V. cholerae is likely linked to changes in environmental conditions such as temperature, salinity, radiation, and nutrients and in the species of phytoplankton and zooplankton.
FIG. 1.
Diversity of environmental V. cholerae isolates from Newport Bay watershed revealed by ERIC-PCR fingerprinting. A total of 37 representative patterns of the 104 unique fingerprints generated from 137 isolates are shown. An O1 classical toxigenic strain (ATCC 11623) and an O1 El Tor nontoxigenic strain were included as references. ctxA- and zot-positive strains were detected by PCR, and the presence of ctxA was further confirmed by internal probe hybridization.
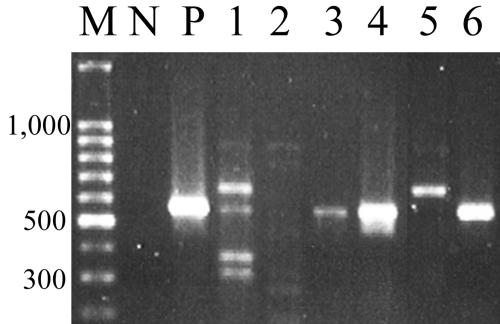
PCR amplification (using primers originally designed by Fields et al.) (12) of ctxA in many cases yielded amplicons that were either bigger or smaller than the expected size or yielded multiple amplicons (Fig. 2). To rule out the possibility of cross-contamination during genomic DNA purification and PCR, ctxA-positive environmental strains were retested multiple times in the absence of the positive-control strains. Hybridizations with a probe internal to a ctxA amplicon confirmed the presence of ctxA in 18 of the 104 strains tested (Table 1). Positive hybridizations were detected only with amplicons of the expected size, and only one amplicon of the expected size failed to hybridize with the internal probe.
FIG. 2.
Amplification (using primers specific for the ctxA gene) of environmental non-O1/O139 V. cholerae isolates from Newport Bay, California. A subset of representative amplicons from environmental isolates is shown. M, molecular mass ladder (in base pairs); N, negative control; P, positive control (ATCC11623). Lane 1, strain BC1299b; lane 2, BC0999b; lane 3, C20200d; lane 4, C20200f; lane 5, C10899a; lane 6, C10200a.
TABLE 1.
Occurrence of ctxA and zot genes among non-01/0139 environmental Vibrio cholerae isolates
| Isolate characteristics | No. of isolates | % of total |
|---|---|---|
| ctxA positive and zot positive | 6 | 5.6 |
| ctxA positive and zot negative | 12 | 11.5 |
| ctxA negative and zot positive | 12 | 11.5 |
| ctxA negative and zot negative | 74 | 71.1 |
Sequencing analysis of two randomly selected ctxA amplicons (GenBank accession numbers AY376267 and AY376268) showed only 1- and 4-bp differences, respectively, from the sequence of El Tor strain N16961 (GenBank accession number AE004224) among 564-bp sequences. Multiple alignments with ctxA genes of environmental isolates from South Korea (GenBank accession AD175708), China (AF516341 and AF516349), and India (AF414369) also showed high degrees of similarity, with differences ranging from 1 to 8 bp. These results suggest that ctxA genes (found among environmental V. cholerae isolates, including isolates from Newport Bay, California) are highly conserved and similar to those found in clinical isolates. However, only one-third of the ctxA-positive strains from Newport Bay were also PCR positive for the CTXΦ structural gene zot, while 12 zot-positive strains were negative for ctxA (Table 1). These results indicate that a portion of the CTXΦ prophage genome may be missing or may have been disrupted by deletion or insertion, suggesting that many CTXΦ genes among the environmental strains are likely defective. Defective prophage genomes are commonly found among all genera of bacteria (29). Approximately 6% of the strains from Newport Bay were positive for both ctxA and zot genes.
It is also interesting that isolates with nearly identical ERIC-PCR fingerprints (i.e., strains C21199d and C31199b and strains BC1299b and BC1299e) are different with respect to the harboring of toxin genes (Fig. 1). This result parallels the results of a previous study using a high-resolution DNA fingerprinting method to show that clinical toxigenic V. cholerae isolates are closely related to nontoxigenic environmental strains (17) and further suggests that CT genes are highly mobile among environmental isolates.
