SUMMARY
Growth differentiation factor 11 (GDF11) is one of the significant genes that control skeletal formation. Knockout of GDF11 function causes abnormal patterning of the anterior/posterior axial skeleton. The mRNA of GDF11 is initially translated to a precursor protein that undergoes a proteolytic cleavage to generate the C-terminal peptide or mature GDF11, and the N-terminal peptide named GDF11 propeptide. The propeptide can antagonize GDF11 activity in vitro. To investigate the effects of GDF11 propeptide on GDF11 function in vivo, we generated transgenic mice that over-express the propeptide cDNA in skeletal tissue. The transgenic mice showed formation of extra ribs on the seventh cervical vertebra (C7) as a result of transformation of the C7 vertebra into a thoracic vertebra. The GDF11 propeptide transgene mRNA was detected in tail tissue in embryos and was highly expressed in tail and calvaria bones after birth. A high frequency of C7 rib formation was noticed in the transgenic mouse line with a high level of transgene expression. The anterior boundaries of Hoxa-4 and Hoxa-5 mRNA in situ expressions showed cranial shifts from their normal prevertebra locations in transgenic embryos. These results demonstrated significant effects of GDF11 propeptide transgene on vertebral formation, which are likely occurring through depressing GDF11 function and altered locations of Hoxa-4 and Hoxa-5 expression.
INTRODUCTION
Bone morphogenetic proteins (BMPs) are known to play significant roles in bone development and remodeling. BMPs were originally identified as protein factors with the capacity to induce ectopic formation of bone when implanted locally in soft tissues (Urist, 1965). Over the past decades, BMPs have been shown to participate in various developmental processes, including embryonic skeleton formation, postnatal bone growth, and metabolism (Kingsley et al., 1992; Luo et al., 1995; Solloway et al., 1998; Daluiski et al., 2001; Kugimiya et al., 2005; Okamoto et al., 2006). Growth differentiation factor 11 (GDF11), which is also called BMP11, is a member of the BMP family (Gamer et al., 1999; Nakashima et al., 1999). GDF11 is not only necessary for formation of the anterior/posterior axial skeleton, but also critical for limb skeleton formation and development. Knockout of GDF11 in mice caused homeotic transformations of vertebrae into more anterior vertebrae (McPherron et al., 1999). Implantation of GDF11 beads into the early wing bud of the chicken embryo result in dramatic shortening of the forelimb (Gamer et al., 2001). In addition, eliminating the functions of both GDF11 and its highly homologous molecule-GDF8/myostatin in knockout mice causes serious defects in the axial skeleton and limb skeletons, further indicating that GDF11 regulates both vertebral and limb formation (McPherron et al., 2009).
GDF11, like the closely related protein myostatin, is initially translated as a precursor protein that undergoes a proteolytic cleavage at the tetrabasic site (RXRR) to generate the mature GDF11 protein and GDF11 propeptide, which corresponds to the C-terminal peptide and the N-terminal peptide, respectively (Ge et al., 2005). Like the myostatin propeptide, whose over-expression in transgenic mice can block the function of myostatin (Lee and McPherron, 2001; Yang et al., 2001; Yang and Zhao, 2006), GDF11 propeptide has been shown to form a latent complex with GDF11 and to antagonize GDF11 activity in vitro (Ge et al., 2005). However, it was not known whether or not GDF11 propeptide could depress GDF11 function in vivo. Besides the abnormal axial skeleton, GDF11-knockout mice have defects in other tissues where GDF11 is expressed, including reduced pancreas size, lack of kidneys, and increased number of olfactory neurons and progenitors (McPherron et al., 2009). To study the specific role of GDF11 function in bone formation, we generated transgenic mice that over-expressed GDF11 propeptide under the control of a bone-specific promoter, namely 2.3 kb α1 type 1 collagen promoter, and analyzed the effects of the GDF11 propeptide transgene on skeletal formation.
RESULTS
Generation of Transgenic Mice
The transgene expression cassette was constructed as shown (Fig. 1A). Transgenic mice were generated by the intracytoplasmic sperm injection (ICSI) technique in combination with piggyBac transposon-mediated gene transfer. A total of 657 mouse oocytes from strain B6D2F1 were injected with B6D2F1 sperm heads that were mixed with the transgene plasmid. A total of 382 injected oocytes developed into 2-cell embryos, which were transferred into CD-1 surrogate mothers. Forty-four mice were delivered; among them, 19 transgenic mice were identified as founders (Fig. 1B). The transgenic mice were viable and fertile. Fourteen lines of transgenic mice were established after breeding the 19 adult transgenic founder mice with wild-type B6D2F1 mice.
Figure 1.
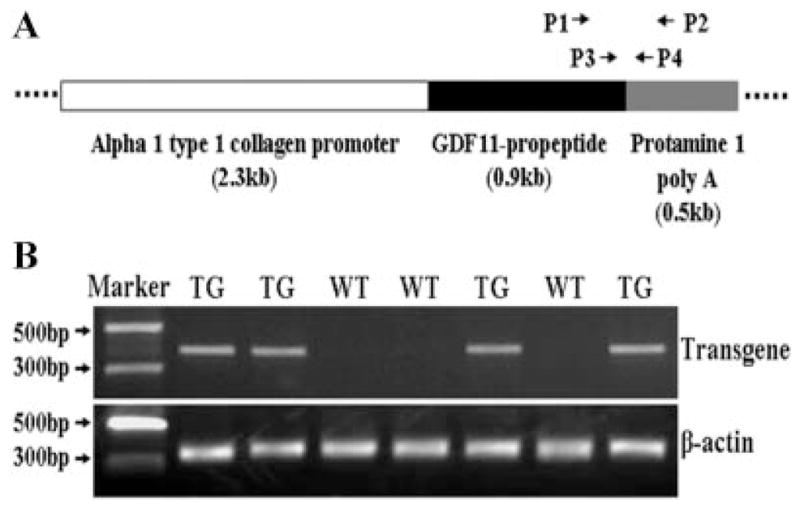
Production of transgenic mice. A: Schematic construct of transgene expression cassette and design of primers for analyzing the transgenic mice. P1 and P2 primers are used for transgene PCR genotyping and reverse-transcription PCR while P3 and P4 primers are used for quantitative real-time PCR. B: The representative results from PCR genotyping. TG, transgenic; WT, wild-type.
