Abstract
Autophagy mediates the degradation of cellular components in lysosomes, assuring removal of altered or dysfunctional proteins and organelles. Autophagy is not only activated in response to cellular damage, but in fact, one of its strongest and better-characterized stimuli is starvation. Activation of autophagy when nutrients are scarce allows cells to reutilize their own constituents for energy. Besides protein breakdown, autophagy also contributes to the mobilization of diverse cellular energy stores. This recently discovered interplay between autophagy and lipid and carbohydrate metabolism reveals the existence of a dynamic feedback between autophagy and cellular energy balance.
Keywords: glycogen, energy, lysosomes, lipid stores, lipolysis, proteolysis
Introduction
Autophagy, or the degradation of intracellular components in lysosomes, has received considerable renewed attention in recent years. Although originally described in the late ‘60s, the first genes and protein products that contribute to autophagy were not identified until almost thirty years later (Mizushima et al., 2008). The discovery of the autophagic molecular machinery has been rapidly followed by numerous studies supporting the occurrence of autophagic alterations in different common human disorders such as cancer, neurodegenerative and muscular diseases, and infectious disorders, among others (Mehrpour et al., 2010; Mizushima et al., 2008). Most of these connections of autophagy with cellular physiology and disease have emphasized the important function of autophagy in quality control and clearance of altered and damaged intracellular proteins and organelles, its contribution to cellular remodeling through degradation of structural components, or its role in cellular defense as part of both the innate and acquired immunity (Mizushima et al., 2008).
This interest in the autophagic process has also recently led to a renewed attention to the interplay between autophagy and cellular metabolism. Interestingly, protein catabolism was the first defined function for autophagy, and the regulation of the autophagic activity by changes in the nutritional status constituted the center of attention of many early studies (Mortimore and Poso, 1987). The recently gained knowledge about the molecular components of autophagy has allowed revisiting under a new light the “old” function of autophagy in cellular metabolism. In this work, we describe the recent advances on the contribution of autophagy to cellular fueling through mobilization of distinct intracellular energy stores. We also comment on the consequences of failure of autophagy on cellular energetic balance and on recently established connections between compromised autophagy and different metabolic disorders.
Autophagic pathways and molecular effectors of the autophagic process
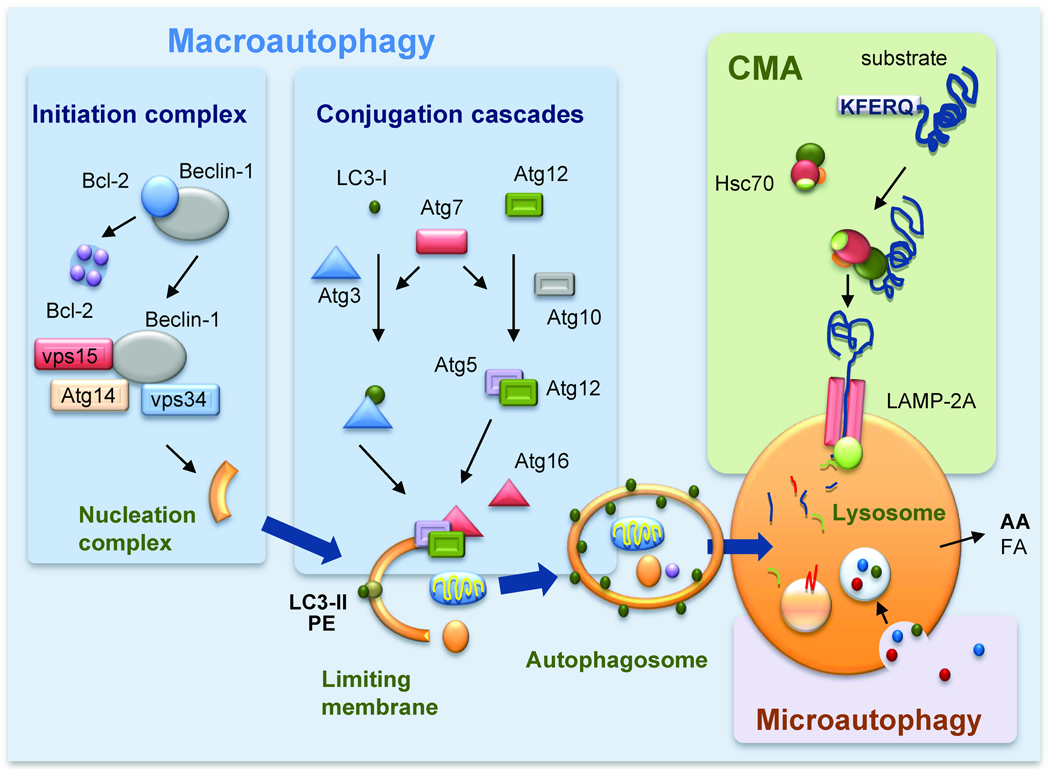
Recent studies have revealed the existence of multiple mechanisms for the delivery of cargo to lysosomes, which give rise to the different types of autophagy (Fig. 1). Cargo can be sequestered selectively or “in bulk” inside de novo formed double membrane vesicles (or autophagosomes) that acquire the hydrolases required for degradation upon fusion with late endosomal and lysosomal compartments. This form of autophagy, known as macroautophagy, also includes very different variants depending on whether the cargo is sequestered “in bulk” (cytosol with any content present in that area) or selectively (mitophagy, ribophagy, lipophagy, aggregophagy, etc.) (Kadandale and Kiger, 2010). Sequestration inside vesicles is also the mechanism used for cargo delivery through another type of autophagy known as microautophagy (Marzella et al., 1981) (Fig. 1). However, in this case the carrier vesicles are single-membraned and originate by invagination of the membrane of lysosomes that then pinch off inside the lumen where they are rapidly degraded by the resident hydrolases. In addition to these types of vesicle-mediated autophagy, single cytosolic proteins can also be delivered to lysosomes for degradation through chaperone-mediated autophagy (CMA) (Arias and Cuervo, 2010). In this type of autophagy, proteins are recognized by a chaperone complex in the cytosol and delivered one-by-one to the lysosomal membrane where they undergo internalization into the lumen through a membrane translocation complex (Fig. 1). In this review, we focus primarily on macroautophagy because it is the pathway for which stronger connections to cellular metabolism have been established.
Figure 1. Molecular components of mammalian autophagic pathways.
Scheme of three different types of autophagy co-existing in mammals. Macroautophagy: A limiting membrane forms de novo engulfing cytosolic components, and seals to form an autophagosome. Degradation occurs when autophagosomes fuse with lysosomes. Initiation of macroautophagy requires assembly and activation of the lipid kinase complex, class III PI3K that serves to recruit components of the two conjugation cascades, the LC3/PE and Atg5/12 cascades to the limiting membrane. Chaperone-mediated autophagy (CMA): When a cytosolic chaperone (hsc70) recognizes a targeting motif (KFERQ) in cytosolic proteins and delivers them to the lysosomal membrane. Upon binding to the lysosome-associated protein type 2A, substrate proteins unfold and cross the lysosomal membrane assisted by a lumenal chaperone. Microautophagy: Invaginations at the lysosomal membrane trap cytosolic cargo that is internalized after the vesicles pinch off into the lysosomal lumen. AA: amino acids; FA: free fatty acids; Atg: autophagy-related protein; PE: phosphatidylethanolamine.
The recent detailed analysis of the molecular components that participate in macroautophagy has identified more than 30 genes and their corresponding proteins (autophagy-related proteins or Atg) (Mizushima et al., 2008). Atgs organize into functional complexes that participate in each of the steps of macroautophagy. Readers are referred to recent comprehensive reviews detailing the intricacies of the macroautophagic machinery and its contribution to each of the different steps in this process (Mizushima et al., 2008). Briefly, the first event of macroautophagy is the formation of the nucleation complex, the structure that will give rise to the limiting membrane of the autophagosome. ER, mitochondria, Golgi and plasma membrane have all been shown to be cellular sources for autophagosome formation (reviewed in (Cuervo, 2010)). The common feature in all these sites of autophagosome formation is the transient association of a kinase complex formed by vps34, beclin and vps15 and different modulatory proteins to the organelle membrane (Fig. 1). The kinase activity of this complex along with the dynamic delivery of lipids to the site of formation by continuous shuttling of Atg9, the only transmembrane Atg, generates the isolation membrane. This membrane elongates to sequester the cargo through the action of two ubiquitin-like conjugation systems, modulated by Atg7, that result in the formation of the Atg5/12 complex and the conjugation of LC3 to phosphatidylethanolamine (Mizushima et al., 1998) (Fig. 1). The limiting membrane eventually seals confining the cargo inside a double membrane vesicle that docks and fuses with secondary lysosomes and endosomes through mechanisms still only partially understood. The infusion of hydrolases, and the acidification of the autophagosome lumen by the proton pump provided by lysosomes results in the complete degradation of cargo into their constituent components that then gain access to the cytosol through permeases at the lysosomal membrane. Formation of the autophagosomes occurs randomly during non-selective in-bulk macroautophagy, but it is fit to surround specific cargo during selective macroautophagy. In the latter case, cargo-recognizing proteins, such as p62 or NBR1 (Lamark et al., 2009), act as bridges between the cargo and the autophagic machinery.
