Abstract
Background/Aims
Several studies have shown that regions of hypoxia develop in the liver during chronic injury. Furthermore, it has been demonstrated that hypoxia stimulates the release of mediators from hepatic stellate cells (HSCs) that may affect the progression of fibrosis. The mechanism by which hypoxia modulates gene expression in HSCs is not known. Recent studies demonstrated that the hypoxia-activated transcription factor, hypoxia-inducible factor-1α (HIF-1α), is critical for the development of fibrosis. Accordingly, the hypothesis was tested that HIF-1α is activated in HSCs and regulates expression of genes important for HSC activation and liver fibrosis.
Methods
HSCs were isolated from mice and exposed to hypoxia. HIF-1α and HIF-2α activation were measured, and gene expression analyzed by gene array analysis. To identify genes regulated by HIF-1α, HSCs were isolated from Control and HIF-1α-Deficient mice.
Results
Exposure of primary mouse HSCs to 0.5% oxygen activated HIF-1α and HIF-2α. mRNA levels of numerous genes were increased in HSCs exposed to 0.5% oxygen, many of which are important for HSC function, angiogenesis, and collagen synthesis. Of the mRNAs increased, Ccr1, Ccr5, macrophage migration inhibitory factor, interleukin-13 receptor α1, prolyl-4-hydroxylase α2 (PHD α2) were completely HIF-1α-dependent. Upregulation of VEGF and placental growth factor were partially HIF-1α-dependent and upregulation of angiopoietin-like 4 and PHD α1 were HIF-1α-independent.
Conclusions
Results from these studies demonstrate that hypoxia, through activation of HIF-1α, regulates expression of genes that may alter the sensitivity of HSCs to certain activators and chemotaxins, and regulates expression of genes important for angiogenesis and collagen synthesis.
Keywords: Hypoxia-inducible factor, hepatic stellate cells, hypoxia
Hepatic stellate cells in the liver play a key role in the development of fibrosis (1). During chronic liver injury caused by alcohol, hepatitis virus, genetic disorders, among others, mediators are released that stimulate hepatic stellate cells to differentiate into myofibroblasts and stimulate them to proliferate and produce collagen, a key component of fibrosis (2). In addition to soluble mediators as important activators of hepatic stellate cells, alterations in the hepatic microenvironment may modulate hepatic stellate cell function. For example, alterations in matrix stiffness and changes in interactions between integrins on hepatic stellate cells and the extracellular matrix may affect the activation state of these cells (3, 4). In addition to these microenvironmental influences, exposure of hepatic stellate cells to hypoxia during chronic injury may be an important stimulus of hepatic stellate cell activation.
Several studies have shown that regions of hypoxia develop in the liver during chronic injury (5–7). Furthermore, it has been demonstrated that hypoxia stimulates the release of a variety of mediators from hepatic stellate cells that may affect the progression of fibrosis. For example, hepatic stellate cells exposed to hypoxia synthesize and release the proangiogenic mediators, vascular endothelial growth factor (VEGF) and angiopoietin-1 (8, 9). Studies have demonstrated that angiogenesis may be important for liver fibrosis development, and hepatic stellate cells may be an important source of proangiogenic mediators (10). In addition, exposure of the human hepatic stellate cell line, LX-2 cells, to hypoxia increased levels of α-smooth muscle actin and type I collagen, suggesting that hypoxia also promotes activation of these cells (11). Similarly exposure of culture activated rat hepatic stellate cells to hypoxia stimulated synthesis of type I collagen (7). Although these studies have demonstrated that hypoxia affects hepatic stellate cell function, what remains unknown is the mechanism by which hypoxia modulates gene expression in these cells.
One mechanism by which hypoxia modulates gene expression is by activating a number of transcription factors, including hypoxia-inducible factors (HIFs). HIFs are composed of an alpha subunit, typically HIF-1α or HIF-2α, and a beta subunit, HIF-1β also called the arylhydrocarbon receptor nuclear translocator (12, 13). In normoxic cells, HIFα subunits are constitutively produced and immediately targeted for proteolytic degradation. When cells become hypoxic, however, the mechanisms that target HIFα subunits for degradation are inhibited allowing HIFα protein levels to increase (14). HIFα subunits then translocate to the nucleus where they heterodimerize with HIF-1β and regulate expression of genes involved in glycolysis, angiogenesis, iron metabolism, pH control, and others functions. Studies have demonstrated that HIF-1α is activated in the transformed hepatic stellate cell line, LX-2 cells (11). Whether HIF-1α is activated in primary hepatic stellate cells and regulates expression of genes important for hepatic stellate cell function and fibrogenesis, however, has not been investigated. Accordingly, in the present studies the hypothesis was tested that HIF-1α is activated in hepatic stellate cells and regulate expression of genes important for hepatic stellate cell activation and liver fibrosis.
Materials and Methods
Mice
For studies utilizing wild-type mice, male C57BL/6 (Harlan, Madison, WI) mice, 8–12 weeks of age, were used. To selectively reduce HIF-1α levels in adult mice, HIF-1αfl/fl mice described in detail previously (15), were crossed with mice expressing Cre recombinase under control of the Mx interferon-inducible promoter (16) (Mx-Cre+/− mice; Jackson Laboratories, Bar Harbor, ME). This approach was used because deletion of HIF-1α in mice causes embryonic lethality (17). Offspring of the HIF-1αfl/fl mouse breeding were HIF-1αfl/fl-Mx-Cre+ (i.e., HIF-1α-Deficient) or HIF-1αfl/fl-Mx-Cre- (i.e., HIF-1α-Control littermate controls). PCR of genomic DNA was used to detect the floxed HIF-1α gene and the Cre transgene as described previously (15). To activate the MxCre promoter, HIF-1αfl/fl-Mx-Cre+ and HIF-1αfl/fl-Mx-Cre- mice were treated with 500 μg of polyinosinic-polycytidylic acid (pIpC, Sigma Chemical Company, St. Louis, MO) dissolved in sterile saline by intraperitoneal injection every three days for a total of three injections. In mice containing the MxCre transgene, this treatment causes a near complete deletion of loxP-containing genes in liver and immune organs and a partial deletion in other tissues (16, 18). In HIF-1αfl/fl-Mx-Cre+ mice, this treatment results in deletion of exons 13–15 in the HIF-1α gene which encode for the COOH-terminal transactivation domain and a portion of the nuclear localization sequence, both of which are essential for hypoxia responsiveness by HIF-1α (15). Hepatic stellate cells were isolated from these mice at least one week after the final pIpC injection.
All mice were maintained on a 12-h light/dark cycle under controlled temperature (18–21°C) and humidity. Food (Rodent Chow; Harlan-Teklad, Madison, WI) and tap water were allowed ad libitum. All procedures on animals were carried out in accordance with the Guide for the Care and Use of Laboratory Animals promulgated by the National Institutes of Health and were approved by the institutional IACUC committee at the University of Kansas Medical Center.
Hepatic Stellate Cell Isolation
Hepatic stellate cells were isolated from the livers of mice by dual collagenase and pronase perfusion as described in detail previously (19). Briefly, the livers were digested by perfusion with Hank's Balanced Salt Solution (HBSS; Sigma Chemical Company, St. Louis, MO) containing Pronase (Roche Diagnostics, Indianapolis, IN) followed by perfusion with HBSS containing Collagenase B (Sigma Chemical Company). Hepatocytes were removed from the digested liver by centrifugation at 50 g for 2 minutes. Hepatic stellate cells were isolated from the remaining nonparenchymal cell fraction by gradient centrifugation with Percoll (Sigma Chemical Company). The nonparenchymal cells were isolated from the supernatant by centrifugation at 250 g for 10 minutes. The resulting pellet was resuspended in 10.5 ml of 1× phosphate-buffered saline (PBS) containing Percoll at a final concentration of 52%. The cells were overlaid with 20 ml of 50% Percoll in 1×PBS. This layer was overlaid with 20 ml of 35% Percoll in 1×PBS followed by a layer containing 5 ml of 1×PBS. The gradient was centrifuged at 900 g for 30 minutes at 4 degrees C. After centrifugation, hepatic stellate cells, contained at the interface of the 35% percoll and 1×PBS layer, were diluted with an equal volume of 1×PBS, and centrifuged at 900 g for 10 minutes at 4 degrees C. The hepatic stellate cell pellet was resuspended in RPMI-1640 (Sigma Chemical Company) containing 10% FBS and penicillin/streptomycin. They were plated in 100 mm dishes at a density of 2.5 × 105 cells/dish. After 24 hours, the medium was removed, the cells washed twice with 1×PBS, and the medium replaced with serum-containing RPMI-1640. The hepatic stellate cells were activated in culture for 7 days before exposure to hypoxia. The medium was changed every 24 hours during this time period. After 7 days, the cells were washed with 1×PBS and cultured in serum-free RPMI-1640 for 24 hours. For studies utilizing unactivated hepatic stellate cells, the cells were plated for 3 hours in serum-containing medium and then cultured for an additional 15 hours in serum-free medium before exposure to room air or hypoxia. The cells were then cultured in an environment containing room air or 0.5% oxygen in a NAPCO CO2 7000 cell culture incubator (NAPCO Precision, Winchester, VA). These environments also contained 5% CO2 and were balanced with nitrogen. The purity of the hepatic stellate cell cultures were greater than 93% as determined by fluorescence of vitamin A-containing lipid droplets.
Analysis of HIF-1α Deletion in Hepatic Stellate Cells
Genomic DNA was isolated from hepatic stellate cells, and PCR was used to determine the degree of deletion of the floxed exons in the HIF-1α gene as described previously (5, 15, 20).