Expression of Transgene in Transgenic Mice
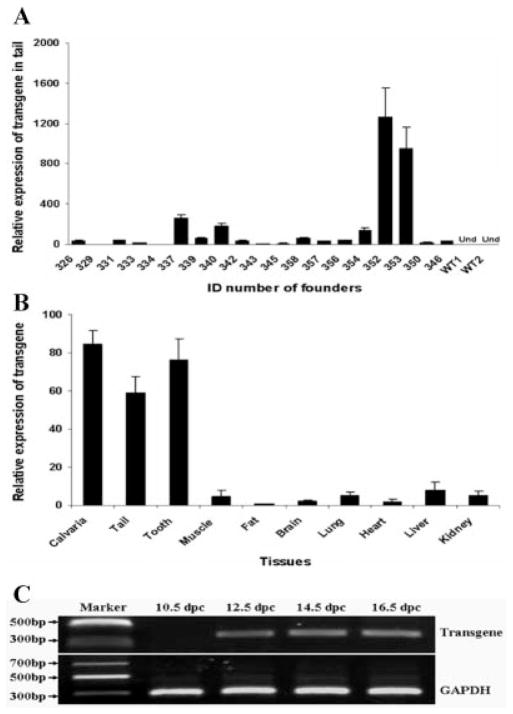
The α1 type 1 collagen promoter used for controlling the transgene expression is highly active in skeleton and other bony tissues. Transgene expression in tail tissue had been used as indicator for α1 type 1 collagen promoter-controlled transgene expression in other skeletal locations (Rossert et al., 1995). Therefore, the mRNA levels of GDF11 propeptide transgene were evaluated in tail tissue. The mRNA levels of GDF11 propeptide transgene were detected by quantitative real-time PCR (qRT-PCR) in all the 19 transgenic founders at the age of 5 weeks. The expression levels of the transgene varied significantly among different transgenic founders (Fig. 2A). In two founder mice (352 and 353), the transgene mRNA abundance was approximately 800–1,200 times that of the founder 334, which had the lowest level of transgene expression in the qRT-PCR data (Fig. 2A). The relative transgene expression level in the offspring was very similar to that detected in their transgenic founder mice. For example, the relative transgene expression level in founder 353, 337, and 339 was 926.08: 221.17: 54.64 (Fig. 2A), which was equal to 16.9: 4.0: 1.0. In the offspring from these three transgenic founders, the relative transgene mRNA level that was measured in tail tissue at 5 weeks of age was 13.4: 4.9: 1.0.
Figure 2.
Expression of transgene in transgenic mice. A: Analysis of transgene expression levels in tail tissues of transgenic founders by relative quantitative real-time PCR at 5 weeks of age. WT, wild-type; Und, undetectable. Real-time PCR was performed three times. Data are presented as mean ± SEM. Expression levels of transgene in all the transgenic founders were normalized to the founder 334, which has the lowest expression of the transgene. B: Analysis of transgene expression levels in different tissues of transgenic mice by relative quantitative real-time PCR at the age of 5 weeks. Real-time PCR was performed on three transgenic littermates of line 353. Data are presented as mean ± SEM. Expression levels of transgene in all tested tissues were normalized to fat tissue, which has the lowest expression of transgene. C: Detections of transgene expression in tail tissue at different embryonic stages by reverse transcription PCR. dpc, days postcoitum.
To investigate tissue-specific transgene expressions, we analyzed transgene expression in bone and several other tissues. A high level of transgene mRNA was detected in calvaria bone compared with muscle, fat, brain, lung, heart, liver, and kidney (Fig. 2B). The GDF11 propeptide transgene also was highly expressed in tail and tooth tissues of transgenic mice (Fig. 2B). These results are consistent with the activities of the promoter used in the transgene construct. The α1 type 1 collagen promoter is highly active in osteoblasts of caudal vertebrae and osteoblast-like odontoblasts (Rossert et al., 1995). In addition, the transgene mRNA was detectable in tail tissue by reverse transcription PCR at 12.5 days postcoitum (dpc) and later times (14.5 and 16.5 dpc); however, transgene expression was not detected at 10.5 dpc (Fig. 2C). These results indicate that the GDF11 propeptide transgene was expressed in the tail tissue from approximately 12.5 dpc, or in the earlier period of time from 10.5 to 12.5 dpc.
