SUMMARY
Cytoplasmic dynein mediates spindle orientation from the cell cortex through interactions with astral microtubules, but neither the mechanism governing its cortical targeting, nor the regulation thereof, are well understood. Here we show that yeast dynein offloads from microtubule plus ends to the daughter cell cortex. Mutants with an engineered peptide inserted between the tail domain and the motor head retain wild-type motor activity but exhibit enhanced offloading and cortical targeting. Conversely, shortening the ‘neck’ sequence between the tail and motor domains precludes offloading from the microtubule plus ends. Furthermore, chimeric mutants with mammalian dynein ‘neck’ sequences rescue targeting and function. These findings provide direct support for an active microtubule-mediated delivery process that appears to be regulated by a conserved masking/unmasking mechanism.
INTRODUCTION
Cytoplasmic dynein is a 1.2 MDa multi-subunit motor complex that powers directional movement of cellular cargoes toward the minus end of microtubule (MT) tracks. This highly conserved motor has been implicated in diverse cellular processes including vesicular transport, centrosome positioning, and directed cell migration (Burakov et al., 2003; Dujardin et al., 2003). How cells regulate dynein activity with precise spatial and temporal control for each of these distinct functions is poorly understood. One way to regulate dynein activity is by spatially deploying the motor to its sites of action. A well-studied example is the regulated targeting of dynein to kinetochores at early prometaphase in mitotic mammalian cells (Whyte et al., 2008). In this case, dynein recruitment to kinetochores – for checkpoint silencing – depends on the phosphorylation state of the dynein intermediate chain, which specifies its interaction with the kinetochore component zw10. Another example of regulated dynein targeting comes from studies in budding yeast where it has been proposed that dynein exploits the dynamic instability of astral MTs for delivery to its cortical receptor, Num1 (Farkasovsky and Kuntzel, 2001; Heil-Chapdelaine et al., 2000; Lee et al., 2003; Sheeman et al., 2003). Although direct evidence for dynein cortical delivery is lacking, mutations that disrupt astral MT plus end localization of dynein result in a drastic reduction in cortical dynein (Markus et al., 2009) and a concomitant spindle misorientation defect (Lee et al., 2003; Sheeman et al., 2003). These data imply that dynein must associate with plus ends before it can be targeted to cortical Num1, and raises the question of how dynein is prevented from being directly recruited to cortical sites in the absence of MT plus end localization.
A clue to the mechanism of cortical targeting in yeast came from analysis of cells expressing truncated fragments of the dynein heavy chain Dyn1 (Markus et al., 2009). While the motor domain fragment of Dyn1 (Dyn1MOTOR) is necessary and sufficient for plus end targeting, the tail domain (Dyn1TAIL) is responsible for interaction with cortical Num1. Association of Dyn1TAIL with Num1 is very robust, more so than the full-length molecule, and importantly, occurs in a manner that is independent of plus end targeting or the presence of MTs. Based on these results, it was proposed that the cortical association domain within the NH2-terminal tail is masked by the motor head, and that targeting of Dyn1 to plus ends unmasks this region, priming the motor for offloading to cortical Num1.
Here we have further examined the mechanism by which the association between cytoplasmic dynein and cortical Num1 is regulated. By inserting peptide linkers between the NH2-terminal tail and COOH-terminal motor domains, we have engineered motility-competent mutants that are capable of bypassing the plus ends for association with cortical Num1. Surprisingly, in addition to observing a plus end-independent targeting mechanism, our analysis of the mutants reveals that they are also actively delivered by the plus ends to the cell cortex. Furthermore, in a genetic background where the dynein-dynactin interaction is enhanced, we observed offloading of wild-type dynein to the cortex, indicating that dynactin is limiting in the offloading process. Our findings support the notion that Dyn1 adopts a folded conformation that negatively regulates its association with cortical sites, and that this mechanism may be conserved throughout evolution.
RESULTS
Generation of a constitutively unmasked dynein
We postulated that, if intramolecular ‘masking’ of the Dyn1 cortical association domain occurs, such masking would likely depend on a carefully calibrated spatial linkage between the tail and motor domains of the protein. We therefore sought to engineer a Dyn1 mutant with a constitutively ‘unmasked’ cortical association domain, by inserting a helix-forming peptide (A(EAAAK)8A; Arai et al., 2001) into the junction between the tail and motor domains, creating Dyn1HL3 (Fig. 1A). We predicted that Dyn1HL3 would exhibit (1) cortical targeting reminiscent of Dyn1TAIL-3GFP, and (2) plus end targeting similar to Dyn1MOTOR-3YFP (Markus et al., 2009). We estimate that the inserted peptide has a length of approximately 11.4 nm (Arai et al., 2001), a distance roughly equivalent to the diameter of the motor head (Burgess et al., 2003). We fused a 3YFP or 13myc tag to the COOH-terminus of Dyn1HL3 for localization and immunoblotting analyses, respectively.
Figure 1. Insertion of a helical peptide (HL3) between the tail and motor domains of Dyn1 enhances its plus end and cortical targeting.
(A) Schematic representation of Dyn1 and the Dyn1HL3 mutant, with domain structure of Dyn1 indicated (blue region, ‘linker’ domain defined by in vitro studies (Reck-Peterson et al., 2006); red regions, six AAA domains (Mocz and Gibbons, 2001); green regions, anti-parallel coiled coils of the stalk; yellow region, MT-binding domain (Gee et al., 1997)). (B) Cells expressing mCherry-Tub1 and either wild-type Dyn1-3YFP (top) or Dyn1HL3-3YFP (bottom). Open arrowheads, SPB foci; closed arrowheads, cortical foci; arrows, plus end foci. Each image is a maximum intensity projection of a 2-μm Z-stack of wide-field images. (C) The percentage of cells that exhibit plus end (top) or cortical (bottom) fluorescent foci is plotted for strains expressing mCherry-Tub1 with Dyn1-3YFP, Dyn1HL3-3YFP, Dyn1MOTOR-3YFP, or Dyn1TAIL-3GFP. Stationary cortical foci and motile plus end foci were identified in two-color movies and scored accordingly. Error bars represent standard error of proportion (n ≥ 120 cells; *p = 0.0278; **p < 0.0001). (D) The percentage of cells exhibiting the indicated number of cortical fluorescent foci is plotted for strains expressing mCherry-Tub1 with Dyn1-3GFP, Dyn1HL3-3YFP or Dyn1TAIL-3GFP.
Like wild-type DYN1-3YFP cells (Fig. 1B, top panel), dyn1HL3-3YFP cells exhibited motile foci associated with spindle pole bodies (SPBs) and MT plus ends, as well as stationary cortical foci (Fig. 1B, bottom panel; 3D reconstructions in Video S1). As predicted, the frequency of Dyn1HL3-3YFP targeting to MT plus ends and the cell cortex was significantly greater than Dyn1-3YFP (Fig. 1C; 2.2- and 3.5-fold respectively; p < 0.0001). Moreover, the number of Dyn1HL3-3YFP cortical foci per cell was elevated with respect to Dyn1-3YFP (Fig. 1D). The frequency of plus end targeting for Dyn1HL3 was lower than Dyn1MOTOR (Fig. 1C; 0.7-fold; p < 0.0001), while that of cortical targeting was slightly higher than Dyn1TAIL (Fig. 1C; 1.2-fold; p = 0.0278). As previously described for DYN1-3GFP and dyn1TAIL-3GFP cells (Markus et al., 2009; Sheeman et al., 2003), plus end and cortical foci in the dyn1HL3-3YFP strain exhibited cell cycle-dependent changes in targeting frequencies, most notably as cells entered anaphase (Fig. S1A), suggesting similar mechanisms underlying the temporal regulation of their targeting. The differences in plus end and cortical targeting between Dyn1HL3 and Dyn1 could not be attributed to altered expression levels or protein stability, as determined by immunoblotting (Fig. S1B). Furthermore, 13myc-tagged Dyn1HL3 and Dyn1 exhibited similar sedimentation profiles in sucrose density gradients (Fig. S1C). Although the gradients were unable to resolve any differences in size and shape, the data suggested that Dyn1HL3 is assembled into a native complex with its accessory polypeptides. In support of this notion, biochemical isolation of TAP-tagged Dyn1HL3 showed that it copurified with the dynein light-intermediate (Dyn3) and intermediate (Pac11) chains to a similar extent as the wild-type TAP-Dyn1 control (Fig. S1D). Taken together, these data demonstrate that the observed targeting phenotype is not a result of improper dynein complex assembly or stability.