Different signaling mechanisms modulate autophagic activity in mammalian cells (Mehrpour et al., 2010). The best-characterized modulators of macroautophagy are those that regulate this catabolic pathway in response to nutritional changes, such as the mammalian target of rapamycin (mTOR). This nutrient sensor kinase complex acts as a hub for the integration of information on the cellular energetic balance and negatively regulates the initiation step of macroautophagy. The mechanisms and essential components of mTOR regulation of macroautophagy will be described in the following sections. Activation of macroautophagy occurs not only in response to nutritional changes, but also as a consequence of intracellular accumulation of toxic protein products, organelle damage, infectious agents and changes in the physical properties of the extracellular media (Mehrpour et al., 2010). Although, some of these stressors activate macroautophagy by modulating mTOR signaling, macroautophagy can also be activated by mechanisms independent of mTOR (Sarkar et al., 2009). Current efforts are focused on discriminating the differences between mTOR-dependent and mTOR-independent macroautophagy and the possible functional interaction between these two regulatory mechanisms.
Activation of autophagy in response to nutrient deprivation
Starvation was recognized as one of the best stimuli of macroautophagy from the very early days, when most studies were done in rodent liver analyzing differences in the size of the autophagic compartment (Mortimore and Poso, 1987). These analyses unveiled a marked inhibitory effect of insulin on hepatic macroautophagy and an opposing stimulatory effect for glucagon (Mortimore and Poso, 1987). When yeast was adapted as a good experimental model to decipher the genetic components involved in macroautophagy, removal of the source of nutrients (nitrogen or carbon) was also found to be a universal stimulus of macroautophagy. In fact, in yeast not only starvation but just a shift in the carbon source is enough to activate macroautophagy. Depletion of amino acids or serum from the culture media, or of glucose to a less extent, activates macroautophagy in mammalian cells in culture (Mitchener et al., 1976).
More recent studies, using a transgenic mouse model expressing a fluorescent marker of macroautophagy, have revealed that starvation activates macroautophagy not only in liver but also in heart, skeletal muscle, exocrine glands and kidney (Mizushima et al., 2004). The fact that autophagosomes, the morphological signature of macroautophagy, were not detected in brain under these conditions, led to the initial conclusion that macroautophagy was not activated in response to starvation in central nervous system, and even questioned the occurrence of autophagy as a whole in brain. Subsequent studies have revealed that macroautophagy can be markedly upregulated pharmacologically in brain (Sarkar et al., 2009). In fact, macroautophagy occurs in neurons and is essential for maintenance of cellular homeostasis (Hara et al., 2006; Komatsu et al., 2006), however, the rapid clearance of autophagosomes limits the number of them visible at a given time (Boland et al., 2008).
An interesting example of extreme starvation that requires proper activation of macroautophagy is the period from birth until newborns gain access to maternal milk (Kuma et al., 2004). Mice with compromised macroautophagy are unable to survive this period, reinforcing the important role of macroautophagy as a source of both amino acids and energy. However, activation of macroautophagy is not limited to extreme nutritional conditions. In fact, in organs such as liver, this process gets activated daily in between meals, contributing both to provide essential components under these conditions and also guaranteeing periodical cellular clearance (Mizushima et al., 2004).
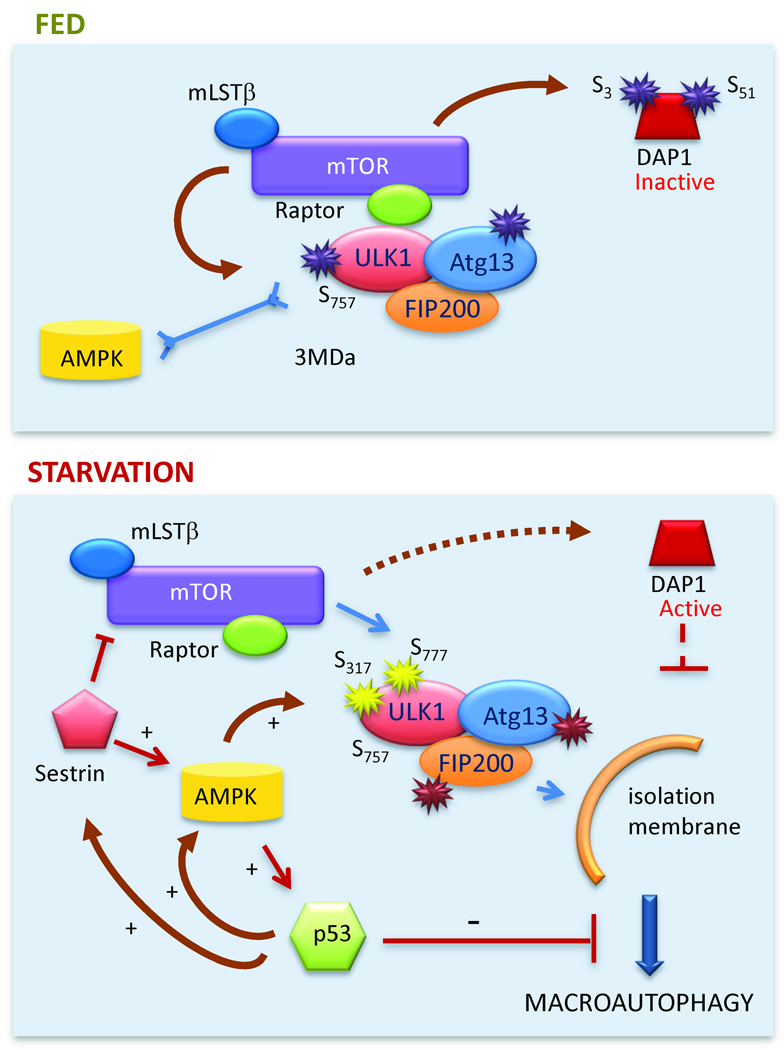
Information on the energy status within a cell is transduced by signaling pathways to effector molecules that promote growth or activate catabolic pathways, such as autophagy. As mentioned in the previous section, mTOR complex 1 (mTORC1) is a critical nutrient sensor intricately linked to macroautophagic regulation (Fig. 2). In yeast, activation of TORC hyperphosphorylates Atg13, preventing its interaction with Atg1 (Kamada et al., 2000). The Atg1-Atg13 complex is required to initiate shuttling of Atg9 to the site of autophagosome formation. Although the mammalian downstream targets of mTOR required to regulate macroautophagy remained elusive for long time, recent studies have revealed that mTORC1 phosphorylates and actively sequesters the mammalian homolog of Atg1, ULK1 in a complex with Atg13 and FIP200 in an inactive state (Chan et al., 2009; Hosokawa et al., 2009; Jung et al., 2009). Activation of a second cellular energy sensor, AMP activated protein kinase (AMPK) during nutrient deprivation inhibits mTOR activity, reducing ULK1 phosphorylation and promoting its release from mTORC1 (Egan et al., 2011; Kim et al., 2011; Lee et al., 2010b; Shang et al., 2011). AMPK-mediated phosphorylation of ULK1 at a different residue favors its mobilization to the region of autophagosome formation and the initiation of macroautophagy (Kim et al., 2011) (Fig. 2). AMPK can also activate autophagy through inhibition of mTOR activity by direct phosphorylation of the Tuberous sclerosis complex 2 (TSC2) and Raptor (Gwinn et al., 2008).
