Abstract
Electron tomography is currently the highest resolution imaging modality available to study the 3-D structures of pleomorphic macromolecular assemblies, viruses, organelles and cells. Unfortunately, the resolution is currently limited to 3–5 nm by several factors including the dose tolerance of biological specimens and the inaccessibility of certain tilt angles. Here we report the first experimental demonstration of equally-sloped tomography (EST) to alleviate these problems. As a proof of principle, we applied EST to reconstructing frozen-hydrated keyhole limpet hemocyanin molecules from a tilt-series taken with constant slope increments. In comparison with weighted back-projection (WBP), the algebraic reconstruction technique (ART) and the simultaneous algebraic reconstruction technique (SART), EST reconstructions exhibited higher contrast, less peripheral noise, more easily detectable molecular boundaries and reduced missing wedge effects. More surprisingly, EST reconstructions including only two-thirds the original images appeared to have the same resolution as full WBP reconstructions, suggesting that EST can either reduce the dose required to reach a given resolution or allow higher resolutions to be achieved with a given dose. EST was also applied to reconstructing a frozen-hydrated bacterial cell from a tilt-series taken with constant angular increments. The results further confirmed the benefits of EST, even when applied to standard tilt-series.
Keywords: Equally-sloped tomography, Electron cryomicroscopy, Iterative algorithm, Radiation dose reduction, Image enhancement
I. Introduction
It has been 4 decades since DeRosier and Klug first reconstructed a negatively stained bacteriophage T4 tail from a single electron microscope (EM) projection (De Rosier and Klug, 1968). Since then a number of important methodological advances have been made. The development of rapid freezing techniques has allowed biological samples to be preserved in a near-native state and decreased their sensitivity to radiation damage (Taylor and Glaeser, 1974; Dubochet et al., 1998). The use of electronic detectors and the implementation of automated low-dose data-acquisition schemes have made it possible to record multiple projections quickly without damaging the specimen unnecessarily (Dierksen et al., 1998). Although electron tomography has already emerged as the most powerful method for obtaining the 3D pleomorphic structure of macromolecular assemblies, organelles and thin cells (Lučić et al., 2005; Jensen and Briegel, 2007; McIntosh, 2001), it still faces two irreconcilable requirements. To obtain high-quality and high-resolution 3D images, both the tilt range and the number of projections have to be as large as possible. On the other hand, the total dose used must be minimized, since biological specimens are gradually destroyed by the high-energy electrons (Henderson, 1995), which limits the number of projections that can be acquired. Furthermore, electron tomography also suffers from the missing wedge problem (i.e. specimens can not be tilted beyond ± 70° and hence the data in the remaining ± 20° projections is missing), low contrast and low signal to noise ratios (Lučić et al., 2005; Jensen and Briegel, 2007; McIntosh, 2001).
Here we report the first experimental demonstration of EST (equally-sloped tomography) to alleviate these limitations. As a proof of principle, we used EST to reconstruct single keyhole limpet hemocyanin (KLH) particles, a 7.9 MDa macromolecule consisting of a double-layered and hollow barrel complex about 30 nm in diameter and 35 nm in length (Mouche et al., 2003). KLH was chosen for the study because a model of this molecule's structure to 12 Å resolution is available. The structural model, obtained by averaging hundreds of projection images (Mouche et al., 2003), allowed us to perform various quantitative tests. In comparison with the standard WBP (Frank, 1992; Radermacher, 1992; Harauz and van Heel, 1986), ART and SART (Marabini et al., 1998; Kak and Slaney, 2001; Natterer and Wubbeling, 2001), EST produces reconstructions with apparently equal resolutions with only two-thirds the dose. Furthermore, EST reconstructions exhibited higher contrast, less peripheral noise, more easily detectable boundaries and reduced missing wedge effects. We also used EST to reconstruct a frozen-hydrated spirillum cell from a tilt-series taken with the traditional constant angular increments, with similar increases in contrast and clarity.