We next quantitated the extent to which plus end and cortical targeting of Dyn1HL3 was dependent on dynein pathway components (Moore et al., 2009). While plus end targeting of Dyn1HL3 required the tip-tracking proteins Pac1 (LIS1 homologue) and Bik1 (CLIP-170 homologue), its association with the cortex required the cortical protein Num1 (Fig. 2A and B; Fig. S2B). These results are consistent with Dyn1 (Lee et al., 2003; Sheeman et al., 2003), Dyn1MOTOR and Dyn1TAIL at each site (Markus et al., 2009). Furthermore, the pattern of Dyn1HL3 targeting observed in mutants lacking the dynein accessory chains (Dyn3 or Pac11) or a component of dynactin (Nip100) is more similar to that of Dyn1MOTOR or Dyn1TAIL than the full-length Dyn1 molecule at each site (see Fig. S2). Most notably, a high percentage of pac1Δ (42.7% ± 3.5%) and bik1Δ (46.0% ± 4.3%) cells exhibited stationary cortical Dyn1HL3-3YFP foci (Fig. 2B and C; 3D reconstruction in Video S2). This finding contrasts with Dyn1, which exhibited a loss of cortical foci in the same mutant strains (Markus et al., 2009). These data indicate that insertion of the helical linker enables Dyn1 to bypass the plus end for targeting to the cell cortex.
Figure 2. Association of Dyn1HL3 with the cell cortex occurs independently of plus end targeting.
(A – B) The percentage of cells that exhibit (A) plus end or (B) cortically associated Dyn1HL3-3YFP foci is plotted for wild-type (WT) and indicated null strains (n ≥ 105 cells). Stationary cortical or motile plus end foci were identified in two-color movies and scored accordingly. Error bars represent standard error of proportion. (C) Representative images of pac1Δ or bik1Δ cells expressing mCherry-Tub1 and Dyn1HL3-3YFP used for quantitation in panels (A) and (B). Closed arrowhead indicates cortical Dyn1HL3-3YFP foci. Each image is a maximum intensity projection of a 2-μm Z-stack of wide-field images.
Direct observation of Dynein offloading to the cell cortex
Although Dyn1HL3 does not require plus end targeting to associate with the cortex, we observed that a significantly greater percentage of cells exhibited cortical Dyn1HL3-3YFP foci when the plus end targeting mechanism was functional (i.e., in WT versus pac1Δ or bik1Δ backgrounds; Fig. 2B; 1.8-fold and 1.6-fold, respectively; p < 0.0001). Furthermore, several mutants with disrupted cortical targeting (num1Δ, pac11Δ, and nip100Δ) exhibited an enhancement of Dyn1HL3-3YFP at MT plus ends (Fig. S2C). Together, these data suggest that, in addition to direct recruitment from the cytosol, Dyn1HL3 might also be actively delivered to the cell cortex from the plus ends of astral MTs. Such delivery (offloading) has been previously proposed (Lee et al., 2005; Lee et al., 2003; Markus et al., 2009; Sheeman et al., 2003), but direct evidence has remained elusive. We investigated whether Dyn1HL3 can be observed undergoing offloading.
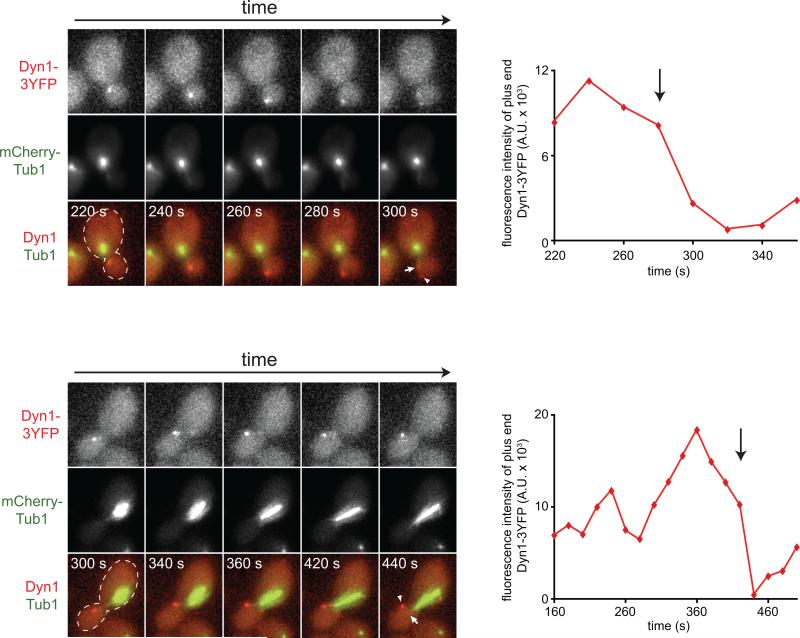
Strikingly, two-color time-lapse imaging of mCherry-Tub1 and Dyn1HL3-3YFP revealed that Dyn1HL3 is indeed offloaded from the plus ends of astral MTs to the cell cortex (Fig. 3A and B, left; Fig. S3C and D; Video S3). Intensity measurements of plus end Dyn1HL3-3YFP at the moments preceding and following the offloading events revealed that the majority of Dyn1HL3-3YFP was delivered to the cell cortex (Fig. 3A, right). Analysis of 27 offloading events revealed that the majority occurred in daughter cells during anaphase (Fig. S3A and B). Furthermore, we noted that immediately following offloading, the majority of astral MTs (96%) underwent catastrophe (Fig. 3B, right; Fig. S3D). These observations show that dynein does in fact utilize an offloading mechanism for cortical targeting. To our knowledge they represent the first demonstration that a cellular motor exploits the dynamic instability of astral MTs for delivery to its site of action.
Figure 3. Direct observation of Dyn1HL3 offloading from MT plus end to the cell cortex.
(A) Representative movie frames of cells expressing Dyn1HL3-3YFP and mCherry-Tub1. Arrowhead, offloaded Dyn1HL3; arrow, MT plus end following offloading event. Graphs depict fluorescence intensity of plus end-associated Dyn1HL3-3YFP at the moments preceding and directly following an offloading event (vertical arrow). Each image is a maximum intensity projection of a 2-μm Z-stack of wide-field images. Also see Video S3 and Fig. S3C. (B) Kymograph depicting a Dyn1HL3 offloading event and a life history plot of the same MT leading up to and immediately following the offloading event (also see Fig. S3D). Merge image in kymograph shows mCherry-Tub1 in green and Dyn1HL3-3YFP in red. MT lengths were measured using ImageJ from two-dimensional projections of 2-μm Z-stacks of wide-field fluorescence images. The time at which offloading occurred is indicated on the kymograph and the life history plot by the vertical arrow. (C) Similar to (A) but with cells expressing Dyn1HL3-3YFP, Bik1-3mCherry and CFP-Tub1. Graph depicts fluorescence intensity of plus end-associated Dyn1HL3-3YFP (red) and Bik1-3mCherry (blue). Also see Video S5, top.
We recently showed that the dynactin complex, which is required for cortical dynein localization (Lee et al., 2003), is limiting at MT plus ends with respect to dynein (1 dynactin to 3 dynein complexes; Markus et al., 2011). We predicted that enhancing the dynactin:dynein ratio at MT plus ends would enable us to visualize the offloading of wild-type dynein to the cell cortex. To this end, we generated a yeast strain lacking a regulator of dynactin-dynein binding at MT plus ends, She1 (Woodruff et al., 2009). Cells lacking She1 exhibit a dynactin:dynein ratio at MT plus ends that is close to 1:1 (Markus et al., 2011). Strikingly, as predicted, time lapse imaging of mCherry-Tub1 and Dyn1-3YFP in she1Δ cells revealed that wild-type Dyn1 is also offloaded from MT plus ends to the cell cortex (Fig. 4; Video S4). Analysis of 16 Dyn1-3YFP offloading events revealed that the majority occurred in daughter cells (15/16), while all events took place during pre-anaphase. These data are consistent with the notion that the association of dynactin with plus end-associated dynein is a limiting step in the offloading process.