Figure 2. Nutritional regulation of macroautophagy.
Top: Under normal nutritional conditions mTOR 1) phosphorylates ULK1 which locks the ULK1-ATG13-FIP200 complex at the TORC1 complex and prevents its interaction with AMPK, and 2) inactivates DAP1 by phosphorylation. Bottom: During starvation, AMPK phosphorylates ULK1 favoring its release from TORC1 and its association to the site of isolation membrane formation. Reduced mTOR activity decreases DAP1 phosphorylation that through its inhibitory effect on macroautophagy prevents abnormal upregulation of this pathway. The positive feedback loops between AMPK, sestrins and p53 are shown. Independent of its transcriptional activity, cytosolic p53 can exert a direct inhibitory effect on macroautophagy.
Recent studies support the existence of feedback mechanisms that prevent both prolonged activation and sustained repression of macroautophagy. The death-associated protein 1 (DAP1) is a negative regulator of macroautophagy that remains inactive through phosphorylation by mTOR. When mTOR is inhibited, for example during starvation, DAP1 is dephosphorylated and contributes to prevent uncontrolled activation of macroautophagy (Koren et al., 2010). In contrast, cells bypass the inhibitory effect of chronic mTOR activation on macroautophagy by upregulating a family of proteins known as sestrins. In addition to sending inhibitory feedback signals to mTOR, sestrins activate AMPK directly leading to the subsequent upregulation of macroautophagy (Lee et al., 2010a). Likewise, the phosphorylation of the insulin receptor substrate 1 (IRS1) by a substrate of mTOR – p70S6K – reduces insulin signaling and allows induction of macroautophagy even in the presence of nutrients to sustain its quality control function.
The number of intracellular signaling pathways with modulatory effects on macroautophagy keeps growing, unveiling the complexity of the regulation of this catabolic process (Mehrpour et al., 2010). Of particular interest are the connections between macroautophagy and p53, a tumor suppressor recently shown to also contribute to regulate energy metabolism (Vousden and Ryan, 2009). During metabolic stress, p53 is directly activated by AMPK-dependent phosphorylation, and sestrins and AMPK are among the many genes activated by p53 (Fig. 2). The effect of p53 on autophagy has revealed to be more complex than initially anticipated. Thus, p53 mediates the transcriptional upregulation of autophagy activators such as DRAM (damage-regulated autophagy modulator) (Crighton et al., 2006) but at the same time cytosolic forms of p53 can directly repress autophagy by mechanisms still poorly characterized (Tasdemir et al., 2008) (Fig. 2). Different stress-activated kinases also link the cellular energy status with macroautophagy. Starvation-induced c-jun N-terminal kinase (JNK) activation leads to Bcl-2 phosphorylation that makes Beclin-1 accessible for macroautophagic activation (Wei et al., 2008). A non-canonical MEK/ERK cascade downstream of AMPK, and the p38α MAPK have also both shown to upregulate macroautophagy (Wang et al., 2009; Webber and Tooze, 2010). Recent studies support that another cellular stress-activated kinase, IκB kinase (IKK) modulates nutrient-regulated macroautophagy in a NF-κB-independent manner by activating AMPK and JNK1 (Criollo et al., 2010). Inhibition of macroautophagy by extracellular cytokines and growth factors is also exerted by multiple signaling cascades, such as MAPK-ERK1/2, Stat3, Akt/Foxo3, and CXCR4/GPCR, supporting that tight regulation is responsible for fine tuning of induction of macroautophagy.
Autophagy in protein catabolism
Although lysosomes contain a broad array of hydrolases (lipases, proteases, glycosidases, nucleotidases) that allows them to degrade all kind of macromolecules, most of the functional studies on autophagy have focused on protein breakdown. In fact, for a long time, changes in the rate of degradation of long-lived proteins were used to monitor autophagy. In liver, autophagy was estimated to degrade from 1.5 to 5% of the total proteome per hour under fed or starved conditions, respectively (Deter et al., 1967). Autophagy was thus responsible for up to 70% of intracellular protein breakdown in this organ, which was later confirmed in mouse models knocked-out for essential autophagy genes in liver (Komatsu et al., 2005).
The purpose of protein breakdown is two-fold: to utilize amino acids for cellular fueling and to replenish the intracellular pool of amino acids required to maintain protein synthesis. In fact, bulk protein synthesis has been shown to be substantially reduced in autophagy-deficient cellular and animal models (Onodera and Ohsumi, 2005). Although amino acids are in general a poor source of energy, the fact that different phenotypes due to impaired macroautophagy could be restored by addition of the cell-permeable tricarboxylic acid cycle substrate methylpyruvate, supports that one of the functions of autophagy is to contribute energy (Lum et al., 2005). However, this fact may need to be revisited in light of the recently discovered ability of macroautophagy to mobilize other types of energy stores (discussed in the following sections). Branched-chain and some dispensable amino acids can be catabolized in peripheral tissues and in fact, neonates with compromised macroautophagy showed marked reduction of these particular amino acids in circulating blood and peripheral tissues when compared to controls (Kuma et al., 2004). However, when the energetic requirements of the autophagic process itself are taken into consideration, the final energy balance would be rather poor if energy were only to be obtained from the amino acids generated from protein breakdown. All autophagy variants require ATP in different steps of the process. Macroautophagy requires ATP for initiation and progression (Schellens et al., 1988); binding of the chaperones to the CMA substrates and their unfolding before lysosomal translocation are also ATP-dependent processes (Arias and Cuervo, 2010), and even for microautophagy the formation of the membrane invaginations that sequester cargo drives on ATP hydrolysis (Sahu et al., 2011). Common to all these pathways is also the need for ATP to maintain lysosomal pH (Van Dyke, 1988).
In addition to energy, amino acids can be utilized early in fasting to provide substrates for gluconeogenesis and ketogenesis (in liver) and to replenish the intracellular pool of amino acids (Fig. 3). The contribution of different proteolytic systems to this replenishment seems to be timed with the duration of the starvation process. Studies in cultured cells have shown that the proteasome system contributes most of the amino acids to this pool in the first hours of starvation (Vabulas and Hartl, 2005), whereas macroautophagy starts right after and reaches a peak at about 6–8h into the starvation period (Deter et al., 1967). In fact, although autophagosomes are still visible up to 24h of starvation, the maximal rates of autophagosome formation are reached around 6h and decline progressively after that. If starvation persists beyond 8h, protein breakdown switches now to CMA that reaches maximal upregulation at 10–12h into starvation and persists at similar rates up to 3 days (Cuervo et al., 1995). It is likely that switching from macroautophagy to CMA may allow cells to spare from degradation essential proteins and organelles under prolonged nutritional stress. Although the molecular mechanisms that regulate this autophagic switch are not fully understood, ketone bodies produced during starvation have been shown to have a stimulatory effect on CMA by inducing an increase in intracellular levels of oxidized proteins, common substrates for this autophagic pathway (Finn and Dice, 2005).