Skeletal Abnormalities in Transgenic Mice
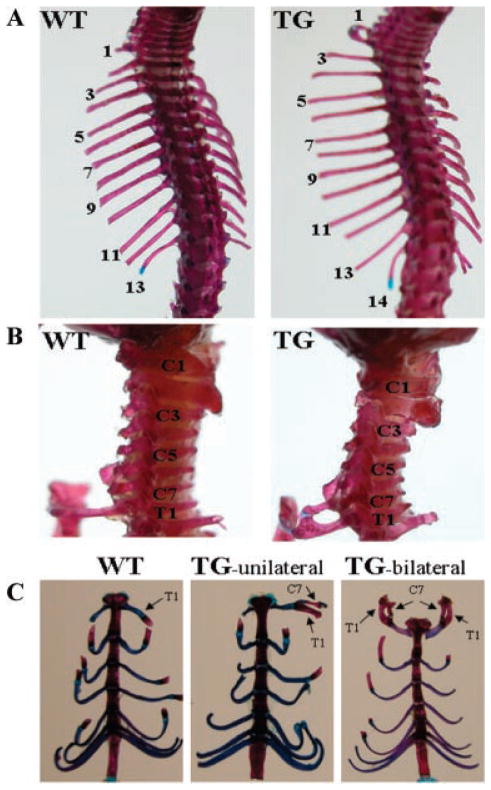
To reveal the effects of the transgene on bone development, we stained the skeletons of both transgenic mice and their wild-type littermates with alizarin red and alcian blue. Transgenic mice have abnormal cervical vertebral development. As shown in Figure 3A, the transgenic mice had 14 pairs of ribs, 1 extra pair of ribs compared with their wild-type littermates. The supernumerary ribs in the transgenic mice were caused by ectopic formation of ribs on the seventh cervical vertebra (C7) instead of inserting an additional thoracic vertebra into the vertebral column (Fig. 3B). The additional rib, also called the C7 rib, did not attach to the sternum independently like the normal vertebrosternal ribs, but fused to the middle of the rib that is articulated with the first thoracic vertebra (T1) in the transgenic mice (Fig. 3B). In some cases, the cervical rib was shown only unilaterally while in other cases it was exhibited bilaterally (Fig. 3C). Among 20 transgenic mice with a unilateral C7 rib from the 3 transgenic lines analyzed, 11 mice showed a C7 rib on the right side and 9 mice showed a C7 rib on the left side (Table 1). These data suggest that C7 rib in the transgenic mice was randomly formed on the either side of the skeleton. Analysis of the lumbar, sacral, and caudal regions of the anterior/posterior axial skeleton indicated that the vertebrae in these regions were normal in the transgenic mice. Furthermore, in order to determine if abnormality exists in the length of the bones of the forelimb in transgenic mice, we measured the length of two forelimb bones, the ulna and the humerus, which were dissected from 10-week-old transgenic and wild-type mice. The lengths of these two forelimb bones in the transgenic mice did not differ from that of their wild-type littermates.
Figure 3.
Abnormalities in the anterior/posterior axial skeleton of transgenic mice. Skeletons from 10-day-old mice were stained with alizarin red and alcian blue. A: Dorsal view of the anterior/posterior axial skeleton in wild-type (WT) and transgenic (TG) mice, showing TG mice have 14 pairs of ribs while WT mice have 13 pairs of ribs. B: Lateral view of cervical vertebrae in WT and TG mice. A supernumerary rib in TG mice is formed on the seventh cervical vertebra (C7). This cervical rib is fused to the middle of the rib that is articulated with the first thoracic vertebra (T1) and is attached to the sternum together at the T1 position in TG mice. C: Ventral view of vertebrosternal ribs in WT and TG mice. The cervical rib (C7 rib), which is fused to the T1 rib. In some cases it is unilateral (TG-unilateral) while it is bilateral (TG-bilateral) in other cases.
TABLE 1.
Skeletal Phenotype of the Transgenic Mice and Their Wild-Type Littermates*
| Line 353
|
Line 337
|
Line 339
|
||||
|---|---|---|---|---|---|---|
| WT | TG | WT | TG | WT | TG | |
| No. of analyzed mice | 24 | 23 | 23 | 21 | 21 | 20 |
| No. of mice with unilateral C7 rib | 0 | 14 | 0 | 4 | 0 | 2 |
| No. of mice with bilateral C7 ribs | 0 | 4 | 0 | 0 | 0 | 1 |
| Percentage of skeletal abnormalities | 0 | 81.8% | 0 | 19.0% | 0 | 15.0% |
Skeleton of 4–5 litters of 1-day-old mice from each line were analyzed by alizarin red and alcian blue staining after PCR genotyping. There are a total of 20 transgenic mice with unilateral C7 rib from the three lines, among them 11 mice showed C7 rib on the right side and 9 mice showed C7 rib on the left side. The relative transgene expression level among lines 353, 337, and 339 can be found in Figure 2A.
The effect of the GDF11 propeptide transgene on the patterning of the anterior/posterior axial skeleton of the transgenic mice was investigated in several lines of the transgenic mice. The frequency (81.8%) of C7 ribs in the high transgene expression line 353 was much higher than other transgenic mouse lines (e.g., lines 337 and 339), which have low transgene expression levels (Table 1).
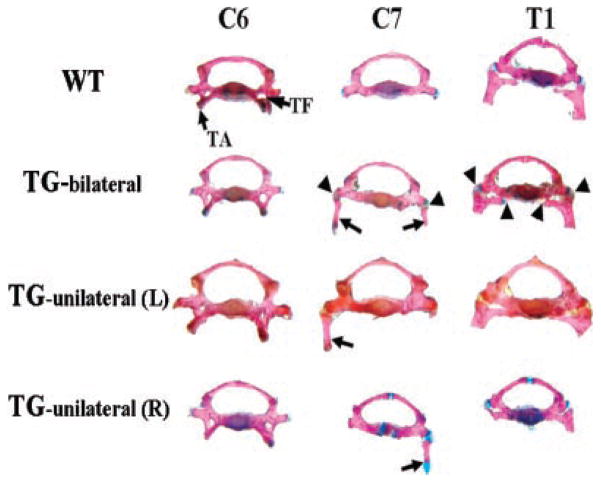
To investigate whether or not the transgene affects the appearance of the C7 vertebral body of the transgenic mice, we analyzed the vertebral bodies of the C7 and two neighbor vertebrae, C6 and T1. In wild-type mice, although the C7 vertebral body does not have the anterior tubercle of the transverse process, or the transverse foramen, which are markers of the C6 vertebral body, the morphology of the C7 vertebral body most resembles that of the C6 vertebral body rather than T1 (Fig. 4). However, in transgenic mice, especially those displaying bilateral C7, the C7 vertebral body tended to transform into a vertebral body most resembling T1 (Fig. 4). The manner of the attachment between the C7 ribs and the C7 vertebral body was different from that between the T1 ribs and the T1 vertebral body. The C7 ribs were articulated with the C7 vertebral body through only one contact point while the T1 ribs were articulated with the T1 vertebral body by two contact points (Fig. 4).
Figure 4.