Figure 4. Direct observation of wild-type Dyn1 offloading from MT plus end to the cell cortex.
(A) Representative movie frames of she1Δ cells expressing Dyn1-3YFP and mCherry-Tub1. Arrowhead, offloaded Dyn1; arrow, MT plus end following offloading event. Graphs depict fluorescence intensity of plus end-associated Dyn1-3YFP at the moments preceding and directly following an offloading event (vertical arrow). Each image is a maximum intensity projection of a 2-μm Z-stack of wide-field images. Also see Video S4.
In budding yeast, cortical dynein drives the sliding of astral MTs along the cell cortex (Adames and Cooper, 2000). However, we did not observe such activity following any of the Dyn1HL3 offloading events. Furthermore, dyn1HL3-3YFP cells had a level of spindle misorientation that was comparable to that of a dynein null strain (Fig. 5), suggesting that motor activity is compromised in this mutant (see more results below).
Figure 5. In vivo functional assessment of Dyn1 neck mutants.
The percentage of cells with a misoriented mitotic spindle in a cold (16°C) spindle position assay (Lee et al., 2005; Li et al., 2005) is plotted for haploid strains carrying DYN1-3YFP (indicated as DYN1), dyn1Δ, dyn1Δ pac1Δ, dyn1HL3-3YFP (indicated as dyn1HL3), dyn1HL3-3YFP pac1Δ, dyn1Δ20-3YFP (indicated as dyn1Δ20), and dyn1Δ20-3YFP pac1Δ. Spindles were visualized using mCherry-Tub1. Strains were imaged after growth at 16°C to mid-log in synthetic defined media lacking methionine (to induce mCherry-Tub1 expression controlled by the MET3 promoter). Error bars represent standard error of proportion (n ≥ 184 cells for each strain). Student's t-test was used to calculate p values.
Plus end-targeting components ectopically colocalize with cortical Dyn1HL3-3YFP
We used functionally-tagged fluorescent proteins (Markus et al., 2011) to assess the localization of dynein pathway components with respect to cortical Dyn1HL3-3YFP. As previously described for cortical Dyn1TAIL-3GFP foci (Markus et al., 2009), cortical Dyn1HL3-3YFP foci colocalized with Num1-mCherry, the dynein intermediate chain Pac11-3mCherry, and the dynactin subunit dynamitin Jnm1-3mCherry (Fig. S4A-C). The frequency with which cortical Pac11-3mCherry and Jnm1-3mCherry foci were observed in dyn1HL3-3YFP cells was enhanced significantly with respect to wild-type DYN1 cells (Jnm1: from 6.6% ± 1.8% to 73.5% ± 6.3%; Pac11: from 13.0% ± 1.8% to 86.8% ± 3.0%; n ≥ 49 cells; p < 0.0001). Since cortical targeting of both Pac11 and dynactin depend on Dyn1 (Lee et al., 2005; Moore et al., 2008), these data indicate that both are recruited to the cortex in complex with Dyn1HL3-3YFP.
Compared to wild-type (DYN1) cells, dyn1HL3-3YFP cells exhibited similar localization patterns and expression levels of Num1 (Fig. S5A and B), indicating that the enhanced cortical targeting of Dyn1HL3 is not due to changes in the dynein cortical receptor. However, we observed an enhanced association between Dyn1HL3 and Num1 as assessed by the bimolecular fluorescence complementation assay (BiFC; Hu et al., 2002). VN-Dyn1HL3 and VC-Num1 (see Fig. S5) expressing cells possessed brighter and a significantly greater number of cortical BiFC foci than those expressing VN-Dyn1 and VC-Num1 (Fig. S5C and D). These observations are consistent with the presence of a higher-order complex of Dyn1HL3 with various dynein and dynactin components at the cell cortex.
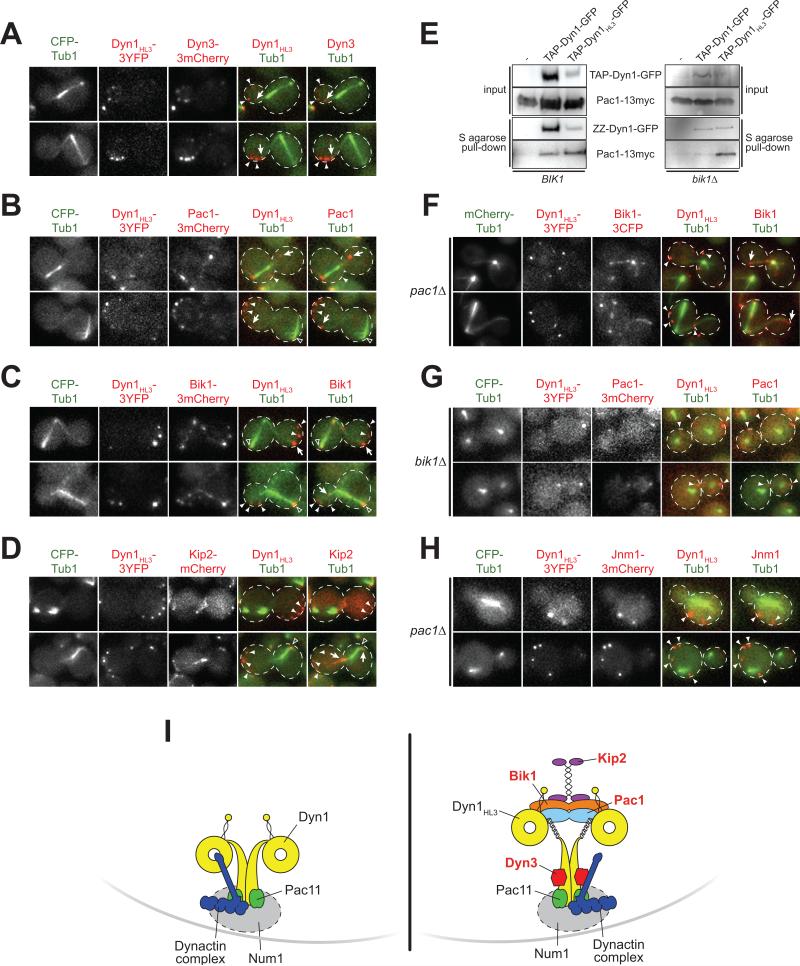
We found that, in contrast to cortical Dyn1 (depicted in Fig. 6I), cortical Dyn1HL3-3YFP colocalized with four components required for dynein plus end-targeting (Carvalho et al., 2004; Lee et al., 2003; Sheeman et al., 2003; S. Markus and W.-L. Lee, unpublished), namely Dyn3-3mCherry, Pac1-3mCherry, Bik1-3mCherry, and the kinesin Kip2-mCherry (Fig. 6A–D and I). However, another tip-tracking protein, Bim1-3mCherry, the EB1 homologue, was not found at the cortex in dyn1HL3-3YFP cells, consistent with its noninvolvement in the budding yeast dynein pathway (Carvalho et al., 2004) (Fig. S4D). Dyn3 associates with the tail domain (Fig. S4E), whereas our previous work (Markus et al., 2009) suggested that Pac1, Bik1, and Kip2 likely associate with the motor domain of Dyn1HL3 (Fig. 6I). Since Pac1 binds with higher affinity to Dyn1MOTOR than to full-length Dyn1 (Markus et al., 2009), we tested if Pac1 would exhibit an enhanced interaction with Dyn1HL3. As expected, and consistent with being unmasked, TAP-tagged Dyn1HL3 pulled down more Pac1-13myc as compared to wild-type TAP-Dyn1 (Fig 6E; 5.6-fold when normalized to levels of purified Dyn1; see Fig. 6E legend). We conclude that all four of the plus end-targeting components (Dyn3, Pac1, Bik1, and Kip2) are recruited to the cortex in complex with Dyn1HL3-3YFP, given that they are found exclusively associated with MTs and are absent from the cortex of wild-type cells (Carvalho et al., 2004; Lee et al., 2005; Lee et al., 2003; Lin et al., 2001).
Figure 6. Dyn1HL3 expressing cells exhibit ectopic cortical Dyn3, Pac1, Bik1 and Kip2.