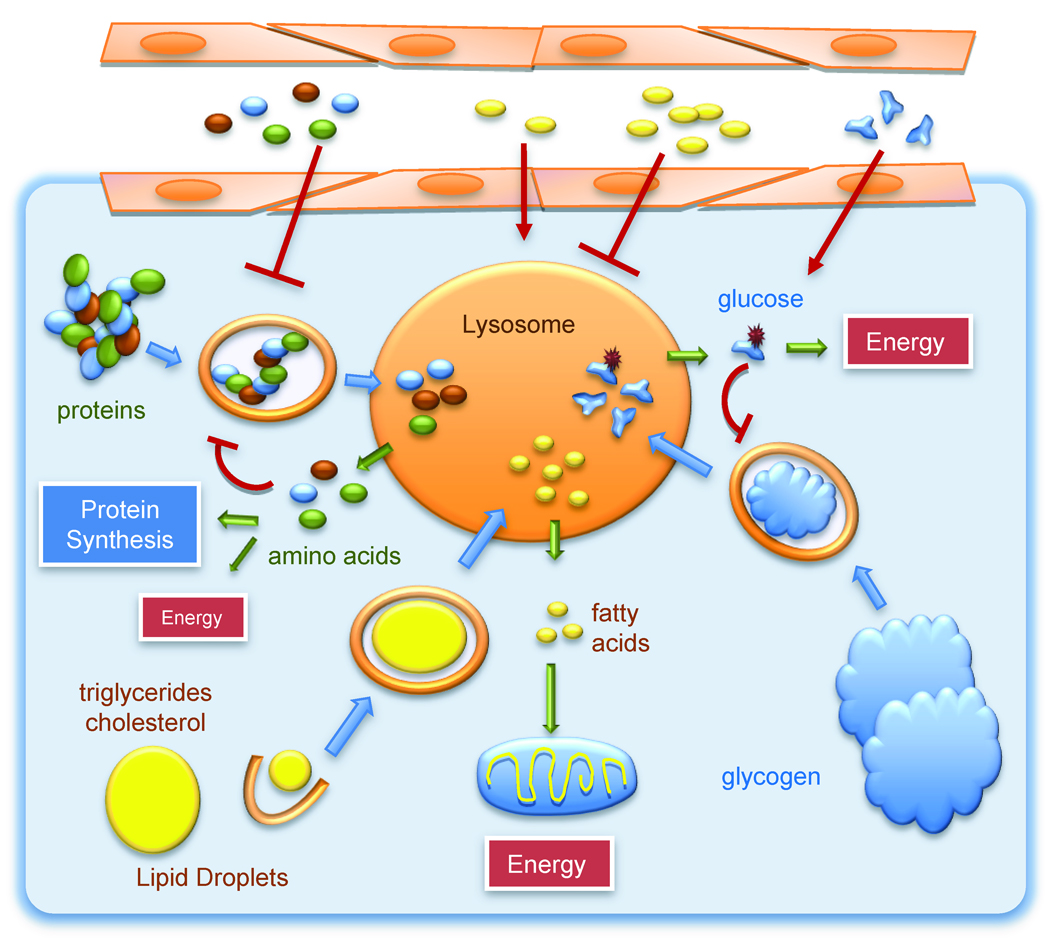
Figure 3. Different catabolic pathways converge in lysosomes.
Macroautophagy contributes to the delivery of proteins, lipid stores and glycogen for breakdown into lysosomes. The constituent components of these macromolecules exit the lysosome and become available for production of energy. In the case of protein breakdown, the resulting amino acids may have less energetic value and be preferentially utilized for the synthesis of new proteins. Levels of amino acids, free fatty acids and sugars circulating in blood or in the extracellular media have a direct impact on intracellular macroautophagy.
As described in previous sections, protein breakdown by autophagy is self-regulated as the products of this breakdown, the constitutive amino acids of the cargo proteins, impose a break on the autophagic process. Signaling mechanisms known to modulate macroautophagy integrate information on both, the availability of extracellular amino acids as well as their levels in the intracellular pool (Mehrpour et al., 2010). The exact mechanism by which amino acids signal through mTOR to downregulate macroautophagy is still unclear, but the contribution of vps34, Ras-related small GTPases (that relocate mTOR to the lysosomal compartment) and a bidirectional transporter that exchanges L-glutamine by essential amino acids have all been involved in this signaling process (Nicklin et al., 2009; Sancak et al., 2010)
Although beyond the scope of this review, degradation of proteins by autophagy also contributes to quality control and prevents proteotoxicity associated with accumulation of abnormal proteins. In fact, defective autophagy often associates with formation of proteins aggregates (Hara et al., 2006; Komatsu et al., 2006) and is likely the basis for protein conformational disorders such as Alzheimer’s and Parkinson’s disease.
Autophagy in lipid metabolism
The contribution of autophagy to lipid metabolism has been recently elucidated through the initial discovery of a process now termed macrolipophagy (Fig. 3) (Singh et al., 2009a). Mobilization of neutral lipids from lipid droplets (LD; main cellular lipid stores) to generate fatty acids for mitochondrial oxidation and energy production or for extracellular secretion was solely attributed to cytosolic or ER-associated lipases. Although lysosomal lipases (acid lipases) were considered to only breakdown endocytosed lipids, we have recently shown that intracellular lipids can also reach the lysosomal lumen through sequestration inside autophagosomes (Singh et al., 2009a). Pharmacological or genetic inhibition of macroautophagy in cultured hepatocytes does not affect lipogenesis or lipid secretion but leads to reduced rates of beta-oxidation and marked lipid accumulation in cytosolic LD (Singh et al., 2009a). In fact, mice knocked-out for an essential autophagy gene in liver (Atg7) display massive accumulation of triglycerides and cholesterol in the form of LD, confirming that macroautophagy is also important for lipolysis in vivo (Singh et al., 2009a). Although further studies are needed to better elucidate the mechanisms behind LD recognition and sequestration by the macroautophagic machinery, preliminary evidence supports that the limiting membrane of the autophagosome forms at the surface of the LD trapping only a discrete region of the droplet. It is possible that specific adaptor proteins may facilitate interactions between lipid droplets and the autophagic components.
Macrolipophagy is not limited to hepatocytes but occurs in almost every cell type investigated till date. In addition to the initial description in cultured hepatocytes and embryonic fibroblasts (Singh et al., 2009a), disruption of macroautophagy leads to intracellular accumulation of lipid stores in endothelial cells, lymphoblasts, dendritic cells, glial cells and even in neurons (Koga et al., 2010), suggesting a generalized function of macroautophagy in cellular lipid mobilization. The accumulation of LD upon blocking macroautophagy even in the absence of any nutritional challenge, supports that macrolipophagy is a constitutive process in many cells (Singh et al., 2009a) (Fig. 4).
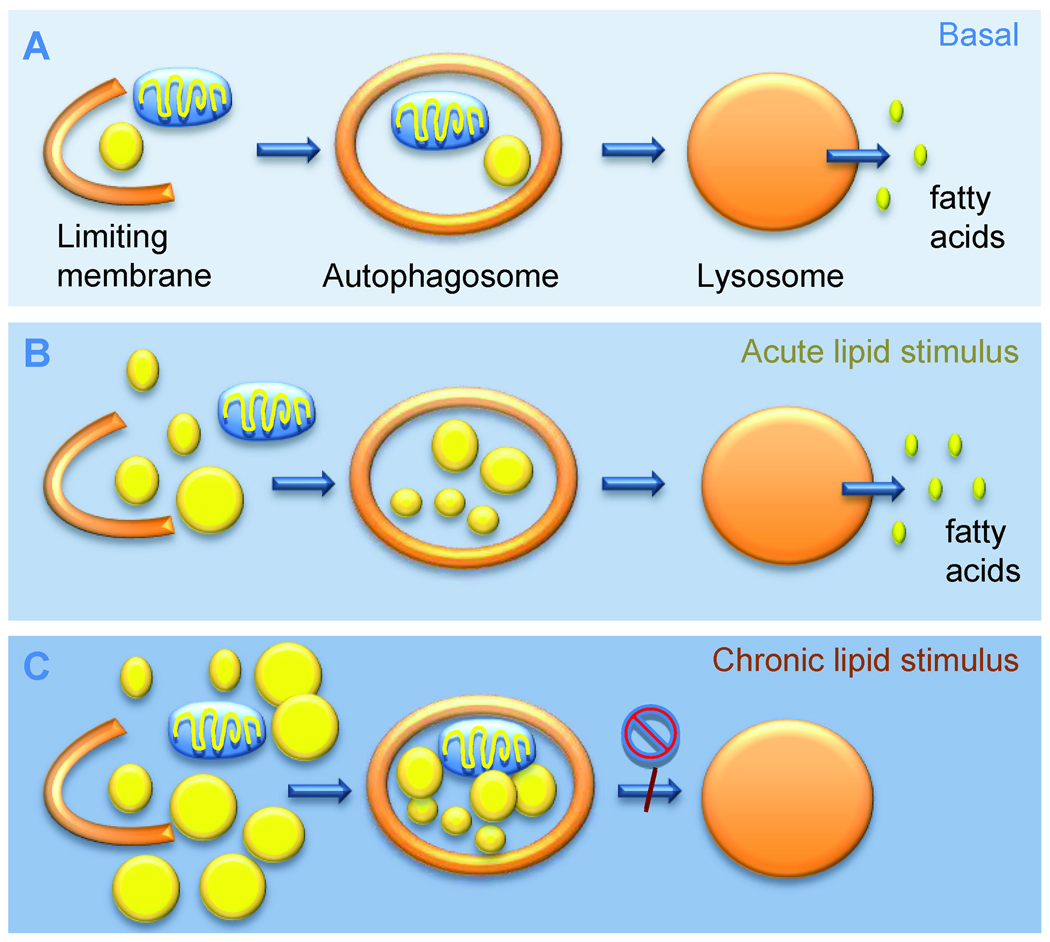
Figure 4. Autophagy in the metabolism of cellular lipid stores.