Real-time PCR
RNA was isolated from hepatic stellate cells using TRI reagent (Sigma Chemical Company, St. Louis, MO), and reverse transcribed into cDNA as described by us previously (21). Real-time PCR was used to quantify vascular endothelial cell growth factor (VEGF), placental growth factor angiopoietin-like 4, macrophage migration inhibitory factor, Ccr1, Ccr2, α2b adrenergic receptor, interleukin receptor α1, prolyl-4-hydroxylase α1, prolyl-4-hydroxylase α2, and 18S and performed on an Applied Biosystems Prism 7300 Real-time PCR Instrument (Applied Biosystems, Foster City, CA) using the SYBR green DNA PCR kit (Applied Biosystems) as described by us previously. The sequences of the primers were as follows: 18S Forward: 5'-TTG ACG GAA GGG CAC CAC CAG-3'; 18S Reverse: 5'-GCA CCA CCA CCC ACG GAA TCG-3'; PDGF-B Forward: 5'-CCC ACA GTG GCT TTT CAT TT-3'; PDGF-B Reverse: 5'-GTG GAG GAG CAG ACT GAA GG-3'; VEGF Forward: 5'-CAT CTT CAA GCC GTC CTG TGT-3'; VEGF Reverse: 5'-CTC CAG GGC TTC ATC GTT ACA-3'; placental growth factor Forward: 5'- GTG TGC CGA TAA AGA CAG CCA-3'; placental growth factor Reverse: 5'-GCA TTC ACA GAG CAC ATC CTG A-3'; angiopoietin-like 4 Forward: 5'- AGG ATA GAG TCC CTG AAG GCC A-3'; angiopoietin-like 4 Reverse: 5'- GGC CAC CTT CTG GAA CAA TTG-3'; macrophage migration inhibitory factor Forward: 5'- CTT TTA GCG GCA CGA ACG AT -3'; macrophage migration inhibitory factor Reverse: 5'- GGC CAC ACA GCA GCT TAC TGT A-3'; Ccr1 Forward: 5'- ACC CAC TGT TGT GTC AAC CCA -3'; Ccr1 Reverse: 5'- GCC ATT TTG CCA GTG GTA TAG C-3'; Ccr5 Forward: 5'- ACA CAC TGC TGC CTA AAC CCT G-3'; Ccr5 Reverse: 5'- CTT GCT GGA AAA TTG AAC ACC G-3'; α2B adrenergic receptor Forward: 5'-TTC TGT ACC TCC TCC ATC GTG C-3'; α2B adrenergic receptor Reverse: 5'- ATT TGA TGC GGC GTG GAG T-3'; interleukin 13 receptor α1 Forward: 5'- AAG TTC AGC CAC CTG TGA CGA A-3'; interleukin 13 receptor α1Reverse: 5'- AGT GCA ATT TGG ACT GGC TCC-3'; prolyl-4-hydroxylase α1 Forward: 5'- GTT TCG CGT GCA AGA CAC CTA C-3'; prolyl-4-hydroxylase α1Reverse: 5'- GCT CAA AGC AGT CCT CAG CTG T-3'; prolyl-4-hydroxylase α2 Forward: 5'-GCA GGT CGC AAA CTA CGG AAT-3'; prolyl-4-hydroxylase α2 Reverse: 5'- ACA CGG TTC CCA GTC CCT AAA C-3'.
Western Blot Analysis
Nuclear extracts were isolated from hepatic stellate cells. Briefly, the hepatic stellate cells were scraped into 10 mM Hepes, pH 7.9 containing 100 mM KCl, 1.5 mM MgCl2, 0.1 mM EGTA, 0.5 mM DTT, 0.5% NP-40, and Halt Protease Inhibitor Cocktail (Pierce Biotechnology, Rockford, IL) and incubated on ice. After 15 minutes, the cells were vortexed and centrifuged for 10 seconds at 10,000 g. The pellet was resuspended in 10 mM Hepes, pH 7.9 containing 420 mM NaCl, 1.5 mM MgCl2, 0.1 mM EGTA, 0.5 mM DTT, 5% glycerol, and Halt Protease Inhibitor Cocktail mixed for 1 hour at 4°C. The samples were centrifuged at 10,000 g for 10 minutes at 4°C and the concentration of protein determined in the supernatant. For western blot analysis, aliquots (15 μg) of nuclear extracts were subjected to 10% SDS-polyacrylamide gel electrophoresis, and proteins were transferred to immobilon polyvinylidene difluoride transfer membranes (Millipore Corporation, Bedford, MA). The membranes were then probed with rabbit polyclonal anti-HIF-1α antibody (NB100-449, Novus Biologicals, Littleton, CO) diluted 1:1000 or rabbit polyclonal anti-HIF-2α (NB100-122, Novus Biologicals) diluted 1:1000 followed by incubation with goat anti-rabbit antibody conjugated to horseradish peroxidase (Santa Cruz Biotechnology). Immunoreactive bands were visualized using the Immune-Star HRP Substrate Kit (Bio-Rad Laboratories, Hercules CA). To detect Ccr5, hepatic stellate cells were lysed in RIPA buffer. Western blot analysis was performed as described above. Membranes were probed with rabbit anti-Ccr5 (ab65850, Abcam, Cambridge, MA) diluted 1:1000 followed by incubation with goat anti-rabbit antibody conjugated to horseradish peroxidase. Bands were detected as described above. β-actin and lamin B1 were detected using rabbit anti-β-actin (Sigma Chemical Company) and mouse anti-lamin B1 (Abcam), respectively.
HIF-1α and HIF-2α Immunostaining
For HIF-1α, HIF-2α, and α-smooth muscle actin immunostaining, hepatic stellate cells were fixed in 4% formalin in phosphate-buffered saline (PBS) for 10 minutes at room temperature. Cells were incubated with rabbit polyclonal anti-HIF-1α (NB100-449, Novus Biologicals, Littleton, CO) or rabbit polyclonal anti-HIF-2α (NB100-122, Novus Biologicals) diluted 1:50 in PBS containing 3% goat serum at room temperature for 3 hours. In addition, the cells were incubated with mouse anti-smooth muscle actin (Biomeda) diluted 1:4 in PBS containing 3% goat serum at room temperature for 3 hours. The sections were washed with PBS, and then incubated with anti-rabbit secondary antibody conjugated to Alexa 594 (red staining; Molecular Probes, Eugene, OR) and anti-mouse secondary antibody conjugated to Alexa 488 (green staining; Molecular Probes). The sections were counterstained with 4',6-diamidino-2-phenylindole (DAPI) to stain DNA.
Gene Array and Data Analysis
RNA was isolated from hepatic stellate cells using TRI reagent. Integrity of the RNA was determined using an Agilent 2100 Bioanalyzer (Agilent Technologies, Inc., Foster City, CA). Gene array was performed using Affymetrix Gene Chip Mouse Genome 430 2.0 arrays. Data from Affymetrix GeneChip CEL files were normalized using GC Robust Multi-array Average (GCRMA) (Wu et al., 2004). Boxplots and Principal Components Analysis (PCA) were employed to assess data quality. Treatment and control comparisons were performed using an empirical Bayes analysis. All statistical analyses were performed in R (v2.8.1) using Bioconductor (2.1).
Statistical Analysis
Results are presented as the mean ± SEM. Data were analyzed by Analysis of Variance (ANOVA). ANOVAs were performed on log X-transformed data in instances in which variances were not homogenous. Comparisons among group means were made using the Student-Newman-Keuls test. The criterion for significance was p < 0.05 for all studies.
Results
Activation of HIF-1α in hypoxic hepatic stellate cells
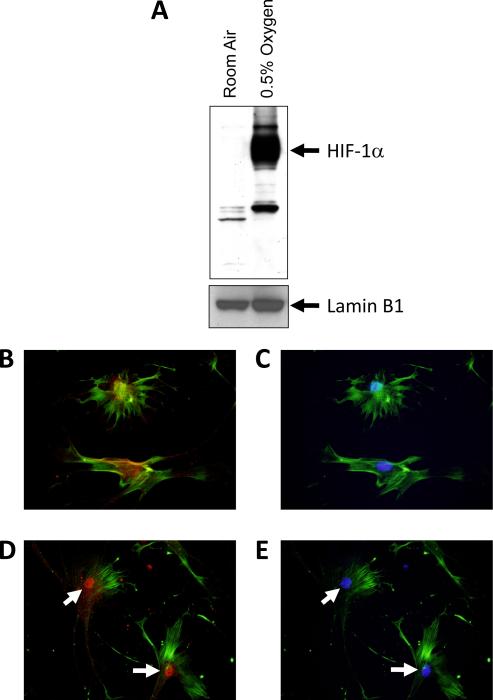
Exposure of culture-activated primary mouse hepatic stellate cells to 0.5% oxygen for 1 hour increased nuclear levels of HIF-1α protein, as detected by western blot analysis (Fig. 1A). HIF-1α immunostaining (red staining) was diffuse and primarily present in the cytoplasm of hepatic stellate cells exposed to room air (Fig. 1B). In hepatic stellate cells exposed to 0.5% oxygen for 1 hour, HIF-1α immunostaining was primarily present in the nucleus (Fig. 1D). Nuclear accumulation of HIF-1α was confirmed by colocalization with DAP-I (blue staining), a nuclear stain (Fig. 1E).
Fig. 1.