Morphological appearances of the seventh cervical vertebra in transgenic mice. Vertebrae were dissected from alizarin red and alcian blue-stained skeleton. In wild-type (WT) mice, the C7 vertebral body resembles the C6 vertebral body instead of the T1 vertebral body without the tuberculi anterior (TA) and the transverse foramen (TF) that mark the C6. In transgenic mice there are ribs on C7, and the C7 vertebral body resembles the T1 vertebral body instead of the C6 vertebral body. This is more obvious in TG-bilateral mice. In TG-bilateral, TG-unilateral (L) and TG-unilateral (R) mice, there is only one joining point between the cervical ribs (indicated by arrows) and the C7 vertebral body while there are two joining points between the T1 ribs and the T1 vertebral body. The joining points between ribs and vertebral bodies are indicated with arrowheads only in TG bilateral but not in TG-unilateral (L) and TG-unilateral (R) mice. TG-bilateral =-transgenic mice showing C7 ribs on both sides; TG-unilateral (L) = transgenic mice showing C7 rib on the left side; TG-unilateral (R) = transgenic mice showing C7 rib on the right side.
Expressions of Hoxa-4 and Hoxa-5 in Transgenic Mice
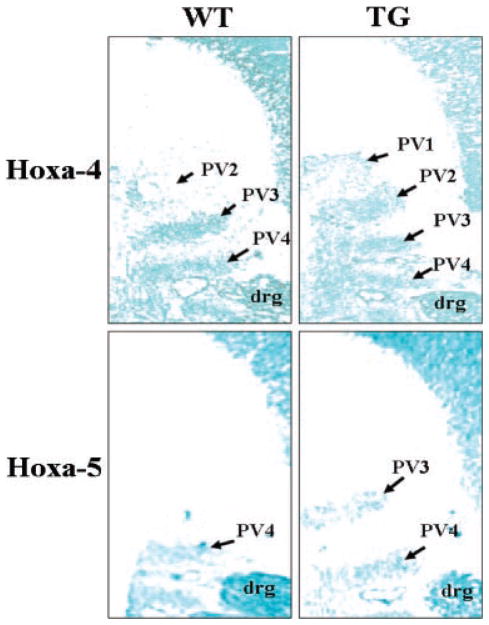
Axial vertebral patterning is thought to be controlled by the spatially restricted expression patterns of Hox genes (Favier and Dolle, 1997; Mark et al., 1997; Wellik, 2007). In the GDF11-knockout mice, it has been shown that GDF11 acts as an upstream molecule to regulate the expression pattern of Hox genes, thereby controlling the pattern of the anterior/posterior axial skeleton (McPherron et al., 1999). Targeted disruption of the mouse homologue of the Drosophila polyhomeotic gene (rae28) demonstrates transformation of the seventh cervical vertebra (C7) into the first thoracic vertebra (T1) in homozygotes (Takihara et al., 1997). Among the tested Hox genes in rae28-deficient mice, anterior expression boundaries of Hoxa-4 and Hoxa-5 genes on the vertebral column are altered (Takihara et al., 1997). To determine the expression patterns of these two Hox genes in the GDF11 propeptide transgenic mice, we examined their expression patterns in 12.5 dpc mice. In situ hybridization analysis showed that the anterior boundaries of Hoxa-4 and Hoxa-5 gene expression were shifted cranially by 1 prevertebra in transgenic embryo compared to their wild-type littermates (Fig. 5). The anterior boundary of Hoxa-4 gene expression was detected on the second prevertebra (PV2) in wild-type mice, however, it was detected on the first prevertebra (PV1) in the transgenic embryos (Fig. 5). There was also a shift of anterior boundary of Hoxa-5 gene expression from the fourth prevertebra (PV4) position in the wild-type embryos to the third prevertebra (PV3) in transgenic embryos (Fig. 5).
Figure 5.
Expression patterns of Hoxa-4 and Hoxa-5 genes along the anterior/posterior axis in transgenic mice. Expressions of Hoxa-4 and Hoxa-5 genes were analyzed by in situ hybridization on sagittal frozen sections of 13-dpc embryos. While the anterior boundary of Hoxa-4 gene expression is located on the first prevertebra (PV1) in transgenic (TG) embryos, it is located on the second prevertebra (PV2) in wild-type (WT) mice. Similarly, the anterior boundary of Hoxa-5 gene expression is located on the third prevertebra (PV3) in TG mice, but it is located on the fourth prevertebra (PV4) in WT mice. drg = dorsal root ganglion. PV1, 2, 3, and 4 corresponds to the final cervical vertebrae C1, C2, C3, and C4, respectively.
DISCUSSION
Although GDF11 propeptide was shown to inhibit GDF11 activity in vitro, its effect on GDF11 function in in vivo models so far has not been reported. The results from this report demonstrated that transgenic over-expression of GDF11 propeptide under the control of a skeleton-specific promoter, α1 type 1 collagen promoter, resulted in supernumerary formation of ribs on C7. The developmental defect on skeleton in the transgenic mice was less severe than that in homozygous GDF11−/− mice, which have homeotic transformation of vertebrae in the lumbar, sacral, and caudal regions (McPherron et al., 1999), and die shortly after birth because of renal agenesis and problems associated with increased numbers of various progenitors (Dichmann et al., 2006). In GDF11−/− mice, vertebral transformation also occurred on C7, which was transformed into the anterior C6 vertebra (McPherron et al., 1999). This is different from the transgenic mice generated from this report, in which the C7 was converted into the more posterior T1 vertebra (Fig. 4). The GDF11 propeptide transgenic mice have normal lumbar, sacral, and caudal vertebrae. The offspring generated from the transgenic founders appear healthy. This is the first report that transgenic mice with over-expressed GDF 11 propeptide showed a transformation of the seventh cervical vertebra into a thoracic vertebra.