A. Autophagy constitutively mobilizes cellular lipid droplets by macrolipophagy. Lipid droplets are sequestered inside autophagosomes and delivered to the lysosomes for degradation, resulting in free fatty acid release for cellular respiration. B. During acute lipid challenge most cells maximally activate macrolipophagy to prevent excessive cellular lipid accumulation. C. Chronically maintained lipid challenges reduce macroautophagic degradation of lipids by decreasing the efficiency of autophagosome-lysosomal fusion, leading to further cellular lipid accumulation.
Energetically, massive intracellular lipid mobilization may be particularly advantageous during nutrient deprivation. In fact, macrolipophagy is activated by starvation and persists beyond the 6–8h limit established for macroautophagy-dependent proteolysis (Singh et al., 2009a). Prolonged starvation results in a switch in the type of cytosolic cargo sequestered by autophagy from a heterogenous mix of cytosolic substrates toward predominant lipid sequestration at later times (Singh et al., 2009a). Besides activation of macrolipophagy during energetically demanding conditions, this process is also used by different cells to handle massive affluence of lipids (Fig. 4). Exposure of cells to acute lipid challenges upregulates macrolipophagy to limit cellular lipid droplet load. However, the stimulatory effect of lipids on macroautophagy is not linear and an exposure of cultured cells to large amounts of lipids or chronic high fat feeding in rodents decreases both macrolipophagy (Singh et al., 2009a) and the macroautophagy of proteins and organelles (Koga et al., 2010). Maturation of autophagosomes into autophagolysosomes appears to be affected by this high lipid content, likely because of reduced vesicular fusion (Fig. 4) (Koga et al., 2010). In vivo, this defective autophagosome-lysosome fusion is partially compensated by fusion of autophagosomes with endosomes (to form amphisomes), which prevents massive accumulation of undegraded vesicles yet results in lower net autophagic balance. Reduced macrolipophagy in a setting of chronic lipid loading promotes cellular lipid accumulation that may set up a vicious circle resulting in inhibition of macroautophagy and exaggerated lipid accumulation that may form the basis for development of fatty liver disease (Fig. 4).
The interplay between autophagy and lipid metabolism has different layers of complexity. Thus, while autophagy serves to regulate cellular lipid droplets, multiple evidences support an important modulatory role of lipids and lipid modifications on autophagy. Lipids, such as phosphatidylinositol 3-phosphate (PI3P) serve as a scaffold for the assembly of the autophagic apparatus required for limiting membrane formation, autophagosome trafficking in microtubules, lysosomal fusion, and possibly even in cargo recognition (reviewed in (Kaushik et al., 2010)). A coordinated balance between the activity of kinases and phosphatases modulates intracellular levels of PI3P (Vergne et al., 2009).
In addition to regulatory lipid molecules, changes in the content of structural lipids in the membrane of the organelles involved in autophagy also affect autophagic activity. For example, the compromise in autophagosome-lysosome fusion in animals exposed to a high fat diet is reproducible in vitro by simply modulating the cholesterol content of any of the vesicular compartments involved in this process (Koga et al., 2010). Changes in lipid composition of lysosomal and late endosomal membranes also affect other forms of autophagy such as CMA (Kaushik et al., 2006) and microautophagy (Sahu et al., 2011).
The function of macroautophagy in the adipose tissue, an organ committed to fat storage, deserves separate mention, since autophagy has been shown to regulate adipose tissue development by modulating adipocyte differentiation (Singh et al., 2009b; Zhang et al., 2009). Compromised macroautophagy in preadipocytes alters adipocyte differentiation that associates with decreased levels of adipogenic transcription factors, and markers of adipocyte differentiation (Singh et al., 2009b). Interestingly, mice deficient in macroautophagy in the white adipose tissue (knock-out for Atg7) show decreased adipose mass that phenocopies brown adipose tissue (Singh et al., 2009b; Zhang et al., 2009). The physiological consequence of this phenotypic switching is a remarkably lean mouse with improved glucose tolerance (Singh et al., 2009b; Zhang et al., 2009). While the molecular mechanisms behind macroautophagy-mediated adipocyte differentiation are unclear, it is possible that accumulation of proteins that modulate adipogenesis or confer a “browning” effect could be behind abnormal adipose differentiation. Although the role of macroautophagy in lipid droplet biogenesis has only been analyzed in adipose tissue, it is possible that formation of lipid droplets in other tissues also requires an intact autophagic system. In support of this idea, reduced content of lipid droplets has been reported in the livers of very young mice knocked out for the essential macroautophagy gene Atg7 (Shibata et al., 2009), which contrasts with the marked accumulation of lipid droplets observed in the same mouse model as the mice reach adulthood (Singh et al., 2009a).
Autophagy in carbohydrate homeostasis
Glycogen degradation by phosphorylase and debranching enzymes occurs primarily in the cytosol, but lysosomal acid glycosidases also contribute to glycogen breakdown (Schworer et al., 1979) (Fig. 3). The acute nutrient deprivation that follows the post-natal period triggers the autophagy of glycogen in liver to provide glucose (Kalamidas and Kotoulas, 2000). Lysosomal mannose 6- and glucose-6-phosphatases modulate the phosphorylation state of glucose favoring its exit from the lysosome. Glycophagy is not limited to liver, and in fact, altered autophagic degradation of glycogen stores may underlie the basis of different muscle disorders, now classified as autophagic vacuolar myopathies, such as Danon disease, X-linked vacuolar myopathy with excessive autophagy and Pompe’s disease (Fukuda et al., 2007; Nishino et al., 2000). Glycogen granules accumulate in muscles of Danon disease patients, resulting in cardiomyopathy, proximal muscle weakness and mental retardation (Nishino et al., 2000). Distinctive of this disease is the fact that despite the accumulation of glycogen, levels of acid maltase activity are normal. Recent studies have revealed that the primary defect is in the ability to deliver the material sequestered in autophagosomes, including glycogen, to lysosomes for degradation (Nishino et al., 2000). Defective autophagy seems also behind the accumulation of polyglucosans, poorly branched glycogen bodies, in organs from patients with Lafora disease (Aguado et al., 2010). In contrast, glycogen accumulates within lysosomes in skeletal muscles in Pompe’s disease patients (Glycogen Storage Disease Type II) due to deficiency of the lysosomal enzyme acid alpha-glucosidase that degrades glycogen (Fukuda et al., 2007). The fact that the primary defect in Pompe’s disease is not in the delivery of glycogen to lysosomes by macroautophagy but in degradation once in this compartment, explains the beneficial effects observed upon muscle-specific inhibition of autophagy (decreased muscle glycogen by half) (Raben et al., 2010). Although macroautophagy has been so far the focus of most studies, it is possible that other forms of autophagy, in particular microautophagy, may also contribute to the delivery of glycogen granules to the lysosomes (Raben et al., 2010).
Autophagy can indirectly contribute to glucose metabolism by modulating pancreatic β-cell mass and function (Ebato et al., 2008; Jung et al., 2008). Mice with β-cell-specific inhibition of macroautophagy reveal progressive β-cell degeneration and decreased insulin secretion (Ebato et al., 2008). Although, many factors could contribute to this reduced insulin secretion in macroautophagy-deficient pancreatic β-cell, the fact that secretion is restored upon antioxidant treatment supports that accumulation of dysfunctional mitochondria that generate reactive oxygen species could be behind impaired secretion.
Regulation of insulin secretion by controlled lysosomal degradation of insulin secretory granules was proposed long ago under the term “crinophagy” that refers to the direct fusion of secretory granules with lysosomes (Marzella et al., 1981). However, recent studies support that macroautophagy can also contribute to the delivery of these secretory granules to lysosomes. For example, in the secretory-deficient Rab3−/− β-cell, intracellular insulin stores are maintained despite decreased secretion because of increased autophagic degradation of insulin (Marsh et al., 2007). Interestingly, macroautophagy-mediated degradation of other secretory granules, such as those containing zymogen in pancreatic acinar cells, may contribute to the activation of the inactive enzymes in lysosomes. In fact, abnormal upregulation of macroautophagy with enhanced processing of these lytic enzymes could be the pathogenic basis for acute pancreatitis (Grasso et al., 2009; Hashimoto et al., 2008).