Hepatic stellate cells were isolated from mice and exposed to room air or 0.5% oxygen for 1 hour. HIF-1α and lamin B1 (i.e., loading control) were then detected in nuclear extracts by western blot. (A) Representative western blot of an n=3. Hepatic stellate cells were exposed to room air (B and C) or 0.5% oxygen (D and E) for 1 hour. Immunohistochemistry was used to detect HIF-1α (red fluorescence; B and D) and α-smooth muscle actin (green fluorescence; BE). The same cells were counterstained with DAPI (blue fluorescence; C and E) to identify the nuclei and show colocalization of HIF-1α immunostaining with nuclear staining. Arrows indicate nuclear HIF-1α immunostaining. Representative of an n=3.
Effect of hypoxia on gene expression in hepatic stellate cells
To identify novel genes upregulated in hypoxic hepatic stellate cells, culture-activated hepatic stellate cells were exposed to room air or 0.5% oxygen for 18 hours and gene expression analyzed by gene array. Hypoxia increased expression of 175 genes (Table 1) and decreased expression of 16 genes (Table 2). Many of the genes increased by hypoxia are known hypoxia-regulated genes, including those involved in energy metabolism (e.g., pyruvate dehydrogenase kinase, phosphofructokinase, triosephosphate isomerase, solute carrier family 2 member 1, aldolase C, lactate dehydrogenase, phosphoglycerate kinase, hexokinase 1), angiogenesis (e.g., VEGF, placental growth factor, angiopoietin-like 4, macrophage migration inhibitory factor), iron metabolism (e.g., transferrin receptor 2), and matrix metabolism (prolyl-4-hydroxylase-α1, prolyl-4-hydroxylase-α2) (Table 1) (13). In addition, mRNA levels of several genes were increased, the protein products of which, are known to affect hepatic stellate cell function, including α2b adrenergic receptor, interleukin 13 receptor α1, chemokine receptor 1, and chemokine receptor 5 (Table 1) (22–25). Many of these have not been described as hypoxia-inducible genes previously.
Table 1.
Gene array analysis of hypoxic hepatic stellate cells-mRNAs increased by hypoxia.
| Gene Name | Fold Change Relative to Room Air | p-value |
|---|---|---|
| arginase, liver (Arg1) | 34.58 | 0.0006995 |
| ovary-specific acidic protein (Osap) | 31.76 | 3.67E-06 |
| selenium binding protein 1 (Selenbp1) | 18.24 | 9.33E-05 |
| regulator of G-protein signaling 4 (Rgs4) | 18.04 | 0.0082626 |
| ankyrin repeat domain 37 (Ankrd37) | 16.73 | 0.0001724 |
| placental growth factor (Pgf) | 15.28 | 0.0001173 |
| EGL nine homolog 3 (Egln3) | 14.22 | 2.75E-06 |
| zinc finger and BTB domain containing 8b (Zbtb8b) | 13.15 | 0.0030468 |
| Adenylate kinase isoenzyme 4, mitochondrial (Ak3l1) | 10.55 | 0.0005776 |
| H-2 class I histocompatibility antigen, Q7 alpha chain (H2-Q7) | 9.96 | 0.0039013 |
| BCL2/adenovirus E1B interacting protein 3 (Bnip3) | 9.90 | 0.0001396 |
| keratin 19 (Krt19) | 9.79 | 0.0002675 |
| Uncharacterized protein C13orf33 homolog (6330406l15Rik) | 8.76 | 0.0001372 |
| N-myc downstream regulated gene 1 (Ndrg1) | 8.50 | 1.13E-05 |
| pyruvate dehydrogenase kinase, isoenzyme 1 (Pdk1) | 8.29 | 5.38E-06 |
| retinoic acid receptor responder (tazarotene induced) 2 (Rarres2) | 8.10 | 0.0761874 |
| solute carrier family 16 (monocarboxylic acid transporters), member 3 (Slc16a3) | 8.01 | 2.69E-05 |
| expressed sequence AU023617 (AU023617) | 7.23 | 0.0056076 |
| predicted gene, ENSMUSG00000079414 (ENSMUSG00000079414) | 6.97 | 0.000996 |
| protein phosphatase 1, regulatory (inhibitor) subunit 3C (Ppp1r3c) | 6.90 | 0.0001335 |
| RIKEN cDNA 6330406I15 gene (6330406l15Rik) | 6.60 | 0.0033789 |
| proline rich 15 (Prr15) | 6.40 | 0.0002953 |
| salt inducible kinase 1 (Sik1) | 5.92 | 0.000474 |
| pyridoxal (pyridoxine, vitamin B6) phosphatase (Pdxp) | 5.85 | 7.54E-05 |
| apolipoprotein L domain containing 1 (Apold1) | 5.83 | 0.0002834 |
| RIKEN cDNA C920025E04 gene (C920025E04Rik) | 5.64 | 0.0021858 |
| RIKEN cDNA A530047J11 gene (A530047J11Rik) | 5.64 | 1.39E-07 |
| vascular endothelial growth factor A (Vegfa) | 5.55 | 0.0003662 |
| histocompatibility 2, class II antigen A, beta 1 (H2-Ab1) | 5.45 | 0.0002173 |
| solute carrier family 2 (facilitated glucose transporter), member 1 (Slc2a1) | 5.42 | 0.0001942 |
| phosphofructokinase, liver, B-type (Pfkl) | 5.26 | 0.0003669 |
| N-myc downstream regulated gene 2 (Ndrg2) | 5.23 | 4.10E-07 |
| PRELI domain containing 2 (Prelid2) | 4.96 | 9.38E-05 |
| N-myc downstream regulated gene 1 (Ndrg1) | 4.93 | 0.0025224 |
| solute carrier family 2 (facilitated glucose transporter), member 1 (Slc2a1) | 4.72 | 0.0012122 |
| tumor necrosis factor (ligand) superfamily, member 9 (Tnfsf9) | 4.54 | 0.0026305 |
| angiopoietin-like 4 (Angptl4) | 4.53 | 0.0016198 |
| ubiquitin specific peptidase 53 (Usp53) | 4.52 | 0.000281 |
| ERO1-like (S. cerevisiae) (Ero1l) | 4.49 | 6.52E-06 |
| triosephosphate isomerase 1 (Tpi1) | 4.45 | 0.0009537 |
| retinoic acid receptor responder (tazarotene induced) 2 (Rarres2) | 4.44 | 0.0901861 |
| membrane bound O-acyltransferase domain containing 2 (Mboat2) | 4.24 | 0.0001866 |
| RIKEN cDNA 1190002H23 gene (1190002H23Rik) | 4.02 | 0.0008036 |
| predicted gene, 100039027 /// RIKEN cDNA 3110057O12 gene (3110057O12Rik) | 4.02 | 3.29E-07 |
| HtrA serine peptidase 3 (Htra3) | 3.95 | 0.0089931 |
| platelet-activating factor acetylhydrolase, isoform 1b, alpha1 subunit (Pafah1b3) | 3.87 | 1.36E-05 |
| family with sequence similarity 162, member A (Fam162a) | 3.80 | 0.0001076 |
| HIG1 domain family member 1A (Higd1a) | 3.74 | 1.74E-05 |
| RIKEN cDNA 9130008F23 gene (9130008F23Rik) | 3.73 | 0.0002627 |
| citrate lyase beta like (Clybl) | 3.70 | 7.67E-07 |
| basic helix-loop-helix family, member e40 (Bhlhe40) | 3.67 | 0.0055876 |
| adrenergic receptor, alpha 2b (Adra2b) | 3.60 | 0.0239358 |
| solute carrier family 4 (anion exchanger), member 4 (Slc4a4) | 3.54 | 0.0011883 |
| very low density lipoprotein receptor (Vldlr) | 3.50 | 0.0040546 |
| Max interacting protein 1 (Mxi1) | 3.48 | 1.50E-05 |
| G0/G1 switch gene 2 (G0s2) | 3.36 | 0.0006106 |
| potassium voltage-gated channel, Isk-related subfamily, gene 3 (Kcne3) | 3.14 | 0.0554731 |
| RIKEN cDNA C230081A13 gene (C230081A13Rik) | 3.08 | 0.0340789 |
| stanniocalcin 2 (Stc2) | 3.08 | 0.0077278 |
| methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like (Mthfd1l) | 3.06 | 0.0013846 |
| synaptopodin 2 (Synpo2) | 3.05 | 0.0032093 |
| RIKEN cDNA 4631426J05 gene (4631426J05Rik) | 3.04 | 0.0504871 |
| RIKEN cDNA D430019H16 gene (D430019H16Rik) | 3.00 | 0.065433 |
| very low density lipoprotein receptor (Vldlr) | 3.00 | 0.0002948 |
| RIKEN cDNA 0610037M15 gene (0610037M15Rik) | 2.99 | 0.0211989 |
| prolactin family 2, subfamily c, member 2 (Prl2c2) | 2.98 | 0.0390782 |
| EGL nine homolog 1 (C. elegans) (Egln1) | 2.97 | 0.0001512 |
| solute carrier family 2 (facilitated glucose transporter), member 3 (Slc2a3) | 2.95 | 0.0001249 |