The different skeletal phenotypes of the transgenic mice with over-expressed GDF11 propeptide and GDF11-knockout mice may be highly related to the timeline of GDF11 and its propeptide transgene expression. The axial skeleton is generated from somites, which are formed during embryonic age of 8–11 dpc in mice (Nagy et al., 2003). GDF11 is highly expressed in the primitive streak and tail bud at 8.5–9.5 dpc and in limb buds at 9.5–10.5 dpc, and later expressed in the mesenchyme between developing skeletal elements (Gamer et al., 1999; McPherron et al., 1999; Nakashima et al., 1999). Mesodermal layer formation and subsequent stem cell migration have been the main developmental events of gastrulation and organogenesis. Skeletal abnormalities, such as anterior homeotic transformations of vertebrae in the GDF11-null mice, is consistent with high levels of GDF11 expression in the primitive streak, presomitic mesoderm, and tail bud (McPherron et al., 1999; Gamer et al., 2001). The time and pattern of GDF11 expression in embryos and the phenotype of GDF11−/− mice suggest that GDF11 acts on mesodermal precursor cells to regulate the patterning of axial vertebrae. In the transgenic mice generated from this study, GDF11 propeptide transgene mRNA was detected in the tail tissue during the mid-gestation period. Therefore, the occurrence of C7 ribs due to over-expressed GDF-11 propeptide probably results from functional blocking of GDF11 expression in the later stages of somatic and tail bud formation. In other words, the product of the transgene was synthesized late in the embryo and failed to depress high-level GDF11 expression during the primitive streak stage and/or early stages of somite formation. Although we did not determine endogenous GDF11 expression in the transgenic mice, we believe that GDF11 and GDF11 propeptide transgene are co-expressed in tail tissue during late stages of embryonic development. Further studies on relative expressions of the transgene and endogenous GDF11 gene during embryonic developmental stages are certainly needed for clarifications of the actual level and specific embryonic stages when GDF11 is depressed by the transgene in the transgenic mice.
GDF11 has 90% identity in amino acid sequences with GDF8/myostatin. The expression of myostatin mRNA is initiated in developing somites by 9.5 dpc, and expands to all somites and the myotome compartment by 10.5 dpc, since then myostatin expression is maintained in all developing muscles during embryonic and postnatal stages (McPherron et al., 1997). Studies show that myostatin function is suppressed by GDF11 propeptide in vitro (Ge et al., 2005). In consideration of the time of GDF11 transgene expression in embryos, there is also the possibility that the over-expressed GDF11 propeptide in late embryonic and postnatal stages may suppress myosatin function. Recent data also suggest that myostatin is redundant to GDF11 in regulating the patterning of axial vertebrae (McPherron et al., 2009). Therefore, the functions of both GDF11 and myostatin may be depressed in the skeleton of transgenic mice by over-expressed GDF11 propeptide.
The activity of chondrogenesis in the cartilage of epiphyseal growth plate is thought to be responsible for determining the length of long bone. The dramatic truncation of the appendicular skeleton in GDF11-treated chicken embryos was believed to be due to the strong negative effects of GDF11 on chondrogenesis (Gamer et al., 2001). In the transgenic mice generated from the present study, expression of the GDF11 propeptide transgene was driven by an osteoblast-specific promoter which is inactive in cartilage. Therefore, the transgene in the transgenic mice might have little effect on chondrogenesis activity in the growth plate of long bone, and thus have no effects on increasing the length of the appendicular skeleton. This probably explains why we did not observe changes in the length of forelimb bones of the GDF11 propeptide transgenic mice.
The expression patterns of two Hox genes in the transgenic mice offered some insights about the downstream regulatory events of the GDF11 propeptide. Precise regulation of the expression boundaries of Hox genes is critical for specifying the segment identities along the anterior/posterior axis (Manak and Scott, 1994). In the GDF11-knockout mice, it has been shown that GDF11 acts as an upstream molecule that regulates the expression pattern of Hox genes, thereby controlling the pattern of the anterior/posterior axial skeleton (McPherron et al., 1999). Consistent with this mechanism, anterior displacements of the anterior expression boundaries of Hoxa-4 and Hoxa-5 genes on the anterior/posterior axis were detected in the GDF11 propeptide transgenic embryos. These results further confirm that altered expression patterns of Hoxa-4 and Hoxa-5 genes are involved in the supernumerary formation of ribs on C7. Hoxa-4 and Hoxa-5 are likely the targets of GDF11 down-stream signaling events in the regulation of vertebral formation.
Mammals are remarkably consistent in that they have seven cervical vertebrae. The lack of variation in cervical vertebral number among different mammalian animals is primarily due to developmental constraints. Any changes in the number of cervical vertebrae are coupled with neural problems and an increased susceptibility to early childhood cancer and stillbirths (Galis, 1999). The formation of ribs on C7, which indicates a partial transformation of the C7 into a thoracic vertebra, reduces the number of cervical vertebrae. In humans, the alteration in number of cervical vertebrae due to the formation of C7 ribs was reported to be associated not only with thoracic outlet syndrome, the compression of the branchial nerves, and blood vessels entering the limbs by cervical ribs (Makhoul and Machleder, 1992; Roos, 1996), but also with increased risk of early childhood cancer (Schumacher et al., 1992; Galis, 1999). The formation of the cervical ribs on C7 in the transgenic mice produced from this study provides a useful animal model for understanding developmental defects of the cervical vertebrae in humans. Further study of GDF11 with these transgenic mice will shed light on understanding causes of skeletal defects occurred in human still-birth and early childhood skeletal developmental problems.
In conclusion, transgenic over-expression of GDF11 propeptide under the control of α1 type 1 collagen promoter resulted in transformation of C7 to a thoracic vertebra. The expression of the GDF11 propeptide transgene appeared to occur in tail tissue during mid-gestation, and is highly expressed in tail and calvaria bones after birth. Locations of Hoxa-4 and -5 gene expression were shifted cranially from their normal prevertebra locations in transgenic embryos. These results suggest that expression of GDF11 propeptide transgene results in significant effects on vertebral formation. The transgenic mouse with over-expressed GDF11 propeptide provides a useful animal model for studying the function of GDF11 and its propeptide in regulation of vertebral formation.