CTXΦ transfer among clinical strains requires the presence of the toxin-coregulated pilus (TCP) as receptor. Since TCP expression is optimal in the gastrointestinal tract, it has been hypothesized that the acquisition of CTXΦ occurs within the human host (4, 26). How ctxA genes are spread in an aquatic environment in an area of nonepidemicity is unclear. It is possible that a different mechanism of gene transfer operates for V. cholerae in aquatic environments. Transfer of CTXΦ via general transduction to TCP-negative strains has been demonstrated in the laboratory (2). More interestingly, Faruque et al. (8, 11) also showed that both a TCP-negative environmental V. cholerae isolate and strains of TCP-negative Vibrio mimicus were susceptible to infection by filamentous phage CTXΦ and formed stable lysogens. Therefore, the evidence presented above supports the hypothesis of the existence of a TCP-independent mechanism for infection by CTXΦ. Furthermore, the spread of CT genes in the environment can be facilitated by the exposure of CTXΦ-positive strains to sunlight (7). Studies have shown both increased rates of phage production and transduction to nontoxigenic strains when induced by sunlight.
It is presently unclear whether the CT genes among these environmental isolates are expressed or what their biological and ecological function is in the aquatic environment. Approximately 6% of isolates from Newport Bay, California, may contain a functional CTXΦ. Since a large portion of the Newport Bay is used as a recreational resort, the occurrence of toxigenic V. cholerae here raises a question regarding potential risk of human exposure. A rare cholera outbreak in Louisiana in the United States in September of 1978 was attributed to environmental strains in shellfish collected from the Gulf of Mexico (1). Therefore, the study presented here (in combination with the increasing body of literature reflecting environmental V. cholerae research) supports the idea that CT has an environmental origin and that the complex aquatic environment can give rise to pathogenic V. cholerae.
Nucleotide sequence accession numbers.
The sequences determined in this work were deposited into the National Center for Biotechnology Information (GenBank) under the following accession numbers: AY376267 and AY376268.
Acknowledgments
Funding for this research was provided by a University of California Young Faculty Career Development Award and the Research and Travel Fund to S.J.
REFERENCES
- 1.Blake, P. A., D. T. Allegra, J. D. Snyder, T. J. Barrett, L. McFarland, C. T. Caraway, J. C. Feeley, J. P. Craig, J. V. Lee, N. D. Puhr, and R. A. Feldman. 1980. Cholera—a possible endemic focus in the United States. N. Engl. J. Med. 302:305-309. [DOI] [PubMed] [Google Scholar]
- 2.Boyd, E. F., and M. K. Waldor. 1999. Alternative mechanism of cholera toxin acquisition by Vibrio cholerae: generalized transduction of CTXΦ by bacteriophage CP-T1. Infect. Immun. 67:5898-5905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chakraborty, S., A. K. Mukhopadhyay, R. K. Bhadra, A. N. Ghosh, R. Mitra, T. Shimada, S. Yamasaki, S. M. Faruque, Y. Takeda, R. R. Colwell, and G. B. Nair. 2000. Virulence genes in environmental strains of Vibrio cholerae. Appl. Environ. Microbiol. 66:4022-4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chiang, S. L., and J. J. Mekalanos. 1999. Horizontal gene transfer in the emergence of virulent Vibrio cholerae, p. 156-169. In E. Rosenberg (ed.), Microbial ecology and infectious disease. American Society for Microbiology, Washington, D.C.
- 5.Colwell, R. R., and W. M. Spira. 1992. The ecology of Vibrio cholerae, p. 107-127. In D. Barua and W. B. Greenough (ed.), Cholera. Plenum, New York, N.Y.
- 6.DePaola, A., M. W. Presnell, M. L. Motes, R. M. McPhearson, R. M. Twedt, R. E. Becker, and S. Zywmo. 1983. Non-O1 Vibrio cholerae in shellfish, sediment and waters of the U.S. Gulf Coast. J. Food Prot. 46:802-806. [DOI] [PubMed] [Google Scholar]
- 7.Faruque, S. M., Asadulghani, M. M. Rahman, M. K. Waldor, and D. A. Sack. 2000. Sunlight-induced propagation of the lysogenic phage encoding cholera toxin. Infect. Immun. 68:4795-4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Faruque, S. M., Asadulghani, M. N. Saha, A. Abdul Alim, M. J. Albert, K. M. Nasirul Islam, and J. J. Mekalanos. 1998. Analysis of clinical and environmental strains of nontoxigenic Vibrio cholerae for susceptibility to CTXΦ: molecular basis for origination of new strains with epidemic potential. Infect. Immun. 66:5819-5825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Faruque, S. M., M. J. Albert, and J. J. Mekalanos. 1998. Epidemiology, genetics, and ecology of toxigenic Vibrio cholerae. Microbiol. Mol. Biol. Rev. 62:1301-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Faruque, S. M., and G. B. Nair. 2002. Molecular ecology of toxigenic Vibrio cholerae. Microbiol. Immunol. 46:59-66. [DOI] [PubMed] [Google Scholar]