2. Equally-Sloped Tomography
2.1. The pseudo-polar fast Fourier transform
Conventional tomography reconstructs a 3D object from a tilt-series of projections with constant angular increments. Since the set of projections are on a polar grid and the object on a Cartesian grid, interpolations have to be performed during the reconstruction process. This is due to the fact that no direct and exact fast Fourier transform exists between the polar and Cartesian grids (Briggs and Henson, 1995). Presently, the most popular 3D image reconstruction method in electron tomography is WBP, in which the interpolations are done in object space (Frank, 1992; Radermacher, 1992; Harauz and van Heel, 1986). However, if the projections are obtained with constant slope increments, it has been shown that there exists a direct and exact fast Fourier transform called the pseudo-polar fast Fourier transform (PPFFT) between a pseudo-polar grid and the Cartesian grid (Mersereau and Oppenheim, 1974; Averbuch et al., 2008). Figure 1 shows a pseudo-polar grid and the PPFFT. For an N × N Cartesian grid, the corresponding pseudo-polar grid is defined by a set of 2N lines, each line consisting of 2N grid points mapped out on N concentric squares. The 2N lines are subdivided into a horizontal group (in blue) defined by y = sx, where s is the slope and |s| ≤ 1, and a vertical group (in red) defined by x = sy, where |s| ≤ 1; the horizontal and vertical groups are symmetric under the interchange of x and y, and Δs = 2/N. When these conditions are met, there exists the PPFFT and its inverse algorithm between a pseudo-polar grid and a Cartesian grid that is mathematically exact and geometrically faithful (Averbuch et al., 2008). Note that The PPFFT and its inverse algorithm was originally developed to interpolate tomographic projections from the polar to the Cartesian grid in Fourier space (Mersereau and Oppenheim, 1974; Averbuch et al., 2008). The idea of acquiring tomographic tilt-series at equal slope increments and then combining PPFFT with iterative algorithms for 3D image reconstructions was first suggested by Miao et al. in 2005.
Figure 1.
Pseudo-polar grid and pseudo-polar fast Fourier transform. For an N × N Cartesian grid where N = 8 in this case, the corresponding pseudo-polar grid is defined by a set of 2N lines, each line consisting of 2N grid points mapped out on N concentric squares. The 2N lines are subdivided into a horizontal group (in blue) and a vertical group (in red) with constant slope increments in each group.
2.1 The EST Reconstruction Algorithm
Compared to other data acquisition schemes (Saxton et al., 1984; Leszczynski et al., 1988), the pseudo-polar grid acquires projections with constant slope increments, and allows the use of the mathematically exact PPFFT. The implementation of the PPFFT, however, requires two stringent conditions: i) the tilt range has to be from −90° to +90° and ii) the number of projections needs to be 2N for an N × N object. These conditions make it impossible to directly apply PPFFT to electron tomography. We overcame these limitations by combining PPFFT with an iterative algorithm (Miao et al., 2005), which was modified from the iterative phase recovery algorithms (Gerchberg and Saxton, 1972; Miao et al., 1999; Miao et al., 2008). Figure 2 shows the schematic layout of the algorithm. We first converted the electron micrograph projections to Fourier slices in the pseudo-polar grid. As illustrated in Fig. 1, the distance between the sampling points on the individual 2N lines of the pseudo-polar grid varies from line to line. In order to calculate the Fourier slices from the electron micrograph projections, the fractional Fourier transform (FrFT) has been used to vary the output sampling distance of the Fourier slices (Miao et al., 2005). The 1D FrFT is defined as
| (1) |
where F(u) is the 1D fractional Fourier slice, ρ(x) is the 1D tomographic projection, x and u are the coordinates in object and Fourier space, and α is a parameter for determining the distance between sampling points in each of the 2N lines (Miao et al., 2005). When α = 1, Eq. (1) simply becomes the 1D FFT.
Figure 2.
Schematic layout of the iterative EST method. The algorithm iterates back and forth between Fourier and object space. In each iteration, the calculated slices are updated with the measured (experimental) slices in Fourier space and the physical constraints are enforced in object space.