MATERIALS AND METHODS
Construction of Transgene Plasmids and Production of Transgenic Mice
The cDNA of mouse GDF11 propeptide (encoding for 1-296 AA) was amplified from a synthetic clone (Gene bank accession No.: BC152721, Openbiosystems, Huntsville, AL) by PCR. A stop codon (TAA) was added to the end of the gene and two BamHI sites were added to the flanking regions of the gene during PCR. Amplified GDF 11 propeptide cDNA was cloned into the TA vector pCR4-TOPO (Invitrogen, Carlsbad, CA) and then the gene was cut out by BamH 1 to replace the lacZ gene between the osteoblast-specific 2.3 kb α1 type 1 collagen promoter (Chen et al., 1998) and mouse protamine 1 poly-A signal in the pJ251 plasmid (a kind gift from Dr. Di Chen, University of Rochester). The transgene expression cassette (promoter + GDF11 propeptide cDNA + poly-A signal sequence) was digested with Kpn 1 and EcoR V and inserted into multiple cloning sites of pENTR1A (Invitrogen) to generate plasmid pENTR1A-mGDF11-pro. After a LR recombination reaction between plasmids pENTR1AmGDF11-pro and pmGENIE-2, which is a helper-independent plasmid for piggyBac transposon-mediated gene transfer (Urschitz et al., 2010), the transgene expression cassette was cloned into the piggyBac transposon in pmGENIE-2 to generate the plasmid pmGENIE-2-mGDF11-pro. Transgenic mice were produced by using ICSI technique as described by using pmGENIE-2-mGDF11-pro plasmid (Moisyadi et al., 2009; Urschitz et al., 2010). Mouse strains used for this project included B6D2F1 (B57BL/6 × DBA/2) as sperm and oocyte donors, and pseudopregnant CD-1 females as embryo recipients. Transgenic mice were backcrossed with B6D2F1 to produce offspring for phenotype evaluations and gene expression analysis.
Identifications of Transgenic Mice
Transgenic mice were identified by PCR amplification of a 381-bp fragment with genomic DNA isolated from tail tissue. The positions of PCR primers (P1 and P2) in the transgene expression cassette are indicated in Figure 1A; their sequences are as follows. P1: 5′-GTCACATCCGTATCCGTTCACT-3′; P2: 5′-AGCCCTCTACTTACTTCGCCTC-3′. The amplified 381 bp DNA fragment covers the junction of GDF-11 propeptide cDNA and protamine 1 poly-A signal sequence. To check the quality of the tail genomic DNA, we also used the same tail genomic DNA samples to run PCR amplifications of a 340 bp fragment of mouse β-actin gene primes (forward: 5′-TTGAGACCTTCAACACCCC-3′; reverse: 5′-TTGCCAATAGTGATGACCTG-3′). Several PCR products that were amplified from the tail genomic DNA of transgenic mice were randomly selected for DNA sequencing. The sequence data were used for validation of their DNA identities in accordance of transgene pmGENIE-2-mGDF11-pro plasmid.
Analysis of Transgene Expression
Real-time PCR was performed as previously described (Li et al., 2009). Briefly, total RNA was extracted from tail tissue of 5-week-old mice, treated with DNase I to eliminate any contaminating genomic DNA, then followed by cDNA synthesis with SuperScript II reverse transcriptase (Invitrogen). Expression level of the transgene was analyzed by real-time PCR amplification of a 105 bp fragment with P3 and P4 primers which are indicated in transgene expression cassette (Fig. 1A, P3: 5′-TTTCATGGAGCTCGAGTCCTAGA-3′; P4: 5′-ATCTGCTCCTGCTTTTGCTGC-3′). Expression level of the housekeeping gene, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was also measured with a set of primers (forward: 5′-GCAGTGGCAAAGTGGAGATTG-3′; reverse: 5′-CCGTGAGTGGAGTCATACTGGAA-3′) during real-time PCR and used as an endogenous control. Calculation of relative expression of transgene was followed by the 2−ΔΔCtmethod as described previously (Suzuki et al., 2008).
For detection of transgene expression in embryo tissue, reverse transcription PCR was used for easy visual observation of the transgene mRNA. Embryos were collected at 10.5, 12.5, 14.5, and 16.5 dpc. Whole tail tissue was collected from different embryonic stages for RNA isolation. Tail tissues from embryos were also used for DNA isolation and genotyping. Expression of the transgene in tail tissue at different stages of embryonic development was detected by reverse transcription PCR with the same amount of cDNA as template and the same set of primers (P1 and P2) that are used for PCR genotyping. GAPDH gene was used as internal control and was amplified by a set of primers (forward: 5′-GTGCTGAGTATGTCGTGGAG-3′; reverse: 5′-GTCTTCTGGGTGGCAGTGAT-3′). The amplification product of GAPDH from the cDNA is 295 bp across from exon 3 to exon 5.
Alizarin Red and Alcian Blue Staining of Skeleton
Skeletons of transgenic mice and their wild-type littermate control mice were stained with alizarin red and alcian blue as previously described (Essalmani et al., 2008). Briefly, mice were euthanized, skinned, eviscerated, and dehydrated in 95% ethanol for 4 days followed by incubation in acetone for 3 days to remove fat. Then, specimens were stained for 3 days in alizarin red and alcian blue staining solution containing 70% ethanol, 8% glacial acetic acid, 0.001% alcian blue, and 0.0005% alizarin red. After staining, specimens were cleared in 1% KOH until skeletons were clearly visible, then were placed in solution containing 1% KOH and 20% glycerol until the surrounding tissues were completely leached. Finally, specimens were transferred to 50% glycerol followed by 80% glycerol over several days and stored in 100% glycerol.