- 11.Faruque, S. M., M. M. Rahman, Asadulghani, K. M. Nasirul Islam, and J. J. Mekalanos. 1999. Lysogenic conversion of environmental Vibrio mimicus strains by CTXΦ. Infect. Immun. 67:5723-5729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fields, P. I., T. Popovic, K. Wachsmuth, and O. Olsvik. 1992. Use of polymerase chain reaction for detection of toxigenic Vibrio cholerae 01 strains from the Latin American cholera epidemic. J. Clin. Microbiol. 30:2118-2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hood, M. A., G. E. Ness, G. E. Rodrick, and N. J. Blake. 1983. Distribution of Vibrio cholerae in two Florida estuaries. Microb. Ecol. 9:65-75. [DOI] [PubMed] [Google Scholar]
- 14.Jiang, S. C. 2001. Vibrio cholerae in recreational beach waters and tributaries of Southern California. Hydrobiologia 460:157-164. [Google Scholar]
- 15.Jiang, S. C., and W. Fu. 2001. Detection of Vibrio cholerae in coastal waters by a 16S-23S intergenic spacer probe. Microb. Ecol. 42:540-548. [DOI] [PubMed] [Google Scholar]
- 16.Jiang, S. C., V. Louis, N. Choopun, A. Sharma, A. Huq, and R. R. Colwell. 2000. Genetic diversity of Vibrio cholerae in Chesapeake Bay determined by amplified fragment length polymorphism fingerprinting. Appl. Environ. Microbiol. 66:140-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang, S. C., M. Matte, G. Matte, A. Huq, and R. R. Colwell. 2000. Genetic diversity of clinical and environmental isolates of Vibrio cholerae determined by amplified fragment length polymorphism fingerprinting. Appl. Environ. Microbiol. 66:148-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaper, J. H., Lockman, R. R. Colwell, and S. W. Joseph. 1979. Ecology, serology, and enterotoxin production of Vibrio cholerae in Chesapeake Bay. Appl. Environ. Microbiol. 37:91-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaufman, L., and P. J. Rousseeuw. 1990. Finding groups in data: an introduction to cluster analysis. Wiley, New York, N.Y.
- 20.Kaysner, C. A., C. Abeyta, Jr., M. M. Wekell, A. DePaola, Jr., R. F. Stott, and J. M. Leitch. 1987. Incidence of Vibrio cholerae from estuaries of the United States West Coast. Appl. Environ. Microbiol. 53:1344-1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kenyon, J. E., D. C. Gillies, D. R. Piexoto, and B. Austin. 1983. Vibrio cholerae (non-O1) isolated from California coastal waters. Appl. Environ. Microbiol. 46:1232-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Portillo-Lopez, A., and M. L. Lizarraga-Partida. 1997. Detection of Vibrio cholerae 01 in different habitats of Todos Santos Bay, Baja California, Mexico. Cinc. Mar. 23:435-447. [Google Scholar]
- 23.Rivera, I. G., M. A. R. Chowdhury, A. Huq, D. Jacobs, M. T. Martins, and R. R. Colwell. 1995. Enterobacterial repetitive intergenic consensus sequences and the PCR to generate fingerprints of genomic DNAs from Vibrio cholerae O1, O139, and non-O1 strains. Appl. Environ. Microbiol. 61:2898-2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rivera, I. N. G., J. Chun, A. Huq, R. B. Sack, and R. R. Colwell. 2001. Genotypes associated with virulence in environmental isolates of Vibrio cholerae. Appl. Environ. Microbiol. 67:2421-2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roberts, N. C., R. J. Siebeling, J. B. Kaper, and H. B. Bradford. 1982. Vibrios in the Louisiana Gulf Coast environment. Microb. Ecol. 8:299-312. [DOI] [PubMed] [Google Scholar]
- 26.Rubin, E. J., M. K. Waldor, and J. J. Mekalanos. 1998. Mobile genetic elements and the evolution of new epidemic strains of Vibrio cholerae, p. 147-161. In R. M. Krause (ed.), Emerging infections. Academic Press, San Diego, Calif.
- 27.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higins. 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Twedt, R. M., J. M. Madden, J. M. Hunt, D. W. Francis, J. T. Peeler, A. P. Duran, W. O. Herbert, S. G. McCay, C. N. Roderick, G. T. Spite, and T. J. Wazenski. 1981. Characterization of Vibrio cholerae isolated from oysters. Appl. Environ. Microbiol. 41:1475-1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Villarreal, L. P. 1999. DNA viruses contribute to host evolution. In E. Domingo (ed.), Origin and evolution of viruses. Academic Press, San Diego, Calif.
- 30.Waldor, M. K., and J. J. Mekalanos. 1996. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science 272:1910-1914. [DOI] [PubMed] [Google Scholar]