Since the tilt range and the number of projections in electron tomography are not large enough to allow the direct use of the PPFFT, we filled in the missing Fourier slices with random values. By applying the inverse PPFFT, we obtained a 3D image in object space. We multiplied the negative-valued voxels and the voxels in the vacuum region (i.e. above and below the specimen) by β, a parameter to control the convergence speed of the iterative algorithm. We usually set β equal to 0.9 in our reconstructions. This step gradually pushed the negative-valued voxels and the voxels in the vacuum region towards zero and generated a modified image. The forward PPFFT was applied to the modified image to obtain a set of calculated Fourier slices. We replaced the corresponding calculated Fourier slices with the measured ones, and the remaining slices kept unchanged. The iterative process was then repeated. Each iteration was monitored by an R-factor (Miao et al., 2005),
| (2) |
where u and v are the coordinates in Fourier space, θ is the tilt angle, and represent the measured and the corresponding calculated Fourier slices. The algorithm was terminated either after it reached a maximum number of iterations or if Rf didn't change after a given number of iterations.
3. Experimental procedures
3.1. Sample Preparation
Megathura crenulata Keyhole Limpet hemocyanin (A.G. Scientific) was diluted in 50 mM sodium phosphate, pH 7.2 to final protein concentration of 8 mg/ml. Tennanometer colloidal gold beads (Ted Pella) were combined with the samples and applied to the grids separately. Protein solutions were rapidly frozen onto R 1.2/1.3 carbon grids (Quantifoil) in liquid ethane using a Vitrobot (FEL, the Netherlands) and the following typical conditions: 100% humidity, 23°C, 4 μl/grid if manually applied, 2.0–3.5 s blot time, −2 to −3 mm blot offset. Hylemonella gracilis cultures were grown in 0.5 g tryptone and 0.5 g yeast extract per litre for two days until they reached an O.D. of 0.05. Cells were then centrifuged and concentrated ten-fold in the same media, then frozen in vitreous ice with a Vitrobot (FEI Company).
3.2. Cryoelectron tomography
Frozen-hydrated grids were loaded into a tilt rotation holder of a 300 kV, FEG G2 Polara FEI TEM, and kept frozen by thermal contact with liquid nitrogen. Tilt series of KLH were collected from −69.4° to +69.4° with constant slope increments. The tilt angles were determined by θ = −arctan((N+2−2n)/N) for n = 1, …, N and θ = π/2 − arctan((3N+2−2n)/N) for n = N+1, …, 2N, where N = 2048 in this case (Miao et al., 2005). For the Hylomonella gracilis cell, a tilt series of 114 projections was collected from −65° to +59° with a constant angular increment of 1.1°. The magnification was 34,000 (6.7 Å/pixel) and the total dose was typically ~78 e−/Å2, distributed according to the 1/cos scheme. The nominal defocus was 12 μm.
4. Results
4.1. 3D Reconstruction of KLH Macromolecules from a Tilt-Series with Constant Slope Increments
First, we used the EST algorithm to reconstruct individual KLH molecules. Purified KLH molecules were rapidly frozen in vitreous ice and imaged using a cryoelectron microscope (Iancu et al, 2006). A set of 105 projections was acquired by tilting the specimen around a single axis from −69.4° to +69.4°, gradually increasing the tilt angle so that the slope of each image plane increased by a constant increment. For comparison, the WBP reconstruction was calculated with IMOD where a low-pass filter with a Gaussian fall-off starting at a particular frequency was applied to reduce noise and enhance image (Mastronarde, 1997). The ART and SART reconstructions were calculated with a software package developed at EMBL (Castano et al., 2007). The EST reconstruction was calculated by a parallel implementation of the iterative algorithm described above. The computational time for each slice (2048 × 2048 pixels) was 5.5 minutes per iteration. Figure 3 shows the convergence of the iterative algorithm. The R-factor (Rf) steadily decreased from 70.2% at the 1st iteration to 8.7% at the 100th iteration whereas the EST reconstruction of the 1st iteration can be considered equivalent to the WBP reconstruction. We also performed a few more independent reconstructions using different initial random values for the missing Fourier slices, and the algorithm always converged to the same result, indicating the final reconstruction is independent of the initial random values.
Figure 3.
Convergence of the iterative algorithm, showing the steady decrease of the R-factor as a function of the number of iteration.