(A – D) Wide-field fluorescence images of dyn1HL3-3YFP cells expressing (A) Dyn3-3mCherry, (B) Pac1-3mCherry, (C) Bik1-3mCherry, or (D) Kip2-mCherry. (E) TAP-tagged Dyn1HL3-GFP pulls down more Pac1-13myc compared to wild-type Dyn1-GFP, in the presence (left; BIK1) and absence (right; bik1Δ) of Bik1. Equal amounts of protein lysate were incubated with S-protein agarose. Bound proteins were released by TEV protease digestion and immunoblotted with rabbit IgG (for ZZ-Dyn1-GFP or ZZ-Dyn1HL3-GFP) or anti-c-Myc (for Pac1-13myc). The yield of ZZ-Dyn1HL3-GFP from cell lysate was consistently less than that for wild-type ZZ-Dyn1-GFP (left, middle and right lanes; also see Fig. S1D). Upon deletion of Bik1 (right) or Pac1 (not shown), recovery of ZZ-Dyn1HL3-GFP was improved to a level comparable to that for wild-type ZZ-Dyn1-GFP. (F – G) Cortical Bik1 is lost in dyn1HL3-3YFP pac1Δ cells, but Pac1 is retained at the cortex in dyn1HL3-3YFP bik1Δ cells. (H) Colocalization of the dynactin subunit dynamitin Jnm1 with cortical Dyn1HL3 in pac1Δ cells. All images are maximum intensity projections of a 2-μm Z-stack of wide-field fluorescence images. Open arrowheads, SPB foci; closed arrowheads, cortical foci; arrows, plus end foci. (I) Schematic drawings of wild-type (left) and Dyn1HL3 (right) cortical dynein complexes. Kip2, Bik1, Pac1, and Dyn3 (labeled in red) are not found at the cell cortex in wild-type cells.
In support of this conclusion, three-color time-lapse imaging of CFP-Tub1, Dyn1HL3-3YFP and either Bik1-3mCherry or Pac1-3mCherry revealed that both Bik1 and Pac1 are offloaded from MT plus ends to the cell cortex together with Dyn1HL3-3YFP (Fig. 3C; Video S5). These data demonstrate that the dynein plus end-targeting components are recruited to the cell cortex in part through offloading as a co-complex with Dyn1HL3-3YFP.
We asked whether the abnormal association of cortical dynein with Pac1, Bik1 and Kip2 accounts for the lack of dynein function observed in the dyn1HL3-3YFP strain. Since deletion of Pac1 resulted in a loss of cortical Bik1 (Fig. 6F; but not vice versa, Fig. 6G) and Kip2 (Fig. S4F), but not cortical dynactin (Fig. 6H), we used a pac1Δ mutant to assess the function of Dyn1HL3-3YFP. Interestingly, the spindle misorientation defect noted in the dyn1HL3-3YFP strain was partially rescued by loss of Pac1 (Fig. 5), suggesting that association of cortical dynein with the plus end-targeting machinery may be a causal factor for defective activity. Since loss of Dyn3 resulted in defective Dyn1HL3-3YFP cortical targeting (Fig. S2A and B), we were unable to determine the consequence of cortical Dyn3 on in vivo dynein function.
Single-molecule analysis reveals that Dyn1HL3 is a processive motor, and that Pac1 reduces dynein velocity
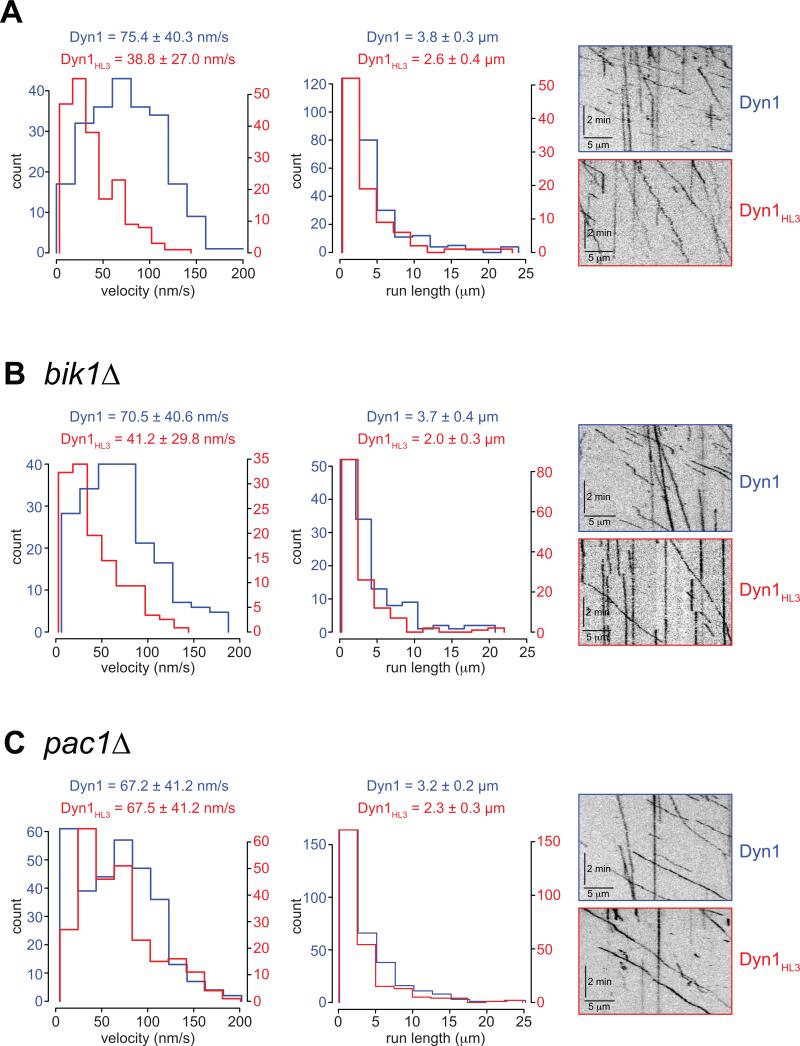
To further investigate the motor function of Dyn1HL3, we purified full-length Dyn1HL3-GFP and wild-type Dyn1-GFP from yeast using an NH2-terminal TAP tag, and examined their motile behavior in vitro at single-molecule resolution using time-lapse total internal reflection fluorescence (TIRF) microscopy (Fig. S6A). We verified that we were observing single molecules by quantitating stepwise photobleaching of purified GFP particles (Fig. S6B and C). The maximum number of bleaching events observed for any fluorescent particle was two, for both Dyn1HL3-GFP and Dyn1-GFP, consistent with the obligatory dimeric nature of motile dynein (Reck-Peterson et al., 2006). Single Dyn1-GFP molecules traveled along MTs with an average velocity of 75.4 nm/sec and an average run length of 3.8 μm (Fig. 7A), values close to that previously described for TMR-labeled yeast cytoplasmic dynein (Cho et al., 2008; Kardon et al., 2009; Reck-Peterson et al., 2006). Interestingly, single molecules of Dyn1HL3-GFP also exhibited processive movement along MTs; however, they moved significantly slower than wild-type Dyn1-GFP (Fig. 7A; 38.8 nm/sec). To determine whether the reduced velocity of Dyn1HL3-GFP was due to an enhanced association with the plus end-targeting machinery (Fig. 6), we purified Dyn1-GFP and Dyn1HL3-GFP from cells lacking either Bik1 or Pac1. Strikingly, Dyn1HL3-GFP isolated from a pac1Δ strain (Fig. 7C; video S7), but not bik1Δ (Fig. 7B; video S6), exhibited a mean velocity (67.5 ± 41.2 nm/s) very similar to wild-type Dyn1-GFP (67.2 ± 41.2 nm/s), with a slightly reduced run length (2.3 ± 0.2 μm versus 3.2 ± 0.2 μm). These data are consistent with the spindle misorientation assay (Fig. 5), and suggest that Pac1, which copurifies with Dyn1HL3 even in the absence of Bik1 (see Fig. 6E), is a potent negative regulator of dynein motility.
Figure 7. Dyn1HL3 isolated from cells lacking Pac1, but not Bik1, exhibits wild-type processive motility.
Histograms of velocities and run lengths of Dyn1 (blue) or Dyn1HL3 (red) isolated from (A) wild-type, (B) bik1Δ or (C) pac1Δ strains are shown with representative kymographs from each. Single molecules of Dyn1-GFP or Dyn1HL3-GFP (see Fig. S6B and C) were visualized on taxol-stabilized rhodamine-labeled MTs using time-lapse TIRF microscopy. Mean velocities ± standard deviation, and run lengths (determined from exponential decay fits) ± standard error are shown for each. Only those dynein motors that moved with a velocity greater than zero were chosen for velocity and run length measurements. Also see Videos S6 and S7.