| serine (or cysteine) peptidase inhibitor, clade B, member 2 (Serpinb2) | 2.90 | 0.0861919 |
| transmembrane protein 45a (Tmem45a) | 2.90 | 0.00799 |
| interferon-stimulated protein (Isg20) | 2.89 | 0.0095545 |
| stanniocalcin 2 (Stc2) | 2.88 | 0.0015753 |
| synaptopodin 2 (Synpo2) | 2.84 | 0.0310473 |
| BCL2/adenovirus E1B interacting protein 3-like (Bnip3I) | 2.83 | 5.81E-05 |
| glycogen synthase 1, muscle (Gys1) | 2.83 | 0.0004906 |
| similar to Lta4h protein /// leukotriene A4 hydrolase (LOC100048056///Lta4h) | 2.83 | 5.69E-05 |
| ring finger protein 217 (Rnf217) | 2.82 | 1.71E-05 |
| receptor accessory protein 1 (Reep1) | 2.82 | 1.23E-05 |
| very low density lipoprotein receptor (Vldlr) | 2.78 | 0.0001273 |
| liver glycogen phosphorylase (Pygl) | 2.76 | 0.0005936 |
| 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 (Pfkfb3) | 2.72 | 0.000311 |
| immediate early response 3 (ler3) | 2.70 | 0.0050945 |
| lymphocyte antigen 6 complex, locus A (Ly6a) | 2.66 | 0.0078143 |
| ubiquitin specific peptidase 53 (Usp53) | 2.65 | 2.45E-05 |
| glyoxylate reductase/hydroxypyruvate reductase (Grhpr) | 2.64 | 0.0006621 |
| fatty acid binding protein 4, adipocyte (Fabp4) | 2.63 | 0.016311 |
| glucan (1,4-alpha-), branching enzyme 1 (Gbe1) | 2.62 | 1.88E-05 |
| ring finger protein 217 (Rnf217) | 2.62 | 3.00E-05 |
| BCL2/adenovirus E1B interacting protein 3-like (Bnip3I) | 2.60 | 1.28E-05 |
| thioredoxin interacting protein (Txnip) | 2.58 | 0.0001261 |
| family with sequence similarity 46, member A (Fam46a) | 2.58 | 0.0031168 |
| galactokinase 1 (Galk1) | 2.58 | 7.80E-05 |
| Max interacting protein 1 (Mxi1) | 2.58 | 0.0055971 |
| proline 4-hydroxylase alpha II polypeptide (P4ha2) | 2.57 | 6.84E-05 |
| synaptonemal complex central element protein 2 (Syce2) | 2.57 | 0.0025006 |
| kinesin family member 21B (Kif21b) | 2.56 | 2.87E-05 |
| ERO1-like (S. cerevisiae) (Ero1l) | 2.55 | 0.0002634 |
| transducin (beta)-like 2 (Tbl2) | 2.54 | 0.0191534 |
| expressed sequence BE136769 (BE136769) | 2.52 | 0.0001937 |
| fatty acid binding protein 4, adipocyte (Fabp4) | 2.52 | 0.0145385 |
| cystin 1 (Cys1) | 2.52 | 0.0035457 |
| potassium voltage-gated channel, Isk-related subfamily, gene 4 (Kcne4) | 2.50 | 0.0063753 |
| aldolase C, fructose-bisphosphate (Aldoc) | 2.50 | 2.71E-05 |
| casein kinase 1, delta (Csnk1d) | 2.49 | 2.03E-05 |
| phosphofructokinase, platelet (Pfkp) | 2.48 | 0.0003496 |
| jumonji domain containing 1A (Jmjd1a) | 2.47 | 3.18E-05 |
| lactate dehydrogenase A (Ldha) | 2.46 | 0.0023058 |
| aldolase A, fructose-bisphosphate (Aldoa) | 2.44 | 0.0009634 |
| cyclin G2 (Ccng2) | 2.44 | 2.29E-05 |
| aldolase A (Aldoa) | 2.43 | 0.0010763 |
| EGL nine homolog 1 (C. elegans) (Egln1) | 2.42 | 9.45E-07 |
| RIKEN cDNA 4631426J05 gene (4631426J05Rik) | 2.39 | 0.0364575 |
| chemokine (C-C motif) receptor 1 (Ccr1) | 2.38 | 0.0009639 |
| protein phosphatase 1, regulatory (inhibitor) subunit 3B (Ppp1r3b) | 2.37 | 0.0008892 |
| cDNA sequence BC023202 (BC023202) | 2.37 | 0.001069 |
| FERM domain containing 3 (Frmd3) | 2.37 | 0.0008735 |
| predicted gene, OTTMUSG00000014994 (OTTMUSG00000014994) | 2.36 | 0.068909 |
| phosphoglycerate kinase 1 (Pgk1) | 2.36 | 0.0002869 |
| pyruvate kinase, muscle (Pkm2) | 2.35 | 0.0053475 |
| adenosine monophosphate deaminase 3 (Ampd3) | 2.35 | 0.0002632 |
| collagen, type XIV, alpha 1 (Col14a1) | 2.33 | 0.0278821 |
| Ras association (RalGDS/AF-6) and pleckstrin homology domains 1 (Raph1) | 2.33 | 0.0081096 |
| cysteinyl leukotriene receptor 1 (Cysltr1) | 2.33 | 0.0071408 |
| phosphoglycolate phosphatase (Pgp) | 2.32 | 0.0018842 |
| BCL2/adenovirus E1B interacting protein 3-like (Bnip3l) | 2.31 | 2.34E-05 |
| cyclin G2 (Ccnfg2) | 2.31 | 1.16E-05 |
| histocompatibility 2, class II antigen A, beta 1 (H2-Ab1) | 2.31 | 0.055698 |
| family with sequence similarity 162, member A (Fam162a) | 2.30 | 0.0059266 |
| hexokinase 1 (Hk1) | 2.30 | 3.46E-05 |
| GPI-anchored HDL-binding protein 1 (Gpihbp1) | 2.29 | 0.0479326 |
| neural cell adhesion molecule 1 (Ncam1) | 2.29 | 0.0075318 |
| phosphoglycerate mutase 1 (Pgam1) | 2.29 | 0.0003186 |
| phosphoglycolate phosphatase (pgp) | 2.28 | 0.0044069 |
| transducin (beta)-like 2 (Tbl2) | 2.28 | 0.0255315 |
| steroid 5 alpha-reductase 1 (Srd5a1) | 2.27 | 0.0004296 |
| Serglycin (Srgn) | 2.25 | 0.000168 |
| RIKEN cDNA A530057A03 gene (A530057A03Rik) | 2.25 | 0.1378197 |
| ATPase, class I, type 8B, member 4 (Atp8b4) | 2.24 | 7.48E-05 |
| neural cell adhesion molecule 1 (Ncam1) | 2.24 | 0.0003728 |
| similar to Ring finger protein 126 /// ring finger protein 126 (LOC100045963 /// Rnf126) | 2.23 | 0.0027228 |
| RIKEN cDNA 2310016C08 gene (2310016C08Rik) | 2.23 | 0.0029255 |
| RIKEN cDNA 3110057O12 gene (3110057O12Rik) | 2.22 | 0.0013277 |
| regulator of G-protein signaling 5 (Rgs5) | 2.21 | 0.1051163 |
| immunoglobulin superfamily, member 6 (Igsf6) | 2.21 | 0.0005853 |
| arsenic (+3 oxidation state) methyltransferase (As3mt) | 2.19 | 0.0051479 |
| complement component 4 binding protein (C4bp) | 2.19 | 0.0156553 |
| prolactin family 3, subfamily a, member 1 (Prl3a1) | 2.19 | 0.2794702 |
| regulator of G-protein signaling 5 (Rgs5) | 2.19 | 0.1286333 |
| C-type lectin domain family 4, member e (Clec4e) | 2.18 | 0.0415139 |
| jumonji domain containing 6 (Jmjd6) | 2.17 | 0.0005009 |
| CD72 antigen (Cd72) | 2.15 | 0.000122 |
| Wiskott-Aldrich syndrome-like (human) (Wasl) | 2.15 | 0.0335297 |
| SAM domain, SH3 domain and nuclear localization signals, 1 (Samsn1) | 2.14 | 0.0086378 |
| adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 2 (Appl2) | 2.14 | 4.87E-05 |
| casein kinase 1, delta (Csnk1d) | 2.14 | 0.0005166 |
| transferrin receptor 2 (Trfr2) | 2.13 | 0.040254 |
| Desmoplakin (Dsp) | 2.13 | 0.0887779 |
| glycogen synthase 1, muscle (Gys1) | 2.12 | 0.0201665 |
| interleukin 13 receptor, alpha 1 (ll13ra1) | 2.12 | 0.1544185 |
| cysteinyl leukotriene receptor 1 (Cysltr1) | 2.11 | 0.0065358 |
| macrophage migration inhibitory factor (Mif) | 2.10 | 0.0028799 |
| similar to nuclear factor, interleukin 3, regulated (LOC100046232 /// Nfil3) | 2.10 | 0.0244131 |
| gene model 22, (NCBI) (Gm22) | 2.10 | 0.017415 |
| cytohesin 1 interacting protein (Cytip) | 2.09 | 0.0034503 |
| UDP-glucose ceramide glucosyltransferase (Ugcg) | 2.08 | 0.0002219 |
| chemokine (C-C motif) receptor 5 (Ccr5) | 2.08 | 0.0027067 |
| cholesterol 25-hydroxylase (Ch25h) | 2.04 | 0.0053625 |
| UDP-glucose ceramide glucosyltransferase (Ugcg) | 2.04 | 0.0006698 |
| phosphoglucomutase 1 /// phosphoglucomutase 2 (Pgm1 /// Pgm2) | 2.04 | 0.0022001 |
| proline 4-hydroxylase alpha 1 polypeptide (P4ha1) | 2.02 | 0.0012325 |
| family with sequence similarity 57, member A (Fam57a) | 2.02 | 0.0102877 |
| endonuclease G (Endog) | 2.02 | 0.0651738 |
| 5' nucleotidase, ecto (Nt5e) | 2.00 | 0.0919347 |
Hepatic stellate cells were exposed to room air or 0.5% oxygen. Eighteen hours later, mRNA levels were analyzed by gene array.