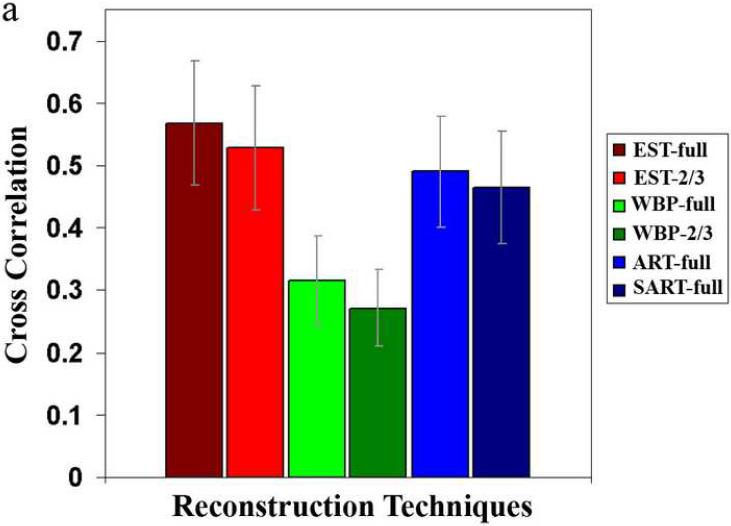
To perform quantitative comparisons, we randomly selected 21 particles from the two reconstructions and compared them with the known high-resolution KLH structure using Bsoft (Heymann, 2001). Figure 4a shows the resulting average cross-correlation coefficients (CCC) and the standard deviations for the 20 EST-, WBP-, ART- and SART-reconstructed particles (labeled “EST-full”, “WBP-full”, “ART-full” and “SART-full”), indicating that EST improved the average CCC by 80% over WBP, 16% over ART, and 22% over SART, respectively. We also calculated Fourier shell correlation (FSC) curves comparing each of the particles to the higher resolution structure and estimated the resolution of each particle based on the FSC = 0.5 criterion (Frank, 2006). Figure 4b shows the average FSC curves for the KLH particles. EST resulted in an average resolution improvement of 5 Å over WBP, 18 Å over ART, and 21 Å over SART, respectively. To highlight the effects of the missing wedge, we calculated the power spectra of a KLH particle reconstructed by EST, WBP, ART and SART, respectively, shown in Fig. 5. Compared to the power spectra of WBP, ART and SART, the EST spectrum is more continuous and smoother as the missing wedge region is filled in with some structural information by the iterative algorithm, suggesting that EST partly alleviates the effects of the missing wedge.
Figure 4.
Quantitative comparisons between the EST, WBP, ART and SART reconstructions. (a) Average cross-correlation coefficients and standard deviations of 20 KLH particles reconstructed by EST of 105 projections (EST-full), EST of 70 projections (EST-2/3), WBP of 105 projections (WBP-full), WBP of 70 projections (WBP-2/3), ART of 105 projections (ART-full) and SART 105 projections (ART-full), respectively. (b) Average Fourier shell correction curves of the reconstructed KLH particles. The EST reconstruction obtained with just two-thirds of the images appears to have essentially the same resolution as the WBP reconstruction.
Figure 5.
Power spectra of a KLH particle reconstructed by (a) EST, (b) WBP, (c) ART and (d) SART, respectively. Compared to the WBP, ART and SART power spectra, the EST spectrum is more continuous and smoother as the missing wedge region is filled in with some structural information by the iterative algorithm. Note that the WBP reconstruction employed a low-pass filter with a Gaussian fall-off starting at a spatial frequency of 0.29 nm−1 (Mastronarde, 1997).
Because the WBP reconstructions exhibit a higher resolution than the ART and SART reconstructions, we performed further quantitative comparisons between WBP and EST. To visualize the difference between the two reconstructions, we generated three orthogonal projections and the iso-surface renderings of a reconstructed particle. Figures 6a and b show three 13.4-nm-thick slices along the XY, YZ and XZ planes of the WBP and EST reconstructions, respectively where the Z-axis corresponds to the direction of the electron beam. These slices show the higher contrast and more easily detectable molecular boundaries generated by EST. Similar clarity could not be achieved with the WBP reconstruction simply by adjusting the contrast limits: instead the EST algorithm made the difference in density of the particle and the surrounding ice larger in comparison with either's variance. To compare iso-surfaces of the particle, we first performed 15 cycles of denoising only to the particle reconstructed by WBP (Frangakis and Hegerl, 2001), implemented in the bnad program of Bsoft (Heymmann, 2001). Figures 7a–d show iso-surface renderings in three different orientations of the model, WBP, denoised WBP and EST, respectively. Compared with WBP before or after denoising, the EST reconstruction shows reduced peripheral noise and more easily detectable molecular boundaries. We also orientated the particles to show the missing wedge effects in object space (the middle column in Fig. 7). While the WBP reconstruction has a few holes in this region as indicated by an arrow in Fig. 7c, the same region in the EST reconstruction (Fig. 7d) is more continuous and smoother, which is consistent with Fig. 5.