Insertion versus removal of amino acids at the tail/motor junction produces opposite dynein targeting phenotypes
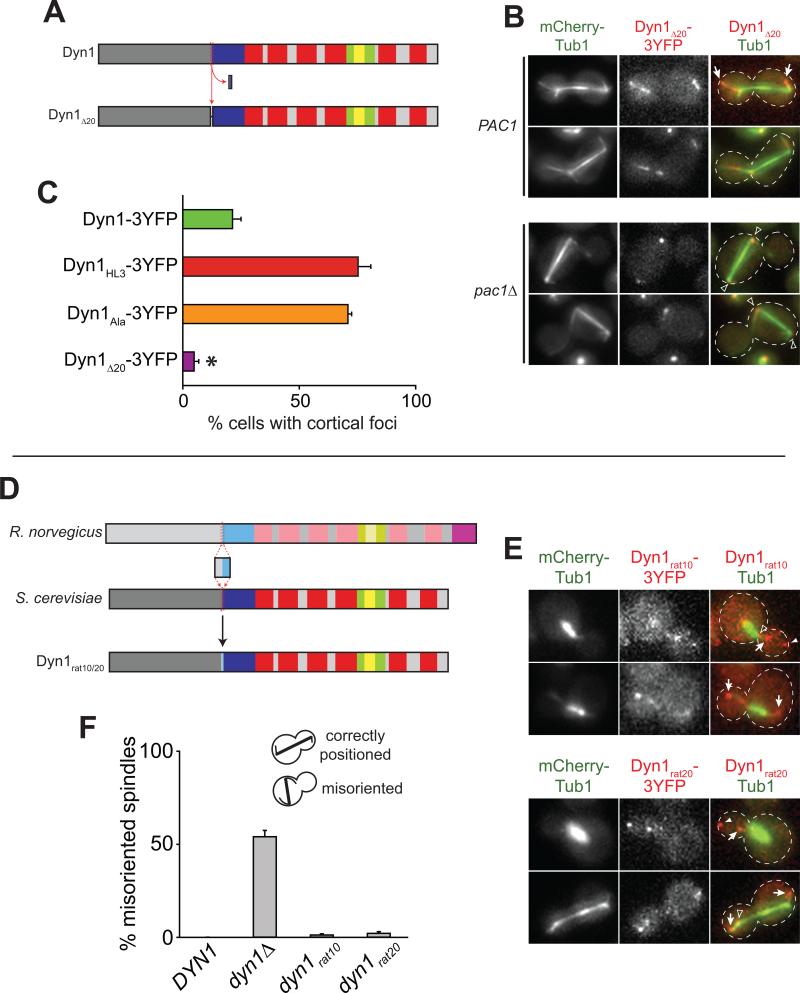
We next asked whether linkers of different lengths and properties could enable Dyn1 to bypass the plus end for association with the cell cortex. We inserted shorter helix-forming or flexible linkers (Arai et al., 2001) into the tail/motor junction along with a COOH-terminal 3YFP tag. To our surprise, all linkers tested, including a single alanine (Dyn1Ala-3YFP) or proline (Dyn1Pro-3YFP) insertion, were sufficient to cause a phenotype consistent with a constitutively unmasked state. Cortical targeting for Dyn1Ala-3YFP was indistinguishable from Dyn1HL3-3YFP in both PAC1 and pac1Δ cells (Fig. 8C and data not shown).
Figure 8. Differential targeting of dynein elicited by peptide insertion, deletion and substitution.
(A) Schematic depicting construction of the Dyn1Δ20 mutant. (B) Representative wide-field fluorescence images of PAC1 (top) or pac1Δ (bottom) cells expressing mCherry-Tub1 and Dyn1Δ20-3YFP. Open arrowheads, SPB foci; arrows, plus end foci. (C) The percentage of cells that exhibit cortical fluorescent foci is plotted for strains expressing mCherry-Tub1 with Dyn1-3YFP, Dyn1HL3-3YFP, Dyn1Ala-3YFP, or Dyn1Δ20-3YFP. Stationary cortical foci were identified in two-color movies and scored accordingly. Error bars represent standard error of proportion (n ≥ 69 cells; *p < 0.0001). (D) Schematic representation of the Dyn1rat10 and Dyn1rat20 mutants (see Fig. 1A for domain structure). (E) Representative wide-field fluorescence images of cells expressing mCherry-Tub1 and either Dyn1rat10-3YFP (top) or Dyn1rat20-3YFP (bottom). Open arrowheads, SPB foci; closed arrowheads, cortical foci; arrows, plus end foci. Each image (in B and E) is a maximum intensity projection of a 2-μm Z-stack of wide-field images. (F) The percentage of cells with a misoriented mitotic spindle in a cold (16°C) spindle position assay is plotted for strains carrying DYN1 (wild-type), dyn1Δ, dyn1rat10-3YFP, or dyn1rat20-3YFP (n ≥ 217 cells for each strain). Error bars represent standard error of proportion.
To test whether the unmasked phenotype is specifically elicited by peptide insertions, we deleted a 20 amino acid sequence spanning the tail/motor junction (Fig. 8A) and determined its effects on Dyn1 localization and function. Like wild-type Dyn1 and the insertion mutants, Dyn1Δ20-3YFP localized to SPBs and astral MT plus ends; however, this mutant also localized along the length of astral MTs (Fig. 8B, top), and was found at the cell cortex in only 4.9% ± 1.6% of cells (compared to 21.4% ± 3.9% for Dyn1-3YFP; Fig. 8C). Furthermore, deletion of Pac1 resulted in a complete loss of Dyn1Δ20-3YFP from plus ends and the cell cortex (n = 141 cells; Fig. 8B, bottom), suggesting that Dyn1Δ20-3YFP could not be directly recruited from the cytosol to the cell cortex, a result consistent with a masked phenotype. Additionally, a dyn1Δ20-3YFP mutant and a dyn1Δ20-3YFP pac1Δ double mutant exhibited levels of spindle misorientation comparable to a dyn1Δ mutant and a dyn1Δ pac1Δ double mutant (Fig. 5). Together, these data suggest that Dyn1Δ20-3YFP exhibits properties indicative of a constitutively masked state.
Conservation of structure-function within the dynein ‘neck’ region
Previous structural analyses of the dynein heavy chain (Burgess et al., 2004; Meng et al., 2006) have revealed a great degree of flexibility – both planar and torsional – at the junction between the tail and motor domains. The pivot point for this flexibility is situated within the ‘neck’, the region targeted for mutagenesis in our study. Secondary structure prediction of this region revealed a high probability of alpha-helical content that is strongly conserved among S. cerevisiae, S. pombe, and R. norvegicus, despite a fairly low similarity in primary sequence (Fig. S7A and B). Moreover, we noted that the number of amino acids within this region is invariant across species, with no gaps observed in a 219 amino acid stretch (Fig. S7C, red underline), suggesting that the length of the region spanning the tail/motor junction is important for dynein function. Since altering this region had little effect on motor activity as demonstrated by the in vitro motility assays, we deduced that this region may be important for the proper targeting of dynein.
Given the highly conserved alpha-helical pattern of the neck region (Fig. S7A), we asked whether a neck sequence from the rat dynein heavy chain could functionally substitute for the corresponding region in yeast Dyn1. We replaced a 10- or 20-amino acid stretch spanning the tail/motor junction of Dyn1 (amino acids 1359-1368 or 1354-1373) with the corresponding rat sequence, generating Dyn1rat10-3YFP and Dyn1rat20-3YFP, respectively (Fig. 8D). We found that the plus end and cortical targeting for Dyn1rat10-3YFP and Dyn1rat20-3YFP was comparable to wild-type Dyn1-3YFP (Fig. 8E). Furthermore, both chimeras fully rescued dynein function as determined by a spindle misorientation assay (Fig. 8F). These data indicate that the secondary structure of the neck, but not the primary sequence per se, is important for Dyn1 targeting and function. They further suggest a conservation of the mechanism regulating the subcellular targeting of the dynein complex.