Table 2.
Gene array analysis of hypoxic hepatic stellate cells-mRNAs decreased by hypoxia.
| Gene Title | Fold Change Relative to Room Air | p-value |
|---|---|---|
| RIKEN cDNA 1700007K13 gene (1700007K13Rik) | 0.50 | 0.00090569 |
| SH3-domain binding protein 5 (BTK-associated) (Sh3bp5) | 0.50 | 0.016978812 |
| F-box and leucine-rich repeat protein 7 (Fbxl7) | 0.50 | 0.000524786 |
| Rab38, member of RAS oncogene family (Rab38) | 0.49 | 0.018670872 |
| cysteine conjugate-beta lyase 2 (Ccbl2) | 0.49 | 0.019470745 |
| ELOVL family member 6, elongation of long chain fatty acids (yeast) (Elovl6) | 0.49 | 0.030557143 |
| wingless-type MMTV integration site 9A (Wnt9a) | 0.48 | 0.002358591 |
| methyl-CpG binding domain protein 1 (Mbd1) | 0.46 | 0.061122083 |
| Rho GTPase activating protein 19 (Arhgap19) | 0.46 | 0.000347739 |
| Rab38, member of RAS oncogene family (Rab38) | 0.44 | 0.003454317 |
| pantothenate kinase 3 (Pank3) | 0.44 | 0.00764511 |
| lipase, endothelial (Lipg) | 0.42 | 0.011722097 |
| cystathionase (cystathionine gamma-lyase) (Cth) | 0.40 | 0.001026255 |
| integrin alpha 1 (Itga1) | 0.39 | 0.098586804 |
| hepatocyte growth factor (Hgf) | 0.38 | 0.009611965 |
| formin homology 2 domain containing 3 (Fhod3) | 0.34 | 0.035392908 |
Hepatic stellate cells were exposed to room air or 0.5% oxygen. Eighteen hours later, mRNA levels were analyzed by gene array.
HIF-1α-dependent regulation of gene expression in hypoxic hepatic stellate cells
Next, we determined the molecular mechanism by which hypoxia increased expression of genes in hypoxic hepatic stellate cells that affect hepatic stellate cell function and promote liver fibrosis. One mechanism by which hypoxia modulates gene expression in cells is by activating hypoxia-inducible factor transcription factors. To evaluate the role of HIF-1α in regulation of gene expression in hypoxic hepatic stellate cells, HIF-1αfl/fl mice were crossed with Mx-Cre+/− mice. Treatment of these mice with pIpC upregulates Cre recombinase in most cell types and deletes essential functional regions of the HIF-1α gene as described in detail previously (5, 15). To confirm that this results in deletion of HIF-1α in hepatic stellate cells, these cells were isolated from pIpC-treated HIF-1αfl/fl-MxCre− and HIF-1αfl/fl-MxCre+ mice and exposed to 0.5% oxygen for 1 hour. Stabilized HIF-1α protein was present in hypoxic hepatic stellate cells isolated from HIF-1αfl/fl-MxCre− mice, but not detected in cells isolated from HIF-1αfl/fl-MxCre+ mice indicating a complete loss of HIF-1α signaling in these hepatic stellate cells (Fig. 2). For the remainder of this manuscript HIF-1αfl/fl-MxCre− mice will be referred to as Control mice and HIF-1αfl/fl-MxCre+ mice will be referred to as HIF-1α Deficient mice.
Fig. 2.
Hepatic stellate cells were isolated from HIF-1αfl/fl MxCre- mice and HIF-1αfl/fl MxCre+ mice and exposed to 0.5% oxygen for 1 hour. HIF-1α and lamin B1 (i.e., loading control) were then detected in nuclear extracts by western blot.
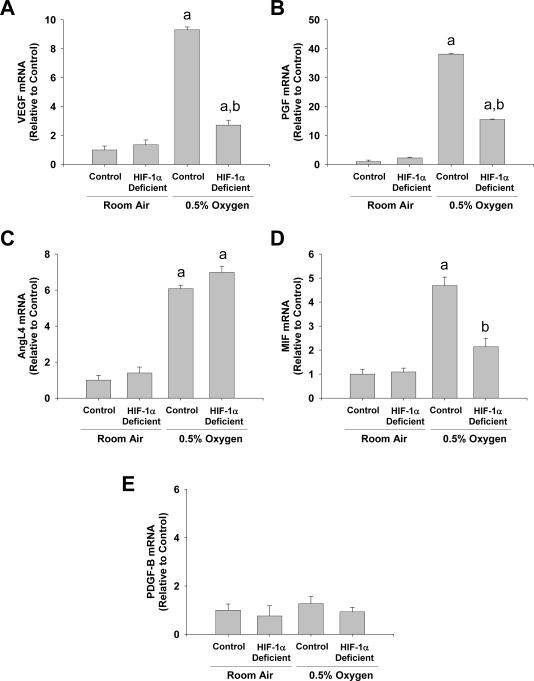
We first determined whether HIF-1α is required from upregulation of proangiogenic mediators by hypoxia. Studies have demonstrated that angiogenesis is important for fibrosis, and gene array analysis identified several proangiogenic mediators upregulated in hypoxic hepatic stellate cells, including VEGF, placental growth factor (PGF), and angiopoietin-like 4 (AngL4), and macrophage migration inhibitory factor (MIF). Exposure of hepatic stellate cells to hypoxia increased mRNA levels of VEGF, PGF, AngL4, and MIF as quantified by real-time PCR (Fig. 3). Upregulation of VEGF and PGF was partially attenuated in hepatic stellate cells isolated from HIF-1α-deficient mice, and upregulation of MIF was completely prevented in hepatic stellate cells isolated from HIF-1α-deficient mice (Fig. 3). Upregulation of AngL4 was not affected by deletion of HIF-1α. Platelet-derived growth factor-B (PDGF-B) is an important proangiogenic mediator and activator of hepatic stellate cells (26). We previously demonstrated that HIF-1α is required for upregulation of PDGF-B in the livers of bile duct-ligated mice (5). Although PDGF-B levels were not increased by gene array analysis, we confirmed these results since HIF-1α is an important regulator of this gene in vivo. Similar to the gene array studies, PDGF-B mRNA levels were not increased in hypoxic hepatic stellate cells (Fig. 3).
Fig. 3.
Hepatic stellate cells were isolated from HIF-1α-Control and HIF-1α-Deficient mice, activated in culture for 7 days, and then exposed to room air or 0.5% oxygen. Eighteen hours later, (A) VEGF, (B) PGF, (C) AngL4, (D) MIF, and (E) PDGF-B mRNA levels were quantified by real-time PCR. aSignificantly different from hepatic stellate cells exposed to room air (p<0.05). bSignificantly different from HIF-1α-Control hepatic stellate cells exposed 0.5% oxygen (p<0.05). Data are expressed as means ± SEM; n = 3.
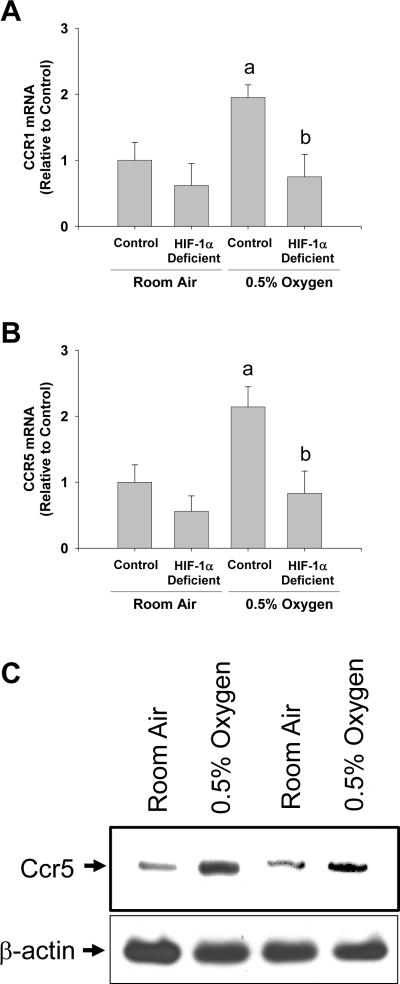
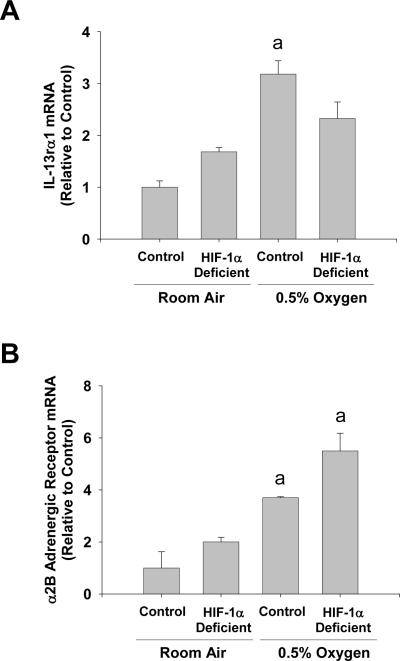
Exposure of hepatic stellate cells to hypoxia increased mRNA levels of several receptors, the ligands of which, are known to affect hepatic stellate cell function, including α2b adrenergic receptor (Adrα2b), interleukin 13 receptor α1 (IL-13rα1), chemokine receptor 1 (Ccr1), and chemokine receptor 5 (Ccr5) (22–25). Upregulation of Ccr1 and Ccr5 by hypoxia was completely prevented in hepatic stellate cells isolated from HIF-1α-deficient mice (Fig. 4A and 4B). Upregulation Ccr5 protein by hypoxia was confirmed by western blot analysis (Fig. 4C). Upregulation of IL-13rα1 was only partially prevented in hepatic stellate cells isolated from HIF-1α-deficient mice (Fig. 5), and deletion of HIF-1α from hepatic stellate cells did not prevent the increase in Adrα2b mRNA levels after exposure to hypoxia (Fig. 5).