Despite the pronounced phenotype of autophagy blockage in β-cells, macroautophagy does not increase in these cells during starvation (Ebato et al., 2008). Interestingly, active autophagosome formation was observed in diet-induced obese mice and in the genetic insulin-resistant db/db mice (Fujitani et al., 2009), suggesting that impaired insulin signaling permits upregulation of β-cell autophagy. Increased β-cell macroautophagy in response to chronic lipid stress is perceived as an adaptive response against insulin resistance since β-cell expansion that occurs in response to high fat feeding is not observed in β-cell-macroautophagy-deficient mice (Ebato et al., 2008).
The interplay between autophagy and carbohydrate metabolism is also bi-directional as growing evidence supports the regulatory effect of different glycidic groups on macroautophagy (Ravikumar et al., 2003). Both glucose deprivation and increased cellular glucose affluence have been shown to enhance macroautophagy, although likely by different mechanisms (Fig. 3). Activation of macroautophagy during glucose deprivation has been proposed to be, in part, a consequence of the associated induction of oxidative stress. In contrast, the enhanced degradation of pathogenic proteins by macroautophagy elicited by increasing intracellular glucose was proposed to result from reduced phosphorylation of Akt, mTOR and S6K1 mediated by glucose 6-phosphate, as the effects were mimicked by 2-deoxyglucose but not 3-O-methyl glucose that cannot be phosphorylated (Ravikumar et al., 2003). Other sugars, such as the disaccharide trehalose, physiologically present in many non-mammalian species, also induce macroautophagy and clearance of pathogenic proteins (Sarkar et al., 2007). Although some of the benefits of trehalose are probably via its chaperone-like chemical properties, trehalose can also activate macroautophagy in an mTOR-independent manner to remove neurotoxic proteins. Aminosugars such as glucosamine can also activate macroautophagy for clearing ubiquitin-conjugated protein aggregates (Shintani et al., 2010).
Metabolic dysfunction and autophagy
Defective autophagy has a negative impact in cell metabolism at very different levels and in fact, recent studies support that it may be an aggravating factor or even underlie the basis of common metabolic disorders.
In almost all tissues, the most direct manifestation of the inability to degrade proteins by autophagy is the accumulation of polyubiquitinated protein inclusions that can become a sink for chaperones and proteases, and an additional source of reactive species (Hara et al., 2006; Komatsu et al., 2006; Komatsu et al., 2005). In contrast to this well characterized function in quality control, there is only sparse information (mainly restricted to the studies in neonates (Kuma et al., 2004)) on the metabolic consequences of deficient protein breakdown by macroautophagy.
A growing number of studies support tight connections between dysfunctional autophagy and lipid and carbohydrate metabolic disorders, although the autophagic changes are revealing themselves to be clearly tissue-dependent. Exciting data from human studies reported alterations in macroautophagy within adipose tissues from obese individuals (Kovsan et al., 2011). Increased macroautophagy markers and faster autophagic clearance have been observed in both, subcutaneous fat depots and, to a larger extent, in omental fat. Autophagy upregulation is even more pronounced in obese individuals that have developed insulin resistance (Kovsan et al., 2011). In contrast to this upregulation of autophagy in adipose tissue and pancreatic β-cells during adiposity, chronic high fat feeding inhibits autophagy in liver (Singh et al., 2009a; Yang et al., 2010). In fact, dietary and genetic models of obesity (ob/ob mouse) display markedly reduced macroautophagy in liver that alters insulin signaling in these models (Yang et al., 2010). Proper insulin signaling can be restored when the autophagic defect is corrected (Yang et al., 2010). Factors independent of insulin levels per se, likely ER stress, may contribute to reduced hepatic macroautophagy in obesity because streptozotocin injections in ob/ob mice failed to restore autophagy in the liver, as opposed to the induction of autophagy in streptozotocin-injected lean mice (Yang et al., 2010). Additional mechanisms such as the chronic upregulation of mTOR signaling observed in obesity (Newgard et al., 2009) may contribute to decreased macroautophagy. It is plausible that reduced hepatic macroautophagy during obesity may set up a vicious circle that furthers insulin resistance secondary to alterations in proteins crucial for insulin signal transduction.
A similar “autophagy-related vicious circle” may contribute to the metabolic syndrome and explain its higher prevalence with age. It is anticipated that the well documented decrease in macroautophagy activity in liver with age (Cuervo, 2008) will reduce the ability of this organ to mobilize intracellular lipids and contribute, at least in part, to altered lipid metabolism (adiposity and dyslipidemia) intrinsic to the metabolic syndrome. Chemical manipulation of macroautophagy presents as an attractive anti-aging therapy that could prevent or delay onset of the metabolic syndrome. In fact, although selective enhancers of macroautophagy are still missing, several multi-action drugs that efficiently increase life-span in vertebrates and health-span in rodents and primates, such as rapamycin, resveratrol or spermidine (reviewed in (Cuervo, 2008; Kaushik et al., 2010)), also exert a stimulatory effect on macroautophagy.
Inhibition of autophagy aimed at reducing cell-intrinsic energy supply may also have therapeutic applicability as a means to promote death in cancer cells. Although the interplay between autophagy and carcinogenesis is highly complex, blockage of macroautophagy has been shown to reduce tumorigenesis of cancer cells that upregulate this catabolic pathway to support the demanding energetic requirements of oncogenesis (readers interested in this topic are encouraged to consult recent reviews on this topic (Rabinowitz and White, 2010)).
Concluding remarks
The multiplicity of mechanisms by which autophagy impacts the cellular energetic balance reveal that this catabolic pathway is an important regulator of energy homeostasis. The efficiency of the system ultimately resides in the confining of a wide array of hydrolases in a single compartment with recycling capabilities, and the development of efficient mechanisms for selective delivery of cargo to this degradative compartment. There are many aspects of this selectivity that still remain elusive. What is the signal that triggers glycogen recognition by the autophagy machinery? What activates the autophagy machinery already associated to LD to initiate the formation of the sequestering membrane in macrolipophagy? Even the identification of the cargo-recognition molecules is still in its early days.
Now that it is accepted that autophagy can contribute to the mobilization of diverse energy stores, understanding the regulation of each of these processes, the feedback mechanisms from other catabolic pathways and the organ-intrinsic differences in these processes are a priority. Continuous cross-communication between organs such as the adipose tissue and the liver, as well as opposing metabolic responses to the same stimulus are well characterized, however the discovery of the role that differences in autophagy regulators or effectors play in this organ individuality is just beginning.
Although most efforts and experimental models have evolved around starvation as a stimulus for autophagy, many other nutritional challenges can affect this catabolic process, and probably its lipolytic, proteolytic and glycolytic capabilities are going to be affected in a different manner depending on the stimuli. As the number of human pathologies for which alterations in autophagy are being revealed keeps growing, it will become important to keep in mind that defective autophagy does not only associate with accumulation of damaged cellular structures but could also lead to severe energetic compromise in cells and organs. In this respect, restoration of normal autophagic activity presents now as an attractive therapeutic approach for different pathologies. Most chemical blockers and upregulators of autophagy used till date do not have the required specificity and often target major regulatory nodes that affect multiple intracellular pathways. This need for chemical modulators of autophagy is the driving force for more than a dozen of currently ongoing drug screenings that should offer a collection of autophagy-specific inhibitors and activators in the coming years.