Figure 6.
Three 13.4-nm-thick slices along the XY, YZ and XZ planes of a KLH particle reconstructed by (a) WBP-full, (b) EST-full, (c) WBP-2/3 and (d) EST-2/3, respectively. The EST reconstructions show higher contrast, more easily detectable molecular boundaries and reduced noise.
Figure 7.
Iso-surfaces shown in 3 different orientations for (a) the known higher resolution model, (b) WBP-full, (c) WBP-full-denoising, (d) EST-full, (e) WBP-2/3, (f) WBP-2/3-denoising and (g) EST-2/3 reconstructions of a KLH particle. Compared with WBP, the EST reconstructions show less peripheral noise, more easily detectable molecular boundaries and more continuous density. The arrows indicate regions where EST has alleviated the missing wedge effect.
To explore the potential of EST to reduce the total radiation dose required, we performed 3D image reconstructions from a smaller number of projections. We discarded every third projection from the KLH tilt-series and re-calculated the reconstructions (i.e. from only 70 projections), again using both the iterative EST algorithm and the WBP algorithm. Using the same 21 particles considered earlier, Figure 4a shows that the average CCC of the reduced-dose EST reconstructions (labeled “EST-2/3”) was 68% and 95% higher than that of the full-dose WBP reconstructions (“WBP-full”) and the reduced-dose WBP reconstructions (“WBP-2/3), respectively. Corresponding FSC curves (Fig. 4b) indicated that the reduced-dose EST reconstructions had the same resolution as the full-dose WBP reconstructions. Orthogonal projections (Figs. 5c and d) and iso-surfaces (Figs. 7e–g) showed that even the reduced-dose EST reconstructions had higher image contrast, more easily detectable molecular boundaries, less peripheral noise, and more continuous densities than any of the WBP reconstructions.
4.2. 3D Reconstruction of an Intact Bacterial Cell from a Tilt-Series with Constant Angular Increments
After exploring the effects of the EST algorithm on reconstructions from tilt-series with constant slope increments, we next tested whether similar improvements could be realized from more traditional tilt-series taken with constant angular increments. Using the same cryoelectron microscope (Iancu et al., 2006), we acquired a set of 114 projections of a frozen-hydrated Hylomonella gracilis cell. The cell specimen was tilted around a single axis from −65° to +59° with a constant angular increment of ~1.1°. During the EST reconstruction, we assigned each projection image to the closest corresponding Fourier plane in the PPFFT construct. For an array size of 1024 × 1024 pixels, the angular difference between each measured (constant angular increment) projection and its closest PPFFT plane is less than 0.088°, which is smaller than the angular uncertainty present in assigning angles to the experimental images. According to our computer simulations, the small angular difference would have a negligible effect on the 3D reconstruction. The EST reconstruction was carried out using 100 iterations. For comparison, a second, traditional WBP reconstruction of the cell was calculated with IMOD (Mastronarde, 1997). Figures 8a–c and 8e–g show three 6.7-nm-thick slices along the XY, XZ and YZ planes of the WBP and EST reconstructions, respectively. The zoom-in view of a nucleoid region of the cell is shown in Figs. 8d and h. Compared with standard WBP reconstruction, the EST reconstruction clearly exhibits higher resolution, higher contrast, less peripheral noise and more easily detectable boundaries.
Figure 8.