Fig. 4.
Hepatic stellate cells were isolated from HIF-1α-Control and HIF-1α-Deficient mice, activated in culture for 7 days, and then exposed to room air or 0.5% oxygen. Eighteen hours later, (A) Ccr1 and (B) Ccr5 mRNA levels were quantified by real-time PCR. aSignificantly different from hepatic stellate cells exposed to room air (p<0.05). bSignificantly different from HIF-1α-Control hepatic stellate cells exposed 0.5% oxygen (p<0.05). (C) Ccr5 and β-actin (i.e., loading control) proteins were detected by western blot. Data are expressed as means ± SEM; n = 3.
Fig. 5.
Hepatic stellate cells were isolated from HIF-1α-Control and HIF-1α-Deficient mice, activated in culture for 7 days, and then exposed to room air or 0.5% oxygen. Eighteen hours later, (A) Adrα2b and (B) IL-13rα1 mRNA levels were quantified by real-time PCR. aSignificantly different from hepatic stellate cells exposed to room air (p<0.05). bSignificantly different from HIF-1α-Control hepatic stellate cells exposed 0.5% oxygen (p<0.05). Data are expressed as means ± SEM; n = 3.
Hepatic stellate cells are an important source of collagen during the genesis of fibrosis. Prolyl-4-hydroxylase-α1 (P4hα1) and P4hα2 are key enzymes in the generation of stable collagen triple helices (27). The mRNA levels of both of these enzymes were increased in hepatic stellate cells exposed to hypoxia as determined by real-time PCR (Fig. 6). Upregulation of P4hα1 by hypoxia was unaffected in hepatic stellate cells isolated from HIF-1α-deficient mice (Fig. 6A). In contrast, upregulation of P4hα2 was completely prevented in hepatic stellate cells isolated from HIF-1α-deficient mice (Fig. 6B).
Fig. 6.
Hepatic stellate cells were isolated from HIF-1α-Control and HIF-1α-Deficient mice, activated in culture for 7 days, and then exposed to room air or 0.5% oxygen. Eighteen hours later, (A) P4hα1 and (B) P4hα2 mRNA levels were quantified by real-time PCR. aSignificantly different from hepatic stellate cells exposed to room air (p<0.05). bSignificantly different from HIF-1α-Control hepatic stellate cells exposed 0.5% oxygen (p<0.05). Data are expressed as means ± SEM; n = 3.
In the studies described above, the hepatic stellate cells were activated in culture for 7 days prior to exposure to hypoxia. To assess whether these genes are similarly upregulated by hypoxia in unactivated hepatic stellate cells, hepatic stellate cells were isolated from mice and exposed to hypoxia 18 hours after isolation. In contrast to culture-activated hepatic stellate cells, only MIF and P4hα2 were significantly increased in unactivated hepatic stellate cells (Table 3).
Table 3.
Effect of Hypoxia on Expression of Genes in Unactivated Hepatic Stellate Cells.
| Gene | Room Air | 0.5% Oxygen |
|---|---|---|
| Vascular Endothelial Growth Factor | 1.2 +/− 0.08 | 2.0 +/− 0.35 |
| Placental Growth Factor | 1.2 +/− 0.10 | 2.4 +/− 0.74 |
| Macrophage Migration Inhibitory Factor | 1.1 +/− 0.03 | 2.2 +/− 0.40a |
| Interleukin-13 Receptor α1 | 1.1 +/− 0.04 | 1.8 +/− 0.42 |
| Prolyl-4-hydroxylase-α1 | 1.1 +/− 0.09 | 2.4 +/− 0.90 |
| Prolyl-4-hydroxylase-α2 | 1.2 +/− 0.07 | 4.6 +/− 1.77a |
| Angiopoietin-like 4 | 1.2 +/− 0.08 | 1.1 +/− 0.10 |
| Ccr1 | 1.4 +/− 0.19 | 1.4 +/− 0.62 |
| Ccr5 | 1.3 +/− 0.24 | 0.9 +/− 0.15 |
| α2b adrenergic receptor | 0.9 +/− 0.11 | 2.9 +/− 1.44 |
Hepatic stellate cells were isolated from mice and allowed to attach for 18 hours. The cells were then exposed to room air or 0.5% oxygen for 18 hours. Levels of the genes indicated were quantified by real-time PCR.
Significantly different from hepatic stellate cells exposed to room air (p<0.05Data are expressed as means ± SEM; n = 4.
Activation of HIF-2α in hypoxic hepatic stellate cells
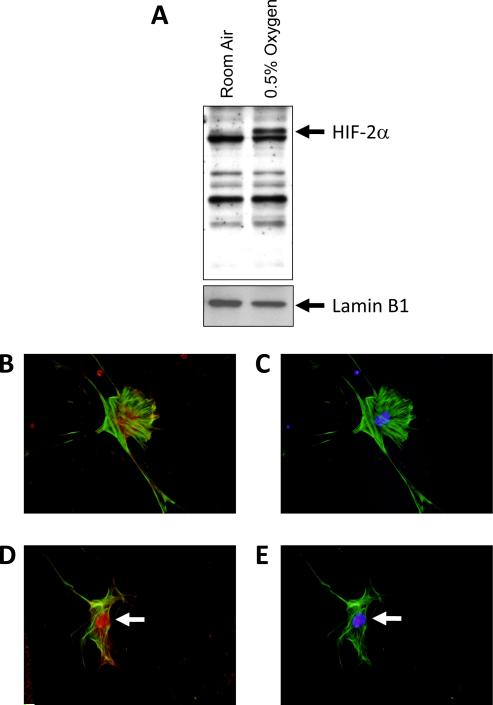
Deletion of HIF-1α from hepatic stellate cells was not sufficient to completely prevent upregulation of some genes by hypoxia (Figs. 3–6). This suggests that other hypoxia-activated transcription factors may contribute to upregulation of these genes. One possibility is HIF-2α. Accordingly, the hypothesis was tested that HIF-2α is activated in hypoxic hepatic stellate cells. Exposure of culture-activated primary mouse hepatic stellate cells to 0.5% oxygen for 1 hour increased nuclear levels of HIF-2α protein (Fig. 7A). Similar to HIF-1α, HIF-2α immunostaining was diffuse and primarily present in the cytoplasm of hepatic stellate cells exposed to room air (Fig. 7B). In hepatic stellate cells exposed to 0.5% oxygen for 1 hour, HIF-2α immunostaining was primarily present in the nucleus (Fig. 7D).
Fig. 7.
Hepatic stellate cells were isolated from mice, activated in culture for 7 days, and then exposed to room air or 0.5% oxygen for 1 hour. HIF-2α and lamin B1 were then detected in nuclear extracts by western blot. (A) Representative western blot of an n=3. Hepatic stellate cells were exposed to room air (B and C) or 0.5% oxygen (D and E) for 1 hour. Immunohistochemistry was used to detect HIF-2α (red fluorescence; B and D) and α-smooth muscle actin (green fluorescence; B–E). The same cells were counterstained with DAPI (blue fluorescence; C and E) to identify the nuclei and show colocalization of HIF-2α immunostaining with nuclear staining. Arrows indicate nuclear HIF-2α immunostaining. Representative of an n=3.
Discussion
During chronic injury, regions of hypoxia develop in the liver (5, 6). Our previous studies demonstrated that hypoxia, through activation of HIF-1α, is an important driving force for the development of liver fibrosis (5). The impact of hypoxia exposure and HIF-1α activation on hepatic stellate cell function, however, has not been fully examined. The present study demonstrated that hypoxia modulates expression of several genes that are important for hepatic stellate cell activation, motility, and collagen production by a HIF-1α-dependent mechanism. In addition, hypoxia stimulated production of proangiogenic mediators by hypoxic hepatic stellate cells by a HIF-1α-dependent mechanism. These studies have not only demonstrated that hypoxia may alter the sensitivity of hepatic stellate cells to certain activators and chemotaxins, but have identified several novel HIF-regulated genes.
Hypoxia increased mRNA levels of several receptors in hepatic stellate cells, the ligands of which affect hepatic stellate cell chemotaxis, proliferation, and collagen production. Two of these, Ccr1 and Ccr5, bind chemokines and direct cell chemotaxis. The ligands for Ccr1 include MCP-1, MIP-1α, MIP-1β, RANTES, MCP-3, MCP-2, Ccl13, Ccl14, and Ccl15 (28). The ligand binding specificity for Ccr5 is similar to Ccr1 (28). Studies have shown that hepatic stellate cells express these receptors and migrate towards chemokines that bind these receptors (23, 24). Furthermore, the levels of several of these chemokines are increased in fibrosis and recent studies have shown that Ccr1 and Ccr5 knockout mice have reduced fibrosis (24). Our studies demonstrate that hypoxia upregulates Ccr1 and Ccr5 in hepatic stellate cells (Fig. 4). This suggests that hepatic stellate cells in hypoxic regions of liver may be more responsive to the chemotactic properties of chemokines. Furthermore, our studies demonstrate for the first time that Ccr1 and Ccr5 are putative HIF-1α target genes.