Acknowledgments
We thank Dr. Susmita Kaushik and Ms. Samantha J. Orenstein for critically reading the manuscript. Work in our groups is supported by NIH grants AG021904, AG031782, DK041918, NS038370, a Hirsch/Weill-Caulier Career Scientist Award (AMC) and NIH grant DK087776 (RS).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aguado C, Sarkar S, Korolchuk VI, Criado O, Vernia S, Boya P, Sanz P, de Cordoba SR, Knecht E, Rubinsztein DC. Laforin, the most common protein mutated in Lafora disease, regulates autophagy. Hum Mol Genet. 2010;19:2867–2876. doi: 10.1093/hmg/ddq190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias E, Cuervo AM. Chaperone-mediated autophagy in protein quality control. Curr Opin Cell Biol, e-pub ahead of publication. 2010 doi: 10.1016/j.ceb.2010.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boland B, Kumar A, Lee S, Platt FM, Wegiel J, Yu WH, Nixon RA. Autophagy induction and autophagosome clearance in neurons: relationship to autophagic pathology in Alzheimer's disease. J Neurosci. 2008;28:6926–6937. doi: 10.1523/JNEUROSCI.0800-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan EY, Longatti A, McKnight NC, Tooze SA. Kinase-inactivated ULK proteins inhibit autophagy via their conserved C-terminal domains using an Atg13-independent mechanism. Mol Cell Biol. 2009;29:157–171. doi: 10.1128/MCB.01082-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crighton D, Wilkinson S, O’Prey J, Syed N, Smith P, Harrison PR, Gasco M, Garrone O, Crook T, Ryan KM. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell. 2006;126:121–134. doi: 10.1016/j.cell.2006.05.034. [DOI] [PubMed] [Google Scholar]
- Criollo A, Senovilla L, Authier H, Maiuri MC, Morselli E, Vitale I, Kepp O, Tasdemir E, Galluzzi L, Shen S, et al. The IKK complex contributes to the induction of autophagy. EMBO J. 2010;29:619–631. doi: 10.1038/emboj.2009.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuervo AM. Autophagy and aging: keeping that old broom working. Trends Genet. 2008;24:604–612. doi: 10.1016/j.tig.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuervo AM. The plasma membrane brings autophagosomes to life. Nature Cell Biology. 2010;12:735–737. doi: 10.1038/ncb0810-735. [DOI] [PubMed] [Google Scholar]
- Cuervo AM, Knecht E, Terlecky SR, Dice JF. Activation of a selective pathway of lysosomal proteolysis in rat liver by prolonged starvation. Am J Physiol. 1995;269:C1200–C1208. doi: 10.1152/ajpcell.1995.269.5.C1200. [DOI] [PubMed] [Google Scholar]
- Deter RL, Baudhuin P, De Duve C. Participation of lysosomes in cellular autophagy induced in rat liver by glucagon. J Cell Biol. 1967;35:11–16. doi: 10.1083/jcb.35.2.c11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebato C, Uchida T, Arakawa M, Komatsu M, Ueno T, Komiya K, Azuma K, Hirose T, Tanaka K, Kominami E, et al. Autophagy is important in islet homeostasis and compensatory increase of beta cell mass in response to high-fat diet. Cell Metab. 2008;8:325–332. doi: 10.1016/j.cmet.2008.08.009. [DOI] [PubMed] [Google Scholar]
- Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W, Vasquez DS, Joshi A, Gwinn DM, Taylor R, et al. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science. 2011;331:456–461. doi: 10.1126/science.1196371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn PF, Dice JF. Ketone bodies stimulate chaperone-mediated autophagy. J Biol Chem. 2005;280:25864–25870. doi: 10.1074/jbc.M502456200. [DOI] [PubMed] [Google Scholar]
- Fujitani Y, Kawamori R, Watada H. The role of autophagy in pancreatic beta-cell and diabetes. Autophagy. 2009;5:280–282. doi: 10.4161/auto.5.2.7656. [DOI] [PubMed] [Google Scholar]
- Fukuda T, Roberts A, Plotz PH, Raben N. Acid alpha-glucosidase deficiency (Pompe disease) Curr Neurol Neurosci Rep. 2007;7:71–77. doi: 10.1007/s11910-007-0024-4. [DOI] [PubMed] [Google Scholar]
- Grasso D, Sacchetti ML, Bruno L, Lo Re A, Iovanna JL, Gonzalez CD, Vaccaro MI. Autophagy and VMP1 expression are early cellular events in experimental diabetes. Pancreatology. 2009;9:81–88. doi: 10.1159/000178878. [DOI] [PubMed] [Google Scholar]
- Gwinn DM, Shackelford DB, Egan DF, Mihaylova MM, Mery A, Vasquez DS, Turk BE, Shaw RJ. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Molecular Cell. 2008;30:214–226. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- Hashimoto D, Ohmuraya M, Hirota M, Yamamoto A, Suyama K, Ida S, Okumura Y, Takahashi E, Kido H, Araki K, et al. Involvement of autophagy in trypsinogen activation within the pancreatic acinar cells. J Cell Biol. 2008;181:1065–1072. doi: 10.1083/jcb.200712156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa N, Hara T, Kaizuka T, Kishi C, Takamura A, Miura Y, Iemura S, Natsume T, Takehana K, Yamada N, et al. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell. 2009;20:1981–1991. doi: 10.1091/mbc.E08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung CH, Jun CB, Ro SH, Kim YM, Otto NM, Cao J, Kundu M, Kim DH. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell. 2009;20:1992–2003. doi: 10.1091/mbc.E08-12-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung HS, Chung KW, Won Kim J, Kim J, Komatsu M, Tanaka K, Nguyen YH, Kang TM, Yoon KH, Kim JW, et al. Loss of autophagy diminishes pancreatic beta cell mass and function with resultant hyperglycemia. Cell Metab. 2008;8:318–324. doi: 10.1016/j.cmet.2008.08.013. [DOI] [PubMed] [Google Scholar]
- Kadandale P, Kiger AA. Role of selective autophagy in cellular remodeling "Self-eating" into shape. Autophagy. 2010;6:1194–1195. doi: 10.4161/auto.6.8.13476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalamidas SA, Kotoulas OB. Glycogen autophagy in newborn rat hepatocytes. Histol Histopathol. 2000;15:1011–1018. doi: 10.14670/HH-15.1011. [DOI] [PubMed] [Google Scholar]
- Kamada Y, Funakoshi T, Shintani T, Nagano K, Ohsumi M, Ohsumi Y. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J Cell Biol. 2000;150:1507–1513. doi: 10.1083/jcb.150.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushik S, Massey AC, Cuervo AM. Lysosome membrane lipid microdomains: novel regulators of chaperone-mediated autophagy. EMBO J. 2006;25:3921–3933. doi: 10.1038/sj.emboj.7601283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushik S, Singh R, Cuervo AM. Autophagic pathways and metabolic stress. Diabetes Obes Metab. 2010;12:4–14. doi: 10.1111/j.1463-1326.2010.01263.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga H, Kaushik S, Cuervo AM. Altered lipid content inhibits autophagic vesicular fusion. FASEB J. 2010;24:3052–3065. doi: 10.1096/fj.09-144519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, Ueno T, Koike M, Uchiyama Y, Kominami E, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, Ezaki J, Mizushima N, Ohsumi Y, Uchiyama Y, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169:425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren I, Reem E, Kimchi A. DAP1, a novel substrate of mTOR, negatively regulates autophagy. Curr Biol. 2010;20:1093–1098. doi: 10.1016/j.cub.2010.04.041. [DOI] [PubMed] [Google Scholar]
- Kovsan J, Bluher M, Tarnovscki T, Kloting N, Kirshtein B, Madar L, Shai I, Golan R, Harman-Boehm I, Schon MR, et al. Altered autophagy in human adipose tissues in obesity. J Clin Endocrinol Metab. 2011;96:E268–E277. doi: 10.1210/jc.2010-1681. [DOI] [PubMed] [Google Scholar]
- Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- Lamark T, Kirkin V, Dikic I, Johansen T. NBR1 and p62 as cargo receptors for selective autophagy of ubiquitinated targets. Cell Cycle. 2009;8:1986–1990. doi: 10.4161/cc.8.13.8892. [DOI] [PubMed] [Google Scholar]