3D reconstructions of a frozen-hydrated spirillum cell. (a), (b) and (c) represent 3 6.7-nm-thick slices in the XY, XZ and YZ plane of the WBP reconstruction. (e), (f) and (g) are the same three orthogonal slices obtained by EST. (d) and (h) are the zoom-in view of a nucleoid region. Compared with the WBP reconstruction, the EST reconstruction exhibits higher resolution, higher contrast, less peripheral noise, more easily detectable boundaries and reduced missing wedge effects.
5. Discussion
By using a novel data acquisition scheme and an iterative image reconstruction algorithm, we carried out first experimental demonstration of the EST method for obtaining the 3D pleomorphic structure of macromolecular assemblies, organelles and cells. Compared with WBP which simply re-projects and adds weighted projections to produce a 3D reconstruction (Frank, 1992; Radermacher, 1992; Harauz and van Heel, 1986), and other non-iterative reconstruction methods (Penczek et al., 2004), EST employs an iterative algorithm to gradually reach a final 3D image. The algorithm is guided towards the minimization of an R-factor defined in Eq. (2). Compared with other iterative techniques such as ART and SIRT (Marabini et al., 1998; Kak and Slaney, 2001; Natterer and Wubbeling, 2001), the EST method iterates back and forth between Fourier and object space based on PPFFT which was proved to be algebraically exact (Mersereau and Oppenheim, 1974; Averbuch et al., 2008). In each iteration, the experimentally measured slices are enforced as constraints in Fourier space and the physical constraints (i.e. positivity and/or the vacuum regions above and below the specimen) are applied in object space. As the direct interpolations between the polar and Cartesian grids are eliminated due to the use of algebraically exact PPFFT, the constraints in Fourier and object space are more naturally enforced in the EST method than other iterative algorithms.
While tilt-series with constant slope increments are theoretically preferred, we showed that the benefits of EST can also be realized with existing, traditional tilt-series taken with constant angular increments, since the PPFFT construct samples Fourier space so finely. Due to the employment of the physical constraints in object space and the measured Fourier slices in Fourier space in the iterative algorithm, the EST method can tolerate the inaccuracies of the tilt angles in practical electron tomography, as evidenced by these experimental results (i.e. Figs. 4–8) and computer simulations (data not shown). Although the iterative algorithm requires more computing power than standard WBP, the high computational demands of EST can be met through parallelization. Furthermore, as general-purpose computation based on graphics processing units is commercially available and promises to improve the FFT speed by 50 – 100 times (www.gpgpu.org), the EST method should be feasible in any typical cryo-EM laboratory.
In our test reconstructions of purified macromolecular complexes and an intact bacterial cell, EST reconstructions with even one-third fewer images exhibited higher contrast, less peripheral noise and smoother molecular densities and more reduced missing wedge effects than WBP reconstructions. While qualitatively similar effects may be obtained by a number of denoising algorithms, it is difficult to tell whether (or when) further denoising begins to erase accurate details, as there is no perpetual constraint maintaining consistency with the original experimental data. In contrast, EST's R-factor allowed the progress of each reconstruction to be tracked, and the quantitative cross-correlation and FSC tests we have performed suggested that the EST reconstructions contained more accurate molecular detail than the WBP reconstructions. Finally as it is general to all tomographic applications, we anticipate that the EST method can be applied to electron tomography (Lučić et al., 2005; Jensen and Briegel, 2007; McIntosh, 2001), X-ray diffractive imaging (Miao et al., 1999; Shapiro et al., 2005; Miao et al., 2008), X-ray microscopy (Larabell and Le Gros, 2004), magnetic resonance imaging (Kak and Slaney, 2001), emission computed tomography (Kak and Slaney, 2001), and importantly, may reduce the radiation doses to the patient in CAT scans (Miao and Fahimian, 2008), which is currently a major issue in medical imaging (Brenner and Hall, 2007).