Another receptor increased in hypoxic hepatic stellate cells was IL-13rα1. Previous studies demonstrated that LI90 cells, a transformed cell line with characteristics similar to hepatic stellate cells, express IL-13rα1 (25). Furthermore, exposure of these cells, primary rat hepatic stellate cells, and a rat hepatic stellate cell line to IL-13 stimulated collagen secretion (25, 29). In support of a role for IL-13 in the development of fibrosis, IL-13-deficient mice showed reduced fibrosis after schistosome infection (30). Recently it was reported that IL-13 levels are increased in liver and serum of hepatitis C infected patients, and the level of IL-13 correlated with the stage of fibrosis (29). Our studies suggest that during chronic liver injury, when regions of hypoxia develop, hepatic stellate cells trapped within these hypoxic regions of liver may be more responsive to the profibrotic effects of IL-13. Our studies also demonstrate that IL-13rα1 is potentially directly regulated by HIF-1α (Fig. 5). Further studies are needed, however, to determine whether functional hypoxia-response elements are present in the promoter of IL-13rα1.
Similar to IL-13rα1, Adrα2b mRNA levels were increased in hypoxic hepatic stellate cells (Fig. 5). Adrα2b is a G protein-coupled receptor that binds catecholamines, such as norepinephrine. Previous studies have shown that exposure of hepatic stellate cells to norepinephrine stimulates proliferation and collagen production (22). It has been proposed that increased sympathetic nerve activity in patients with chronic liver injury may be an important driving force for fibrogenesis by activating adrenergic receptors on hepatic stellate cells. Our studies demonstrate that Adrα2b levels are increased in hypoxic hepatic stellate cells, and similar to IL-13rα1, Ccr1, and Ccr5, hepatic stellate cells in hypoxic regions may be more responsive to stimulatory effects of agonists of this receptor. In contrast to IL-13rα1, Ccr1, and Ccr5, upregulation of Adrα2b occurred independently of HIF-1α, suggesting that this receptor is regulated in hypoxic cells by other mechanisms. One possibility is HIF-2α. Our results demonstrate that in addition to HIF-1α, HIF-2α is activated in hypoxic hepatic stellate cells (Fig. 7). It is possible that HIF-2α may also be an important regulator of genes, such as Adrα2b, in these cells.
As expected, hypoxia increased mRNA levels of a number of proteins that are important mediators of angiogenesis, including VEGF, PGF, AngL4, and MIF (Fig. 3) (31–33). As discussed, angiogenesis is thought to be an essential component of fibrosis. Levels of VEGF, PGF, and MIF are increased in patients with chronic liver disease (34–36). Our data suggest that hypoxia may be an important modulator of these mediators in liver during the development of liver fibrosis. All of these mediators are known HIF-1α-target genes. Interestingly, deletion of HIF-1α had no effect on upregulation of AngL4 by hypoxia. This suggests that other hypoxia-regulated transcription factors, such as HIF-2α, may be important for upregulation of AngL4 in hepatic stellate cells. In addition to MIF being an important mediator of angiogenesis and regulator of immune cell function, recent studies have demonstrated that MIF is an important regulator of cellular senescence. In these studies, it was demonstrated that HIF-1α-dependent release of MIF from cultured fibroblasts delays senescence of these cells (37). Interestingly, it was recently demonstrated that hepatic stellate cells are also subject to senescence in vivo during the development of liver fibrosis, and inhibition of senescence leads to more severe fibrosis (38). If MIF is able to inhibit senescence of hepatic stellate cells, it is possible that hepatic stellate cells within hypoxic regions of liver secrete MIF which delays the senescence program in these cells leading to more severe fibrosis. In future studies, it would be interesting to study the influence of hypoxia and MIF on hepatic stellate cell senescence, and also determine the impact of HIF-1α deletion on hepatic stellate cell senescence in vivo during the development of fibrosis.
Lastly, exposure of hepatic stellate cells to hypoxia increased mRNA levels of both P4hα1 and P4hα2 (Fig. 6). Prolyl-4-hydroxylases are key enzymes in the generation of stable collagen triple helices (27). Our studies suggest that upregulation of these enzymes in hepatic stellate cells in hypoxic regions of liver may lead to the generation of a more stable extracellular matrix in the liver. Both P4hα1 and P4hα2 are known HIF-1α-regulated genes (39). Our studies indicate that in hypoxic hepatic stellate cells, P4hα2 is regulated by HIF-1α, whereas P4hα1 is not. It is possible that in hepatic stellate cells P4hα1 may be preferentially regulated by HIF-2α. Further studies are needed, however, to determine the importance of HIF-2α to regulation of gene expression in hypoxic hepatic stellate cells.
Interestingly, although hypoxia increased expression of several genes in hypoxic activated hepatic stellate cells, including VEGF, PGF, AngL4, MIF, Ccr1, Ccr5, IL-13rα1, α2B adrenergic receptor, PHD α1, and PHD α2, only MIF and PHD α2 were increased in hypoxic unactivated hepatic stellate cells. This suggests that the process of activation sensitizes hepatic stellate cells to the effects of hypoxia. The mechanism is unclear but it is possible that the levels of various coactivators and/or corepressors that affect HIF function may be altered during the process of hepatic stellate cell activation. Further studies are needed to evaluate this possibility.
Results from these studies indicate that hypoxia, through activation of HIFs, may be an important modulator of genes that affect hepatic stellate cell function, angiogenesis, and matrix deposition. In addition, these studies have identified potential novel targets of HIF regulation, including Ccr1, Ccr5, and IL-13rα1. Further studies are needed, however, to evaluate the importance of HIFs for regulation of gene expression in hepatic stellate cells in vivo during the development of liver fibrosis, since studies have demonstrated that the repertoire of genes upor down-regulated in activated hepatic stellate cells in vitro differs from those changes that occur in vivo (40). Results from these studies, however, suggest that HIFs may be a novel target of therapy in patients with fibrosis.
Acknowledgements
This study was supported by National Institutes of Health grants DK073566 (B.L.C.) and COBRE (Center of Biomedical Research Excellence) P20 RR021940 as well as the Molecular Biology Core and the Histology Core supported by the COBRE grant. In addition, this work was supported by a Lied Endowed Basic Science Pilot Research Grant (B.L.C.) and grant number P20 RR016475 from the National Center for Research Resources (NCRR). The authors thank Dr. Frank J. Gonzalez at the National Cancer Institute for providing the HIF-1αfl/fl-Mx-Cre+ and HIF-1αfl/fl-Mx-Cre- mice for these studies.
References
- 1.Friedman SL, Roll FJ, Boyles J, Bissell DM. Hepatic lipocytes: the principal collagen-producing cells of normal rat liver. Proc Natl Acad Sci U S A. 1985;82(24):8681–5. doi: 10.1073/pnas.82.24.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Friedman SL. Mechanisms of hepatic fibrogenesis. Gastroenterology. 2008;134(6):1655–69. doi: 10.1053/j.gastro.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Georges PC, Hui JJ, Gombos Z, Mccormick ME, Wang a Y, Uemura M, et al. Increased stiffness of the rat liver precedes matrix deposition: implications for fibrosis. Am J Physiol Gastrointest Liver Physiol. 2007;293(6):G1147–54. doi: 10.1152/ajpgi.00032.2007. [DOI] [PubMed] [Google Scholar]
- 4.Zhou X, Murphy FR, Gehdu N, Zhang J, Iredale JP, Benyon RC. Engagement of alphavbeta3 integrin regulates proliferation and apoptosis of hepatic stellate cells. J Biol Chem. 2004;279(23):23996–4006. doi: 10.1074/jbc.M311668200. [DOI] [PubMed] [Google Scholar]
- 5.Moon JO, Welch TP, Gonzalez FJ, Copple BL. Reduced liver fibrosis in hypoxiainducible factor-1alpha-deficient mice. Am J Physiol Gastrointest Liver Physiol. 2009;296(3):G582–92. doi: 10.1152/ajpgi.90368.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rosmorduc O, Wendum D, Corpechot C, Galy B, Sebbagh N, Raleigh J, et al. Hepatocellular hypoxia-induced vascular endothelial growth factor expression and angiogenesis in experimental biliary cirrhosis. Am J Pathol. 1999;155(4):1065–73. doi: 10.1016/S0002-9440(10)65209-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Corpechot C, Barbu V, Wendum D, Kinnman N, Rey C, Poupon R, et al. Hypoxia-induced VEGF and collagen I expressions are associated with angiogenesis and fibrogenesis in experimental cirrhosis. Hepatology. 2002;35(5):1010–21. doi: 10.1053/jhep.2002.32524. [DOI] [PubMed] [Google Scholar]