DISCUSSION
In summary, we have characterized the mechanism of Dyn1 cortical targeting via offloading from microtubule plus ends. Importantly, the length of the ‘neck’ linking the motor head and cortex-targeting tail domains is a critical determinant of this mode of Dyn1 targeting. Increasing neck length not only promotes plus end Dyn1 offloading to the cortex, and indeed permits cortical binding independent of prior localization to plus ends, it also enhances the association of cortical Dyn1 with plus end protein partners. These findings suggest that the increase in neck length unmasks the heavy chain of yeast cytoplasmic dynein to permit promiscuous association with diverse partners, including Pac1 and Num1. Since shortening the neck conversely stabilizes Dyn1 at plus ends and precludes its offloading to the cortex, we propose that Dyn1 normally utilizes this masking/unmasking mechanism to regulate its subcellular localization. In vitro motility assays revealed that amino acid insertion into the dynein neck region, which is invariant in length across species, does not disrupt MT binding or motor activity, suggesting that the observed in vivo cortical targeting phenotype is not due to compromised motor activity, but that this region instead has a specific role in regulating dynein targeting. Furthermore, the ability of a neck sequence from a vertebrate dynein heavy chain to functionally substitute for the corresponding region in Dyn1 suggests a conservation of structure and function across species, and suggests that a similar mechanism may exist to regulate dynein targeting in higher eukaryotes.
The regulation of Dyn1 targeting to the cell cortex is likely crucial to spatially and temporally restrict dynein activity. Consistent with this idea, we previously showed that association of Dyn1 with the cell cortex is enhanced as cells approach anaphase (Markus et al., 2009). Similarly, cortical targeting of dynein-dynactin in mammalian cells appears to be temporally restricted to prometaphase and metaphase (Busson et al., 1998; Kobayashi and Murayama, 2009), suggesting a similar mechanism may be in place to regulate cortical dynein. Our observations define a strategy by which dynein can restrict its own spatial targeting and support an emerging view that dynein activity can be regulated by its spatial deployment. Since purified dynein is active for processive movement along MTs, it has been proposed that its specificity of action is accomplished by cofactor-mediated inhibition, rather than activation (Kardon and Vale, 2009); however, no such inhibitor has yet been identified. By precisely restricting dynein targeting to its site of action, the need to regulate its motor activity is minimized. It is interesting to note that the majority of Dyn1 offloading events occurred within the daughter cell, regardless of whether the protein was mutated. This bias may be a result of upstream events regulating plus end recruitment of dynein (Grava et al., 2006), and may have implications for the targeting of polarity factors during asymmetric cell division.
Using purified Dyn1TAIL and Dyn1MOTOR, we were unable to detect an interaction between the two domains in solution, as they migrated independently in a sucrose gradient (S. Markus and W.-L. Lee, unpublished). Since the putative unmasking process can be triggered by the insertion of a single amino acid (i.e., Dyn1Ala-3YFP and Dyn1Pro-3YFP), it seems plausible that any potential interaction may be weak, and thus difficult to detect. Additionally, it is possible that the interaction is either inhibited by a copurifying factor, or mediated by a cofactor that is absent from the purification. As an example of the latter, an interaction between the Dam1 and Ndc80 kinetochore complexes could only be detected in the presence of MTs (Lampert et al., 2010). Our data, however, are not consistent with the tail-motor interaction being mediated by MTs, since disruption of Dyn1 plus end association (i.e., in pac1Δ or bik1Δ strains; Markus et al., 2009) results in a loss of cortical dynein due to the adoption of a masked state. Alternatively, it is possible that there is no direct interaction between these two domains. Rather, the masked state may be mediated by a folded conformation resulting from a bending within the neck of the heavy chain. Previous structural analyses of dynein have revealed the potential for such conformational changes as a result of the flexibility situated within the neck region (Burgess et al., 2004; Meng et al., 2006).
Other motors, such as kinesin (Cai et al., 2007; Coy et al., 1999; Friedman and Vale, 1999; Hackney et al., 1992; Seiler et al., 2000; Stock et al., 1999; Verhey et al., 1998) and myosin (Krementsov et al., 2004; Pasternak et al., 1989; Stoffler and Bahler, 1998; Wang et al., 2004) undergo intramolecular interactions to modulate their enzymatic activity. In both cases, the COOH-terminal tail domain inhibits the ATPase activity of the NH2-terminal motor head. Upon cargo binding (or Ca2+, in the case of myosin), the motor head exhibits enhanced ATPase activity and becomes activated for track binding. Nishiura et al. (Nishiura et al., 2004) found that a dynein motor domain construct from Dictyostelium possessed a significantly higher MT-stimulated ATPase activity than full-length bovine dynein. Whether this is attributable to species-specific variation, or to the monomeric (motor fragment) versus dimeric (full-length) states is unknown. However, the authors proposed the possibility that the tail domain may suppress the ATPase cycle of the dynein motor. Here, we provide evidence that a similar, yet distinct process is taking place, that the motor domain is precluding the tail domain from binding to cortical Num1.
The specific event that triggers unmasking is unknown, but may involve the association of dynactin with plus end-bound dynein. Although dynein is targeted to MT plus ends independently of dynactin in yeast, the association of dynein with the cell cortex is dependent upon dynactin (Lee et al., 2003; Moore et al., 2008; Sheeman et al., 2003). Furthermore, we recently showed that dynactin at plus ends is limiting with respect to dynein (1 dynactin complex per ~3 dynein complexes; Markus et al., 2011), and work from another lab showed that She1, a regulator of dynein activity, may actively preclude this association (Woodruff et al., 2009). In support of this hypothesis, we have observed here that wild-type Dyn1 offloads to the cell cortex in cells lacking She1. In she1Δ cells, the dynactin:dynein ratio is increased to 1:1 (Markus et al., 2011). As a result, she1Δ cells have been seen to exhibit hyper-cortical dynein activity (Markus et al., 2011; Woodruff et al., 2009). These data are consistent with the notion that the binding of dynactin to plus end dynein triggers the unmasking of the cortical association domain situated within the dynein tail domain.
The differential motile properties of Dyn1HL3-GFP purified from bik1Δ and pac1Δ strains indicate that the LIS1 homolog, Pac1, which copurifies with Dyn1HL3 even in the absence of Bik1 (see Fig. 6E), is likely responsible for reducing the velocity of this mutant. These data are consistent with the partial rescue of spindle misorientation we observed for Dyn1HL3 in pac1Δ cells (see Fig. 5). These data are also consistent with two recent in vitro studies, which demonstrated that LIS1 reduces the net velocity of dynein (McKenney et al., 2010; Torisawa et al., 2011). The significance of the Pac1/LIS1-mediated reduction of dynein velocity is not known; however, it is tempting to speculate that Pac1 may allow dynein to accumulate at MT plus ends by keeping it in an ‘off’ state, thereby allowing MT-dependent delivery to the cell cortex to consequently occur.
EXPERIMENTAL PROCEDURES
Plasmid construction
A series of plasmids were generated to integrate various peptide sequences between the tail and motor domains of DYN1 (between amino acids 1363 and 1364) at the native genomic locus. The motor domain defined by this junction corresponds to the Dyn1314 kDa construct shown to display functional motility in previous in vitro studies (Reck-Peterson et al., 2006). For a detailed discussion of the specific steps used, please see the Supplemental Experimental Procedures.
Media and strain construction
Strains were either derived from the protease-deficient background YWL29 (a.k.a., BJ5457; Jones, 1990), or from YWL36 or YWL37 (Vorvis et al., 2008) and are available upon request. We transformed yeast strains using the lithium acetate method (Knop et al., 1999). Strains carrying null mutations or fluorescently tagged components were constructed by PCR product-mediated transformation (Longtine et al., 1998) or by mating followed by tetrad dissection. Transformants were clonally purified by streaking to individual colonies on selective media. Proper tagging was confirmed by PCR. At least two independent transformants were chosen from each tagging and disruption procedure for subsequent experiments. Yeast synthetic defined (SD) media was obtained from Sunrise Science Products (San Diego, CA). A yeast genomic DNA isolation kit was obtained from Zymo Research (Orange, CA). For details of strain construction methods, please see the Supplemental Experimental Procedures.