In Situ Hybridization
Embryos at 13 dpc were collected from pregnant wild-type female mice that were mated with transgenic male mice. A piece of tail tissue (50 mg) was taken from each embryo to isolate DNA for PCR genotyping. Embryos were fixed in 4% paraformaldehyde, embedded in OCT and sagittal cryostat sections (10 μm) were taken. Digoxigenin-labeled Hoxa-4 and Hoxa-5 riboprobes were synthesized using the DIG RNA labeling kit (Roche, Indianapolis, IN) and the template plasmids (Kawazoe et al., 2002) that were kindly provided by Dr. Mastake Araki at Kumamoto University, Japan. In situ hybridization was carried out using the digoxigenin labeled riboprobes on frozen sections as described (Takihara et al., 1997). After stopping the color reaction, sections were counter-stained with fast green and mounted with paramount and observed under microscopy.
Statistical Analysis
Data were analyzed by SAS 9.2 program (SAS Institute, Cary, NC), and the two samples t-test for means program was used for mean comparisons in Table 1, PFigure 2. Data are present as mean ± standard error of mean (SEM). Significant difference of means between two different groups was determined at * < 0.05 or **P < 0.01.
Acknowledgments
Supported by USDA-TSTAR (2008-34135-19322) and NIH (5P20RR024206-01A1).
This work is funded by USDA-TSTAR programs (grant # 2008-34135-19322) and NIH COBRE Grant (5P20RR024206-01A1). We are grateful to Dr. Yong Soo Kim, Dr. Scott Lozanoff, Dr. Ching Yuan Hu, Dr. Ashley Stokes, and Dr. Dian Dooley for their comments on the research project and the manuscript.
References
- Chen D, Ji X, Harris MA, Feng JQ, Karsenty G, Celeste AJ, Rosen V, Mundy GR, Harris SE. Differential roles for BMP receptor type IB and IA in differentiation and specification of mesenchymal precursor cells to osteoblast and adipocyte lineages. J Cell Biol. 1998;142:295–305. doi: 10.1083/jcb.142.1.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daluiski A, Engstrand T, Bahamonde ME, Gamer LW, Agius E, Stevenson SL, Cox K, Rosen V, Lyons KM. Bone morphogenetic protein-3 is a negative regulator of bone density. Nat Genet. 2001;27:84–88. doi: 10.1038/83810. [DOI] [PubMed] [Google Scholar]
- Dichmann DS, Yassin H, Serup P. Analysis of pancreatic endocrine development in GDF11-deficient mice. Dev Dyn. 2006;235:3016–3025. doi: 10.1002/dvdy.20953. [DOI] [PubMed] [Google Scholar]
- Essalmani R, Zaid A, Marcinkiewicz J, Chamberland A, Pasquato A, Seidah NG, Prat A. In vivo functions of the proprotein convertase PC5/6 during mouse development: Gdf11 is a likely substrate. Proc Natl Acad Sci USA. 2008;105:5750–5755. doi: 10.1073/pnas.0709428105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favier B, Dolle P. Developmental functions of mammalian Hox genes. Mol Hum Reprod. 1997;3:115–131. doi: 10.1093/molehr/3.2.115. [DOI] [PubMed] [Google Scholar]
- Galis F. Why do almost all mammals have seven cervical vertebrae? Developmental constraints, Hox genes, and cancer. J Exp Zool. 1999;285:19–26. [PubMed] [Google Scholar]
- Gamer LW, Wolfman NM, Celeste AJ, Hattersley G, Hewick R, Rosen V. A novel BMP expressed in developing mouse limb, spinal cord, and tail bud is a potent mesoderm inducer in Xenopus embryos. Dev Biol. 1999;208:222–232. doi: 10.1006/dbio.1998.9191. [DOI] [PubMed] [Google Scholar]
- Gamer LW, Cox KA, Small C, Rosen V. Gdf11 is a negative regulator of chondrogenesis and myogenesis in the developing chick limb. Dev Biol. 2001;229:407–420. doi: 10.1006/dbio.2000.9981. [DOI] [PubMed] [Google Scholar]
- Ge G, Hopkins DR, Ho WB, Greenspan DS. GDF11 forms a bone morphogenetic protein activated latent complex that can modulate nerve growth factor induced differentiation of PC12 cells. Mol Cell Biol. 2005;25:5846–5858. doi: 10.1128/MCB.25.14.5846-5858.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawazoe Y, Sekimoto T, Araki M, Takagi K, Araki K, Yamamura K. Region-specific gastrointestinal Hox code during murine embryonal gut development. Dev Growth Differ. 2002;44:77–84. doi: 10.1046/j.1440-169x.2002.00623.x. [DOI] [PubMed] [Google Scholar]
- Kingsley DM, Bland AE, Grubber JM, Marker PC, Russell LB, Copeland NG, Jenkins NA. The mouse short ear skeletal morphogenesis locus is associated with defects in a bone morphogenetic member of the TGF beta superfamily. Cell. 1992;71:399–410. doi: 10.1016/0092-8674(92)90510-j. [DOI] [PubMed] [Google Scholar]
- Kugimiya F, Kawaguchi H, Kamekura S, Chikuda H, Ohba S, Yano F, Ogata N, Katagiri T, Harada Y, Azuma Y, Nakamura K, Chung UI. Involvement of endogenous bone morphogenetic protein (BMP) 2 and BMP6 in bone formation. J Biol Chem. 2005;280:35704–35712. doi: 10.1074/jbc.M505166200. [DOI] [PubMed] [Google Scholar]
- Lee SJ, McPherron AC. Regulation of myostatin activity and muscle growth. Proc Natl Acad Sci USA. 2001;98:9306–9311. doi: 10.1073/pnas.151270098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Cao B, Zhao B, Yang X, Fan MZ, Yang J. Decreased expression of calpain and calpastatin mRNA during development is highly correlated with muscle protein accumulation in neonatal pigs. Comp Biochem Physiol A Mol Integr Physiol. 2009;152:498–503. doi: 10.1016/j.cbpa.2008.12.004. [DOI] [PubMed] [Google Scholar]