Acknowledgements
We thank H. Jiang for the help with the figures. JM thanks W. Chiu and Z.H. Zhou for stimulating discussions. This work was supported in part by the U.S. Department of Energy, Office of Basic Energy Sciences under the contract number DE-FG02-06ER46276, the U.S. National Science Foundation, Division of Materials Research (DMR-0520894) and the Alfred P. Sloan foundation. GJJ acknowledges funding from the NIH (R01 AI067548 and P50 GM082545), DOE (grant DE-FG02-04ER63785), a Searle Scholar Award to GJJ, the Beckman Institute at Caltech, and gifts to Caltech from the Gordon and Betty Moore Foundation and Agouron Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Averbuch A, Coifman R, Donoho D, Israeli M, Shkolnisky Y. A Framework for Discrete Integral Transformations I – the Pseudopolar Fourier Transform. SIAM J. Sci. Comput. 2008;30:764–784. [Google Scholar]
- Brenner DJ, Hall EJ. Computed Tomography — An Increasing Source of Radiation Exposure. N. Eng. J. Med. 2007;357:2277–2284. doi: 10.1056/NEJMra072149. [DOI] [PubMed] [Google Scholar]
- Briggs WL, Henson VE. The DFT: An Owners' Manual for the Discrete Fourier Transform. SIAM; Philadelphia: 1995. [Google Scholar]
- Castano Diez D, Mueller H, Frangakis AS. Implementation and performance evaluation of reconstruction algorithms on graphics processors. J. Struct. Biol. 2007;157:288–295. doi: 10.1016/j.jsb.2006.08.010. [DOI] [PubMed] [Google Scholar]
- De Rosier DJ, Klug A. Reconstruction of Three Dimensional Structures from Electron Micrographs. Nature. 1968;217:130–134. doi: 10.1038/217130a0. [DOI] [PubMed] [Google Scholar]
- Dubochet J, Adrian M, Chang JJ, Homo JC, Lepault J, McDowall AW, Schultz P. Cryo-electron microscopy of vitrified specimens. Q. Rev. Biophys. 1998;21:129–228. doi: 10.1017/s0033583500004297. [DOI] [PubMed] [Google Scholar]
- Dierksen K, Typke D, Hegerl R, Baumeister W. Towards automatic electron tomography II. Implementation of autofocus and low-dose procedures. Ultramicroscopy. 1993;49:109–120. [Google Scholar]
- Frangakis AS, Hegerl R. Noise reduction in electron tomographic reconstructions using nonlinear anisotropic diffusion. J. Struct. Biol. 2001;135:239–250. doi: 10.1006/jsbi.2001.4406. [DOI] [PubMed] [Google Scholar]
- Frank J, editor. Electron Tomography. Plenum; New York, USA: 1992. [Google Scholar]
- Frank J. Three-Dimensional Electron Microscopy of Macromolecular Assemblies. Oxford University Press; USA: 2006. [Google Scholar]
- Gerchberg RW, Saxton WO. A practical algorithm for the determination of phase from image and diffraction plane pictures. Optik. 1972;35:237–246. [Google Scholar]
- Harauz G, van Heel M. Exact filters for general geometry three dimensional reconstruction. Optik. 1986;73:146–156. [Google Scholar]
- Henderson R. The potential and limitations of neutrons, electrons and X-rays for atomic resolution microscopy of unstained biological molecules. Q. Rev. Biophys. 1995;28:171–193. doi: 10.1017/s003358350000305x. [DOI] [PubMed] [Google Scholar]
- Heymann JB. Bsoft: image and molecular processing in electron microscopy. J. Struct. Biol. 2001;133:156–169. doi: 10.1006/jsbi.2001.4339. [DOI] [PubMed] [Google Scholar]
- Iancu CV, Wright ER, Heymann JB, Jensen GJ. A comparison of liquid nitrogen and liquid helium as potential cryogens for electron cryotomography. J. Struct. Biol. 2006;153:231–240. doi: 10.1016/j.jsb.2005.12.004. [DOI] [PubMed] [Google Scholar]
- Jensen GJ, Briegel A. How electron cryotomography is opening a new window into prokaryotic ultrastructure. Curr. Opin. Struct. Biol. 2007;17:260–267. doi: 10.1016/j.sbi.2007.03.002. [DOI] [PubMed] [Google Scholar]
- Kak AC, Slaney M. Principles of Computerized Tomographic Imaging. SIAM; Philadelphia: 2001. [Google Scholar]