Image acquisition and motility assays
Yeast cultures were grown to mid-log phase at 30°C and analyzed on an agarose pad containing nonfluorescent SD media or 50 mM potassium phosphate buffer, pH 7. Wide-field fluorescence images were collected using a 1.49 NA 100X objective on a Nikon 80i upright microscope equipped with piezo Z-control (Physik Instrumente), electronically controlled SmartShutter (Sutter Instrument), motorized filter cube turret, and a cooled EM-CCD Cascade-II camera (Photometrics). Microscope system was controlled by NIS-Elements software (Nikon). Step size of 1 μm was used to acquire Z-stack images 2 μm thick. Sputtered/ET filter cube sets (Chroma Technology) were used for imaging CFP (49001), GFP (49002), YFP (49003), and mCherry (49008) fluorescence. Confocal images (Videos S1-S2) were acquired at the UMass microscope facility using a 1.49 NA 100X objective on an inverted Nikon Ti-E microscope equipped with a Perkin Elmer UltraVIEW VoX and 488 nm/561 nm lasers. Step size of 0.2 μm was used to acquire Z-stack images 7.2 μm thick. 3D image reconstruction was performed using ImageJ software.
The motility assay was modified from a previously described protocol (Reck-Peterson et al., 2006). Flow chambers were constructed using slides and silanized coverslips (Repel-Silane ES, GE Healthcare) attached with double-sided adhesive tape. The flow chamber was coated with anti-tubulin antibody (8 μg/ml, YL1/2; Accurate Chemical & Scientific Corporation) and then blocked with 5% Pluronic F-127 (Fisher Scientific). Taxol-stabilized MTs assembled from unlabeled and X-rhodamine-labeled bovine tubulin (10:1 ratio; Cytoskeleton) were introduced into the chamber. Following a 15-minute incubation, the chamber was washed with dynein lysis buffer (see Supplemental Experimental Procedures) supplemented with 20 μM taxol, and then either Dyn1-GFP or Dyn1HL3-GFP was added to the chamber. After a 2-minute incubation the chamber was washed again and motility buffer (30 mM HEPES, pH 7.2, 50 mM potassium acetate, 2 mM magnesium acetate, 1 mM EGTA, 1 mM DTT, 1 mM Mg-ATP) supplemented with 0.05% Pluronic F-127, 20 μM taxol and an oxygen-scavenging system (1.5% glucose, 1 U/μl glucose oxidase, 125 U/μl catalase) was added. TIRF images were collected using a 1.49 NA 100X TIRF objective on a Nikon Ti-E inverted microscope equipped with 488 nm and 561 nm 50 mW diode lasers (Coherent), a motorized TIRF illumination unit, a Perfect Focus unit with motorized nosepiece and filter cube turret (Nikon), an electronically controlled emission filter wheel (Sutter Instrument), and an iXON+ EMCCD 888 camera (Andor Technology). Microscope system was controlled by NIS-Elements software (Nikon). We used a multi-pass quad filter cube set (C-TIRF for 405/488/561/638 nm; Chroma) and emission filters mounted in the filter wheel (525/50 nm and 600/50 nm; Chroma) for imaging GFP fluorescence in the TIRF field. To collect movies of individual dynein molecules moving on MTs, we acquired frames at 2 s intervals for 8 min. Velocity and run length were determined from kymographs generated using the MultipleKymograph plugin for ImageJ. For photobleaching experiments, imaging was conducted as above, except the oxygen-scavenging system was omitted, and there was no delay between exposures.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Vladimir I. Gelfand for valuable discussion and experimental suggestions on the characterization of Dyn1HL3. We thank Patricia Wadsworth for critical reading of the manuscript. We thank Jennifer Ross for generously offering the ultracentrifuge in her lab for the sucrose gradient sedimentation analysis. We are very grateful to Juan Daniel Diaz-Valencia and Leslie Conway for their help in preparing taxol-stabilized MTs and flow chambers used in the single molecule assays. The authors wish to specially thank the editor for suggestions on editing the manuscript. This work was supported by an NIH/NIGMS grant (1R01GM076094) to W.-L. L.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Adames NR, Cooper JA. Microtubule interactions with the cell cortex causing nuclear movements in Saccharomyces cerevisiae. J Cell Biol. 2000;149:863–874. doi: 10.1083/jcb.149.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai R, Ueda H, Kitayama A, Kamiya N, Nagamune T. Design of the linkers which effectively separate domains of a bifunctional fusion protein. Protein Eng. 2001;14:529–532. doi: 10.1093/protein/14.8.529. [DOI] [PubMed] [Google Scholar]
- Burakov A, Nadezhdina E, Slepchenko B, Rodionov V. Centrosome positioning in interphase cells. J Cell Biol. 2003;162:963–969. doi: 10.1083/jcb.200305082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess SA, Walker ML, Sakakibara H, Knight PJ, Oiwa K. Dynein structure and power stroke. Nature. 2003;421:715–718. doi: 10.1038/nature01377. [DOI] [PubMed] [Google Scholar]
- Burgess SA, Walker ML, Sakakibara H, Oiwa K, Knight PJ. The structure of dynein-c by negative stain electron microscopy. J Struct Biol. 2004;146:205–216. doi: 10.1016/j.jsb.2003.10.005. [DOI] [PubMed] [Google Scholar]
- Busson S, Dujardin D, Moreau A, Dompierre J, De Mey JR. Dynein and dynactin are localized to astral microtubules and at cortical sites in mitotic epithelial cells. Curr Biol. 1998;8:541–544. doi: 10.1016/s0960-9822(98)70208-8. [DOI] [PubMed] [Google Scholar]
- Cai D, Hoppe AD, Swanson JA, Verhey KJ. Kinesin-1 structural organization and conformational changes revealed by FRET stoichiometry in live cells. J Cell Biol. 2007;176:51–63. doi: 10.1083/jcb.200605097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho P, Gupta ML, Jr., Hoyt MA, Pellman D. Cell cycle control of kinesin-mediated transport of Bik1 (CLIP-170) regulates microtubule stability and dynein activation. Dev Cell. 2004;6:815–829. doi: 10.1016/j.devcel.2004.05.001. [DOI] [PubMed] [Google Scholar]
- Cho C, Reck-Peterson SL, Vale RD. Regulatory ATPase sites of cytoplasmic dynein affect processivity and force generation. J Biol Chem. 2008;283:25839–25845. doi: 10.1074/jbc.M802951200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coy DL, Hancock WO, Wagenbach M, Howard J. Kinesin's tail domain is an inhibitory regulator of the motor domain. Nat Cell Biol. 1999;1:288–292. doi: 10.1038/13001. [DOI] [PubMed] [Google Scholar]
- Dujardin DL, Barnhart LE, Stehman SA, Gomes ER, Gundersen GG, Vallee RB. A role for cytoplasmic dynein and LIS1 in directed cell movement. J Cell Biol. 2003;163:1205–1211. doi: 10.1083/jcb.200310097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkasovsky M, Kuntzel H. Cortical Num1p interacts with the dynein intermediate chain Pac11p and cytoplasmic microtubules in budding yeast. J Cell Biol. 2001;152:251–262. doi: 10.1083/jcb.152.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman DS, Vale RD. Single-molecule analysis of kinesin motility reveals regulation by the cargo-binding tail domain. Nat Cell Biol. 1999;1:293–297. doi: 10.1038/13008. [DOI] [PubMed] [Google Scholar]
- Gee MA, Heuser JE, Vallee RB. An extended microtubule-binding structure within the dynein motor domain. Nature. 1997;390:636–639. doi: 10.1038/37663. [DOI] [PubMed] [Google Scholar]
- Grava S, Schaerer F, Faty M, Philippsen P, Barral Y. Asymmetric recruitment of dynein to spindle poles and microtubules promotes proper spindle orientation in yeast. Dev Cell. 2006;10:425–439. doi: 10.1016/j.devcel.2006.02.018. [DOI] [PubMed] [Google Scholar]
- Hackney DD, Levitt JD, Suhan J. Kinesin undergoes a 9 S to 6 S conformational transition. J Biol Chem. 1992;267:8696–8701. [PubMed] [Google Scholar]