- Luo G, Hofmann C, Bronckers AL, Sohocki M, Bradley A, Karsenty G. BMP-7 is an inducer of nephrogenesis, and is also required for eye development and skeletal patterning. Genes Dev. 1995;9:2808–2820. doi: 10.1101/gad.9.22.2808. [DOI] [PubMed] [Google Scholar]
- Makhoul RG, Machleder HI. Developmental anomalies at the thoracic outlet: An analysis of 200 consecutive cases. J Vasc Surg. 1992;16:534–542. [PubMed] [Google Scholar]
- Manak JR, Scott MP. A class act: Conservation of homeodomain protein functions. Dev Suppl. 1994;1994:61–77. [PubMed] [Google Scholar]
- Mark M, Rijli FM, Chambon P. Homeobox genes in embryogenesis and pathogenesis. Pediatr Res. 1997;42:421–429. doi: 10.1203/00006450-199710000-00001. [DOI] [PubMed] [Google Scholar]
- McPherron AC, Lawler AM, Lee SJ. Regulation of skeletal muscle mass in mice by a new TGF-β superfamily member. Nature. 1997;387:83–90. doi: 10.1038/387083a0. [DOI] [PubMed] [Google Scholar]
- McPherron AC, Lawler AM, Lee SJ. Regulation of anterior/posterior patterning of the axial skeleton by growth/differentiation factor 11. Nat Genet. 1999;22:260–264. doi: 10.1038/10320. [DOI] [PubMed] [Google Scholar]
- McPherron AC, Huynh TV, Lee SJ. Redundancy of myostatin and growth/differentiation factor 11 function. BMC Dev Biol. 2009;9:24–31. doi: 10.1186/1471-213X-9-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moisyadi S, Kaminski JM, Yanagimachi R. Use of intracytoplasmic sperm injection (ICSI) to generate transgenic animals. Comp Immunol Microbiol Infect Dis. 2009;32:47–60. doi: 10.1016/j.cimid.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy A, Gertsenstein M, Vintersten K, Behringer R. Manipulating the mouse embryo: A laboratory manual third edition. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory; 2003. pp. 31–140. [Google Scholar]
- Nakashima M, Toyono T, Akamine A, Joyner A. Expression of growth/differentiation factor 11, a new member of the BMP/TGFbeta superfamily during mouse embryogenesis. Mech Dev. 1999;80:185–189. doi: 10.1016/s0925-4773(98)00205-6. [DOI] [PubMed] [Google Scholar]
- Okamoto M, Murai J, Yoshikawa H, Tsumaki N. Bone morphogenetic proteins in bone stimulate osteoclasts and osteoblasts during bone development. J Bone Miner Res. 2006;21:1022–1033. doi: 10.1359/jbmr.060411. [DOI] [PubMed] [Google Scholar]
- Roos DB. Historical perspectives and anatomic considerations. Thoracic outlet syndrome. Semin Thorac Cardiovasc Surg. 1996;8:183–189. [PubMed] [Google Scholar]
- Rossert J, Eberspaecher H, de Crombrugghe B. Separate cis-acting DNA elements of the mouse pro-alpha 1(I) collagen promoter direct expression of reporter genes to different type I collagen-producing cells in transgenic mice. J Cell Biol. 1995;129:1421–1432. doi: 10.1083/jcb.129.5.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher R, Mai A, Gutjahr P. Association of rib anomalies and malignancy in childhood. Eur J Pediatr. 1992;151:432–434. doi: 10.1007/BF01959357. [DOI] [PubMed] [Google Scholar]
- Solloway MJ, Dudley AT, Bikoff EK, Lyons KM, Hogan BL, Robertson EJ. Mice lacking Bmp6 function. Dev Genet. 1998;22:321–339. doi: 10.1002/(SICI)1520-6408(1998)22:4<321::AID-DVG3>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Suzuki ST, Zhao B, Yang J. Enhanced muscle by myostatin propeptide increases adipose tissue adiponectin, PPAR-α and PPAR-γ expressions. Biochem Biophys Res Commun. 2008;369:767–773. doi: 10.1016/j.bbrc.2008.02.092. [DOI] [PubMed] [Google Scholar]
- Takihara Y, Tomotsune D, Shirai M, Katoh-Fukui Y, Nishii K, Motaleb MA, Nomura M, Tsuchiya R, Fujita Y, Shibata Y, Higashinakagawa T, Shimada K. Targeted disruption of the mouse homologue of the Drosophila polyhomeotic gene leads to altered anteroposterior patterning and neural crest defects. Development. 1997;124:3673–3682. doi: 10.1242/dev.124.19.3673. [DOI] [PubMed] [Google Scholar]
- Urist MR. Bone: Formation by autoinduction. Science. 1965;150:893–899. doi: 10.1126/science.150.3698.893. [DOI] [PubMed] [Google Scholar]
- Urschitz J, Kawasumi M, Owens J, Morozumi K, Yamashiro H, Stoytchev I, Marh J, Dee JA, Kawamoto K, Coates CJ, Kaminski JM, Pelczar P, Yanagimachi R, Moisyadi S. Helper-independent piggyBac plasmids for gene delivery approaches: Strategies for avoiding potential genotoxic effects. Proc Natl Acad Sci USA. 2010;107:8117–8122. doi: 10.1073/pnas.1003674107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellik DM. Hox patterning of the vertebrate axial skeleton. Dev Dyn. 2007;236:2454–2463. doi: 10.1002/dvdy.21286. [DOI] [PubMed] [Google Scholar]
- Yang J, Zhao B. Postnatal expression of myostatin propeptide cDNA maintained high muscle growth and normal adipose tissue mass in transgenic mice fed a high-fat diet. Mol Reprod Dev. 2006;73:462–469. doi: 10.1002/mrd.20452. [DOI] [PubMed] [Google Scholar]
- Yang J, Ratovitski T, Brady JP, Solomon MB, Wells KD, Wall RJ. Expression of myostatin pro domain results in muscular transgenic mice. Mol Reprod Dev. 2001;60:351–361. doi: 10.1002/mrd.1097. [DOI] [PubMed] [Google Scholar]