- 8.Ankoma-Sey V, Wang Y, Dai Z. Hypoxic stimulation of vascular endothelial growth factor expression in activated rat hepatic stellate cells. Hepatology. 2000;31(1):141–8. doi: 10.1002/hep.510310122. [DOI] [PubMed] [Google Scholar]
- 9.Novo E, Cannito S, Zamara E, Valfre Di Bonzo L, Caligiuri A, Cravanzola C, et al. Proangiogenic cytokines as hypoxia-dependent factors stimulating migration of human hepatic stellate cells. Am J Pathol. 2007;170(6):1942–53. doi: 10.2353/ajpath.2007.060887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taura K, De Minicis S, Seki E, Hatano E, Iwaisako K, Osterreicher CH, et al. Hepatic stellate cells secrete angiopoietin 1 that induces angiogenesis in liver fibrosis. Gastroenterology. 2008;135(5):1729–38. doi: 10.1053/j.gastro.2008.07.065. [DOI] [PubMed] [Google Scholar]
- 11.Shi YF, Fong CC, Zhang Q, Cheung PY, Tzang CH, Wu RS, et al. Hypoxia induces the activation of human hepatic stellate cells LX-2 through TGF-beta signaling pathway. FEBS letters. 2007;581(2):203–10. doi: 10.1016/j.febslet.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 12.Coleman ML, Ratcliffe PJ. Oxygen sensing and hypoxia-induced responses. Essays Biochem. 2007;43:1–15. doi: 10.1042/BSE0430001. [DOI] [PubMed] [Google Scholar]
- 13.Gaber T, Dziurla R, Tripmacher R, Burmester GR, Buttgereit F. Hypoxia inducible factor (HIF) in rheumatology: low O2! See what HIF can do! Ann Rheum Dis. 2005;64(7):971–80. doi: 10.1136/ard.2004.031641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cash TP, Pan Y, Simon MC. Reactive oxygen species and cellular oxygen sensing. Free Radic Biol Med. 2007;43(9):1219–25. doi: 10.1016/j.freeradbiomed.2007.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tomita S, Ueno M, Sakamoto M, Kitahama Y, Ueki M, Maekawa N, et al. Defective brain development in mice lacking the Hif-1alpha gene in neural cells. Mol Cell Biol. 2003;23(19):6739–49. doi: 10.1128/MCB.23.19.6739-6749.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuhn R, Schwenk F, Aguet M, Rajewsky K. Inducible gene targeting in mice. Science. 1995;269(5229):1427–9. doi: 10.1126/science.7660125. [DOI] [PubMed] [Google Scholar]
- 17.Iyer NV, Kotch LE, Agani F, Leung SW, Laughner E, Wenger RH, et al. Cellular and developmental control of O2 homeostasis by hypoxia-inducible factor 1 alpha. Genes Dev. 1998;12(2):149–62. doi: 10.1101/gad.12.2.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tomita S, Sinal CJ, Yim SH, Gonzalez FJ. Conditional disruption of the aryl hydrocarbon receptor nuclear translocator (Arnt) gene leads to loss of target gene induction by the aryl hydrocarbon receptor and hypoxia-inducible factor 1alpha. Molecular endocrinology (Baltimore, Md. 2000;14(10):1674–81. doi: 10.1210/mend.14.10.0533. [DOI] [PubMed] [Google Scholar]
- 19.Vrochides D, Papanikolaou V, Pertoft H, Antoniades a A, Heldin P. Biosynthesis and degradation of hyaluronan by nonparenchymal liver cells during liver regeneration. Hepatology. 1996;23(6):1650–5. doi: 10.1002/hep.510230648. [DOI] [PubMed] [Google Scholar]
- 20.Copple BL, Bustamante JJ, Welch TP, Kim ND, Moon JO. Hypoxia-inducible factor-dependent production of profibrotic mediators by hypoxic hepatocytes. Liver Int. 2009;29(7):1010–21. doi: 10.1111/j.1478-3231.2009.02015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim ND, Moon JO, Slitt a L, Copple BL. Early growth response factor-1 is critical for cholestatic liver injury. Toxicol Sci. 2006;90(2):586–95. doi: 10.1093/toxsci/kfj111. [DOI] [PubMed] [Google Scholar]
- 22.Oben JA, Yang S, Lin H, Ono M, Diehl a M. Norepinephrine and neuropeptide Y promote proliferation and collagen gene expression of hepatic myofibroblastic stellate cells. Biochemical and biophysical research communications. 2003;302(4):685–90. doi: 10.1016/s0006-291x(03)00232-8. [DOI] [PubMed] [Google Scholar]
- 23.Schwabe RF, Bataller R, Brenner DA. Human hepatic stellate cells express CCR5 and RANTES to induce proliferation and migration. Am J Physiol Gastrointest Liver Physiol. 2003;285(5):G949–58. doi: 10.1152/ajpgi.00215.2003. [DOI] [PubMed] [Google Scholar]
- 24.Seki E, De Minicis S, Gwak GY, Kluwe J, Inokuchi S, Bursill CA, et al. CCR1 and CCR5 promote hepatic fibrosis in mice. J Clin Invest. 2009;119(7):1858–70. doi: 10.1172/JCI37444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sugimoto R, Enjoji M, Nakamuta M, Ohta S, Kohjima M, Fukushima M, et al. Effect of IL-4 and IL-13 on collagen production in cultured LI90 human hepatic stellate cells. Liver Int. 2005;25(2):420–8. doi: 10.1111/j.1478-3231.2005.01087.x. [DOI] [PubMed] [Google Scholar]
- 26.Friedman SL, Arthur MJ. Activation of cultured rat hepatic lipocytes by Kupffer cell conditioned medium. Direct enhancement of matrix synthesis and stimulation of cell proliferation via induction of platelet-derived growth factor receptors. J Clin Invest. 1989;84(6):1780–5. doi: 10.1172/JCI114362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Myllyharju J. Prolyl 4-hydroxylases, the key enzymes of collagen biosynthesis. Matrix Biol. 2003;22(1):15–24. doi: 10.1016/s0945-053x(03)00006-4. [DOI] [PubMed] [Google Scholar]
- 28.Murphy PM, Baggiolini M, Charo IF, Hebert CA, Horuk R, Matsushima K, et al. International union of pharmacology. XXII. Nomenclature for chemokine receptors. Pharmacological reviews. 2000;52(1):145–76. [PubMed] [Google Scholar]
- 29.Weng HL, Liu Y, Chen JL, Huang T, Xu LJ, Godoy P, et al. The etiology of liver damage imparts cytokines transforming growth factor beta1 or interleukin-13 as driving forces in fibrogenesis. Hepatology. 2009;50(1):230–43. doi: 10.1002/hep.22934. [DOI] [PubMed] [Google Scholar]
- 30.Fallon PG, Richardson EJ, Mckenzie GJ, Mckenzie a N. Schistosome infection of transgenic mice defines distinct and contrasting pathogenic roles for IL-4 and IL-13: IL-13 is a profibrotic agent. J Immunol. 2000;164(5):2585–91. doi: 10.4049/jimmunol.164.5.2585. [DOI] [PubMed] [Google Scholar]
- 31.Cao Y, Linden P, Shima D, Browne F, Folkman J. In vivo angiogenic activity and hypoxia induction of heterodimers of placenta growth factor/vascular endothelial growth factor. J Clin Invest. 1996;98(11):2507–11. doi: 10.1172/JCI119069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chesney J, Metz C, Bacher M, Peng T, Meinhardt A, Bucala R. An essential role for macrophage migration inhibitory factor (MIF) in angiogenesis and the growth of a murine lymphoma. Molecular medicine (Cambridge, Mass. 1999;5(3):181–91. [PMC free article] [PubMed] [Google Scholar]
- 33.Le Jan S, Amy C, Cazes A, Monnot C, Lamande N, Favier J, et al. Angiopoietin-like 4 is a proangiogenic factor produced during ischemia and in conventional renal cell carcinoma. Am J Pathol. 2003;162(5):1521–8. doi: 10.1016/S0002-9440(10)64285-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kumagi T, Akbar F, Horiike N, Onji M. Increased serum levels of macrophage migration inhibitory factor in alcoholic liver diseases and their expression in liver tissues. Clinical biochemistry. 2001;34(3):189–93. doi: 10.1016/s0009-9120(01)00214-4. [DOI] [PubMed] [Google Scholar]
- 35.Akiyoshi F, Sata M, Suzuki H, Uchimura Y, Mitsuyama K, Matsuo K, et al. Serum vascular endothelial growth factor levels in various liver diseases. Dig Dis Sci. 1998;43(1):41–5. doi: 10.1023/a:1018863718430. [DOI] [PubMed] [Google Scholar]
- 36.Salcedo Mora X, Sanz-Cameno P, Medina J, Martin-Vilchez S, Garcia-Buey L, Borque MJ, et al. Association between angiogenesis soluble factors and disease progression markers in chronic hepatitis C patients. Rev Esp Enferm Dig. 2005;97(10):699–706. doi: 10.4321/s1130-01082005001000003. [DOI] [PubMed] [Google Scholar]
- 37.Welford SM, Bedogni B, Gradin K, Poellinger L, Broome Powell M, Giaccia a J. HIF1alpha delays premature senescence through the activation of MIF. Genes Dev. 2006;20(24):3366–71. doi: 10.1101/gad.1471106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krizhanovsky V, Yon M, Dickins RA, Hearn S, Simon J, Miething C, et al. Senescence of activated stellate cells limits liver fibrosis. Cell. 2008;134(4):657–67. doi: 10.1016/j.cell.2008.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hofbauer KH, Gess B, Lohaus C, Meyer HE, Katschinski D, Kurtz A. Oxygen tension regulates the expression of a group of procollagen hydroxylases. Eur J Biochem. 2003;270(22):4515–22. doi: 10.1046/j.1432-1033.2003.03846.x. [DOI] [PubMed] [Google Scholar]
- 40.De Minicis S, Seki E, Uchinami H, Kluwe J, Zhang Y, Brenner DA, et al. Gene expression profiles during hepatic stellate cell activation in culture and in vivo. Gastroenterology. 2007;132(5):1937–46. doi: 10.1053/j.gastro.2007.02.033. [DOI] [PubMed] [Google Scholar]