- Heil-Chapdelaine RA, Oberle JR, Cooper JA. The cortical protein Num1p is essential for dynein-dependent interactions of microtubules with the cortex. J Cell Biol. 2000;151:1337–1344. doi: 10.1083/jcb.151.6.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu CD, Chinenov Y, Kerppola TK. Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol Cell. 2002;9:789–798. doi: 10.1016/s1097-2765(02)00496-3. [DOI] [PubMed] [Google Scholar]
- Jones EW. Vacuolar proteases in yeast Saccharomyces cerevisiae. Methods Enzymol. 1990;185:372–386. doi: 10.1016/0076-6879(90)85033-k. [DOI] [PubMed] [Google Scholar]
- Kardon JR, Reck-Peterson SL, Vale RD. Regulation of the processivity and intracellular localization of Saccharomyces cerevisiae dynein by dynactin. Proc Natl Acad Sci U S A. 2009;106:5669–5674. doi: 10.1073/pnas.0900976106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardon JR, Vale RD. Regulators of the cytoplasmic dynein motor. Nat Rev Mol Cell Biol. 2009;10:854–865. doi: 10.1038/nrm2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M, Siegers K, Pereira G, Zachariae W, Winsor B, Nasmyth K, Schiebel E. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast. 1999;15:963–972. doi: 10.1002/(SICI)1097-0061(199907)15:10B<963::AID-YEA399>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Murayama T. Cell cycle-dependent microtubule-based dynamic transport of cytoplasmic dynein in mammalian cells. PLoS One. 2009;4:e7827. doi: 10.1371/journal.pone.0007827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krementsov DN, Krementsova EB, Trybus KM. Myosin V: regulation by calcium, calmodulin, and the tail domain. J Cell Biol. 2004;164:877–886. doi: 10.1083/jcb.200310065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampert F, Hornung P, Westermann S. The Dam1 complex confers microtubule plus end-tracking activity to the Ndc80 kinetochore complex. J Cell Biol. 2010;189:641–649. doi: 10.1083/jcb.200912021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee WL, Kaiser MA, Cooper JA. The offloading model for dynein function: differential function of motor subunits. J Cell Biol. 2005;168:201–207. doi: 10.1083/jcb.200407036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee WL, Oberle JR, Cooper JA. The role of the lissencephaly protein Pac1 during nuclear migration in budding yeast. J Cell Biol. 2003;160:355–364. doi: 10.1083/jcb.200209022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Lee WL, Cooper JA. NudEL targets dynein to microtubule ends through LIS1. Nat Cell Biol. 2005;7:686–690. doi: 10.1038/ncb1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, de Carvalho P, Kho D, Tai CY, Pierre P, Fink GR, Pellman D. Polyploids require Bik1 for kinetochore-microtubule attachment. J Cell Biol. 2001;155:1173–1184. doi: 10.1083/jcb.200108119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Markus SM, Plevock KM, St Germain BJ, Punch JJ, Meaden CW, Lee WL. Quantitative analysis of Pac1/LIS1-mediated dynein targeting: Implications for regulation of dynein activity in budding yeast. Cytoskeleton (Hoboken) 2011;68:157–174. doi: 10.1002/cm.20502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markus SM, Punch JJ, Lee WL. Motor- and tail-dependent targeting of dynein to microtubule plus ends and the cell cortex. Curr Biol. 2009;19:196–205. doi: 10.1016/j.cub.2008.12.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenney RJ, Vershinin M, Kunwar A, Vallee RB, Gross SP. LIS1 and NudE induce a persistent dynein force-producing state. Cell. 2010;141:304–314. doi: 10.1016/j.cell.2010.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, Samso M, Koonce MP. A flexible linkage between the dynein motor and its cargo. J Mol Biol. 2006;357:701–706. doi: 10.1016/j.jmb.2006.01.028. [DOI] [PubMed] [Google Scholar]
- Mocz G, Gibbons IR. Model for the motor component of dynein heavy chain based on homology to the AAA family of oligomeric ATPases. Structure. 2001;9:93–103. doi: 10.1016/s0969-2126(00)00557-8. [DOI] [PubMed] [Google Scholar]
- Moore JK, Li J, Cooper JA. Dynactin function in mitotic spindle positioning. Traffic. 2008;9:510–527. doi: 10.1111/j.1600-0854.2008.00710.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JK, Stuchell-Brereton MD, Cooper JA. Function of dynein in budding yeast: mitotic spindle positioning in a polarized cell. Cell Motil Cytoskeleton. 2009;66:546–555. doi: 10.1002/cm.20364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiura M, Kon T, Shiroguchi K, Ohkura R, Shima T, Toyoshima YY, Sutoh K. A single-headed recombinant fragment of Dictyostelium cytoplasmic dynein can drive the robust sliding of microtubules. J Biol Chem. 2004;279:22799–22802. doi: 10.1074/jbc.M313362200. [DOI] [PubMed] [Google Scholar]
- Pasternak C, Flicker PF, Ravid S, Spudich JA. Intermolecular versus intramolecular interactions of Dictyostelium myosin: possible regulation by heavy chain phosphorylation. J Cell Biol. 1989;109:203–210. doi: 10.1083/jcb.109.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reck-Peterson SL, Yildiz A, Carter AP, Gennerich A, Zhang N, Vale RD. Single-molecule analysis of dynein processivity and stepping behavior. Cell. 2006;126:335–348. doi: 10.1016/j.cell.2006.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiler S, Kirchner J, Horn C, Kallipolitou A, Woehlke G, Schliwa M. Cargo binding and regulatory sites in the tail of fungal conventional kinesin. Nat Cell Biol. 2000;2:333–338. doi: 10.1038/35014022. [DOI] [PubMed] [Google Scholar]
- Sheeman B, Carvalho P, Sagot I, Geiser J, Kho D, Hoyt MA, Pellman D. Determinants of S. cerevisiae dynein localization and activation: implications for the mechanism of spindle positioning. Curr Biol. 2003;13:364–372. doi: 10.1016/s0960-9822(03)00013-7. [DOI] [PubMed] [Google Scholar]
- Stock MF, Guerrero J, Cobb B, Eggers CT, Huang TG, Li X, Hackney DD. Formation of the compact confomer of kinesin requires a COOH-terminal heavy chain domain and inhibits microtubule-stimulated ATPase activity. J Biol Chem. 1999;274:14617–14623. doi: 10.1074/jbc.274.21.14617. [DOI] [PubMed] [Google Scholar]
- Stoffler HE, Bahler M. The ATPase activity of Myr3, a rat myosin I, is allosterically inhibited by its own tail domain and by Ca2+ binding to its light chain calmodulin. J Biol Chem. 1998;273:14605–14611. doi: 10.1074/jbc.273.23.14605. [DOI] [PubMed] [Google Scholar]
- Torisawa T, Nakayama A, Furuta K, Yamada M, Hirotsune S, Toyoshima YY. Functional dissection of LIS1 and NDEL1 towards understanding the molecular mechanism of cytoplasmic dynein regulation. J Biol Chem. 2011;286:1959–1965. doi: 10.1074/jbc.M110.169847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhey KJ, Lizotte DL, Abramson T, Barenboim L, Schnapp BJ, Rapoport TA. Light chain-dependent regulation of Kinesin's interaction with microtubules. J Cell Biol. 1998;143:1053–1066. doi: 10.1083/jcb.143.4.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorvis C, Markus SM, Lee WL. Photoactivatable GFP tagging cassettes for protein-tracking studies in the budding yeast Saccharomyces cerevisiae. Yeast. 2008;25:651–659. doi: 10.1002/yea.1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Thirumurugan K, Stafford WF, Hammer JA, 3rd, Knight PJ, Sellers JR. Regulated conformation of myosin V. J Biol Chem. 2004;279:2333–2336. doi: 10.1074/jbc.C300488200. [DOI] [PubMed] [Google Scholar]
- Whyte J, Bader JR, Tauhata SB, Raycroft M, Hornick J, Pfister KK, Lane WS, Chan GK, Hinchcliffe EH, Vaughan PS, et al. Phosphorylation regulates targeting of cytoplasmic dynein to kinetochores during mitosis. J Cell Biol. 2008;183:819–834. doi: 10.1083/jcb.200804114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodruff JB, Drubin DG, Barnes G. Dynein-driven mitotic spindle positioning restricted to anaphase by She1p inhibition of dynactin recruitment. Mol Biol Cell. 2009;20:3003–3011. doi: 10.1091/mbc.E09-03-0186. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.