- Larabell CA, Le Gros MA. X-ray tomography generates 3-D reconstructions of the yeast, Saccharomyces cerevisiae, at 60-nm resolution. Mol. Biol. Cell. 2004;15:957–962. doi: 10.1091/mbc.E03-07-0522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leszczynski KW, Franaszczuk PJ, Penczek PA. Alternative reconstruction algorithm for NMR imaging. Med. & Biol. Eng. & Comput. 1988;26:186–192. doi: 10.1007/BF02442262. [DOI] [PubMed] [Google Scholar]
- Lučić V, Förster F, Baumeister W. Structural Studies by Electron Tomography: From Cells to Molecules. Annu. Rev. Biochem. 2005;74:833–865. doi: 10.1146/annurev.biochem.73.011303.074112. [DOI] [PubMed] [Google Scholar]
- Marabini R, Herman GT, Carazo JM. 3D reconstruction in electron microscopy using ART with smooth spherically symmetric volume elements (blobs) Ultramicroscopy. 1998;72:53–65. doi: 10.1016/s0304-3991(97)00127-7. [DOI] [PubMed] [Google Scholar]
- Mastronarde DN. Dual-axis tomography: an approach with alignment methods that preserve resolution. J. Struct. Biol. 1997;120:343–352. doi: 10.1006/jsbi.1997.3919. [DOI] [PubMed] [Google Scholar]
- McIntosh JR. Electron microscopy of cells: a new beginning for a new century. J. Cell Biol. 2001;153:25–32. doi: 10.1083/jcb.153.6.f25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mersereau RM, Oppenheim AV. Digital reconstruction of multidimensional signals from their projections, Proc. IEEE. 1974;62:1319–1338. [Google Scholar]
- Miao J, Fahimian BP. Iterative Methods for Dose Reduction and Image Enhancement in Tomography. United States Patent and Trademark Office, App. No. PCT/US07/75220. 2008 [Google Scholar]
- Miao J, Charalambous P, Kirz J, Sayre D. Extending the methodology of X-ray crystallography to allow imaging of micrometre-sized non-crystalline specimens. Nature. 1999;400:342–344. [Google Scholar]
- Miao J, Foster F, Levi O. Equally Sloped Tomography with Oversampling Reconstruction. Phy. Rev. B. 2005;72:052103. [Google Scholar]
- Miao J, Ishikawa T, Earnest T, Shen Q. Extending the Methodology of X-ray Crystallography to Allow Structure Determination of Non-Crystalline Materials, Whole Cells and Single Macromolecular Complexes. Annu. Rev. Phys. Chem. 2008;59:387–409. doi: 10.1146/annurev.physchem.59.032607.093642. [DOI] [PubMed] [Google Scholar]
- Mouche E, Zhu Y, Pulokas J, Potter CS, Carragher B. Automated three-dimensional reconstruction of keyhole limpet hemocyanin type 1. J. Struct. Biol. 2003;144:301–312. doi: 10.1016/j.jsb.2003.09.001. [DOI] [PubMed] [Google Scholar]
- Natterer F, Wubbeling F. Mathematical Methods in Image Reconstruction. SIAM; Philadelphia: 2001. [Google Scholar]
- Penczek PA, Renka R, Schomberg H. Gridding-based direct Fourier inversion of the three-dimensional ray transform. J. Opt. Soc. Am. A. 2004;21:499–509. doi: 10.1364/josaa.21.000499. [DOI] [PubMed] [Google Scholar]
- Radermacher M. Weighted Back-Projection Methods. In: Frank J, editor. Electron Tomography. Plenum; New York: 1992. pp. 91–115. [Google Scholar]
- Saxton WO, Baumeister W, Hahn M. Three-dimensional reconstruction of imperfect two-dimensional crystals. Ultramicroscopy. 1984;13:57–70. doi: 10.1016/0304-3991(84)90057-3. [DOI] [PubMed] [Google Scholar]
- Shapiro D, Thibault P, Beetz T, Elser V, Howells M, Jacobsen C, Kirz J, Lima E, Miao H, Neiman AM, Sayre D. Biological imaging by soft X-ray diffraction microscopy. Proc. Natl. Acad. Sci. USA. 2005;102:15343–46. doi: 10.1073/pnas.0503305102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor KA, Glaeser RM. Electron diffraction of frozen, hydrated protein crystals. Science. 1974;186:1036–1038. doi: 10.1126/science.186.4168.1036. [DOI] [PubMed] [Google Scholar]