- Lee JH, Budanov AV, Park EJ, Birse R, Kim TE, Perkins GA, Ocorr K, Ellisman MH, Bodmer R, Bier E, et al. Sestrin as a feedback inhibitor of TOR that prevents age-related pathologies. Science. 2010a;327:1223–1228. doi: 10.1126/science.1182228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JW, Park S, Takahashi Y, Wang HG. The association of AMPK with ULK1 regulates autophagy. PloS one. 2010b;5:e15394. doi: 10.1371/journal.pone.0015394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lum JJ, Bauer DE, Kong M, Harris MH, Li C, Lindsten T, Thompson CB. Growth factor regulation of autophagy and cell survival in the absence of apoptosis. Cell. 2005;120:237–248. doi: 10.1016/j.cell.2004.11.046. [DOI] [PubMed] [Google Scholar]
- Marsh BJ, Soden C, Alarcon C, Wicksteed BL, Yaekura K, Costin AJ, Morgan GP, Rhodes CJ. Regulated autophagy controls hormone content in secretory-deficient pancreatic endocrine beta-cells. Mol Endocrinol. 2007;21:2255–2269. doi: 10.1210/me.2007-0077. [DOI] [PubMed] [Google Scholar]
- Marzella L, Ahlberg J, Glaumann H. Autophagy, heterophagy, microautophagy and crinophagy as the means for intracellular degradation. Virchows Archiv. B. Cell Pathology. 1981;36:219–234. doi: 10.1007/BF02912068. [DOI] [PubMed] [Google Scholar]
- Mehrpour M, Esclatine A, Beau I, Codogno P. Overview of macroautophagy regulation in mammalian cells. Cell Res. 2010;20:748–762. doi: 10.1038/cr.2010.82. [DOI] [PubMed] [Google Scholar]
- Mitchener JS, Shelburne JD, Bradford WD, Hawkins HK. Cellular autophagocytosis induced by deprivation of serum and amino acids in Hela cells. Am J Pathol. 1976;83:485–491. [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Noda T, Yoshimori T, Tanaka Y, Ishii T, George MD, Klionsky DJ, Ohsumi M, Ohsumi Y. A protein conjugation system essential for autophagy. Nature. 1998;395:395–398. doi: 10.1038/26506. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. 2004;15:1101–1111. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimore GE, Poso AR. Intracellular protein catabolism and its control during nutrient deprivation and supply. Annu Rev Nutr. 1987;7:539–564. doi: 10.1146/annurev.nu.07.070187.002543. [DOI] [PubMed] [Google Scholar]
- Newgard CB, An J, Bain JR, Muehlbauer MJ, Stevens RD, Lien LF, Haqq AM, Shah SH, Arlotto M, Slentz CA, et al. A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab. 2009;9:311–326. doi: 10.1016/j.cmet.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicklin P, Bergman P, Zhang B, Triantafellow E, Wang H, Nyfeler B, Yang H, Hild M, Kung C, Wilson C, et al. Bidirectional transport of amino acids regulates mTOR and autophagy. Cell. 2009;136:521–534. doi: 10.1016/j.cell.2008.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino I, Fu J, Tanji K, Yamada T, Shimojo S, Koori T, Mora M, Riggs J, Oh S, Koga Y, et al. Primary LAMP-2 deficiency causes X-linked vacuolar cardiomyopathy and myopathy (Danon disease) Nature. 2000;406:906–910. doi: 10.1038/35022604. [DOI] [PubMed] [Google Scholar]
- Onodera J, Ohsumi Y. Autophagy is required for maintenance of amino acid levels and protein synthesis under nitrogen starvation. J Biol Chem. 2005;280:31582–31586. doi: 10.1074/jbc.M506736200. [DOI] [PubMed] [Google Scholar]
- Raben N, Schreiner C, Baum R, Takikita S, Xu S, Xie T, Myerowitz R, Komatsu M, Van der Meulen JH, Nagaraju K, et al. Suppression of autophagy permits successful enzyme replacement therapy in a lysosomal storage disorder--murine Pompe disease. Autophagy. 2010;6:1078–1089. doi: 10.4161/auto.6.8.13378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinowitz JD, White E. Autophagy and metabolism. Science. 2010;330:1344–1348. doi: 10.1126/science.1193497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravikumar B, Stewart A, Kita H, Kato K, Duden R, Rubinsztein DC. Raised intracellular glucose concentrations reduce aggregation and cell death caused by mutant huntingtin exon 1 by decreasing mTOR phosphorylation and inducing autophagy. Hum Mol Genet. 2003;12:985–994. doi: 10.1093/hmg/ddg109. [DOI] [PubMed] [Google Scholar]
- Sahu R, Kaushik S, Cannizzo E, Scharf B, Follenzi A, Clement C, Potolicchio I, Nieves E, Cuervo A, Santambrogio L. Microautohagy of cytosolic proteins by late endosomes. Dev Cell. 2011;20:131–139. doi: 10.1016/j.devcel.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141:290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S, Davies JE, Huang Z, Tunnacliffe A, Rubinsztein DC. Trehalose, a novel mTOR-independent autophagy enhancer, accelerates the clearance of mutant huntingtin and alpha-synuclein. J Biol Chem. 2007;282:5641–5652. doi: 10.1074/jbc.M609532200. [DOI] [PubMed] [Google Scholar]
- Sarkar S, Ravikumar B, Floto RA, Rubinsztein DC. Rapamycin and mTOR-independent autophagy inducers ameliorate toxicity of polyglutamine-expanded huntingtin and related proteinopathies. Cell Death Differ. 2009;16:46–56. doi: 10.1038/cdd.2008.110. [DOI] [PubMed] [Google Scholar]
- Schellens JP, Vreeling-Sindelarova H, Plomp PJ, Meijer AJ. Hepatic autophagy and intracellular ATP. A morphometric study. Exp Cell Res. 1988;177:103–108. doi: 10.1016/0014-4827(88)90028-6. [DOI] [PubMed] [Google Scholar]
- Schworer CM, Cox JR, Mortimore GE. Alteration of lysosomal density by sequestered glycogen during deprivation-induced autophagy in rat liver. Biochem Biophys Res Commun. 1979;87:163–170. doi: 10.1016/0006-291x(79)91661-9. [DOI] [PubMed] [Google Scholar]
- Shang L, Chen S, Du F, Li S, Zhao L, Wang X. Nutrient starvation elicits an acute autophagic response mediated by Ulk1 dephosphorylation and its subsequent dissociation from AMPK. Proc Natl Acad Sci U S A. 2011;108:4788–4793. doi: 10.1073/pnas.1100844108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata M, Yoshimura K, Furuya N, Koike M, Ueno T, Komatsu M, Arai H, Tanaka K, Kominami E, Uchiyama Y. The MAP1-LC3 conjugation system is involved in lipid droplet formation. Biochem Biophys Res Commun. 2009;382:419–423. doi: 10.1016/j.bbrc.2009.03.039. [DOI] [PubMed] [Google Scholar]
- Shintani T, Yamazaki F, Katoh T, Umekawa M, Matahira Y, Hori S, Kakizuka A, Totani K, Yamamoto K, Ashida H. Glucosamine induces autophagy via an mTOR-independent pathway. Biochem Biophys Res Commun. 2010;391:1775–1779. doi: 10.1016/j.bbrc.2009.12.154. [DOI] [PubMed] [Google Scholar]
- Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, Tanaka K, Cuervo AM, Czaja MJ. Autophagy regulates lipid metabolism. Nature. 2009a;458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Xiang Y, Wang Y, Baikati K, Cuervo AM, Luu YK, Tang Y, Pessin JE, Schwartz GJ, Czaja MJ. Autophagy regulates adipose mass and differentiation in mice. J Clin Invest. 2009b;119:3329–3339. doi: 10.1172/JCI39228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasdemir E, Maiuri MC, Galluzzi L, Vitale I, Djavaheri-Mergny M, D'Amelio M, Criollo A, Morselli E, Zhu C, Harper F, et al. Regulation of autophagy by cytoplasmic p53. Nat Cell Biol. 2008;10:676–687. doi: 10.1038/ncb1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vabulas RM, Hartl FU. Protein synthesis upon acute nutrient restriction relies on proteasome function. Science. 2005;310:1960–1963. doi: 10.1126/science.1121925. [DOI] [PubMed] [Google Scholar]
- Van Dyke RW. Proton pump-generated electrochemical gradients in rat liver multivesicular bodies. Quantitation and effects of chloride. J Biol Chem. 1988;263:2603–2611. [PubMed] [Google Scholar]
- Vergne I, Roberts E, Elmaoued RA, Tosch V, Delgado MA, Proikas-Cezanne T, Laporte J, Deretic V. Control of autophagy initiation by phosphoinositide 3-phosphatase Jumpy. EMBO J. 2009;28:2244–2258. doi: 10.1038/emboj.2009.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vousden KH, Ryan KM. p53 and metabolism. Nat Rev Cancer. 2009;9:691–700. doi: 10.1038/nrc2715. [DOI] [PubMed] [Google Scholar]
- Wang J, Whiteman MW, Lian H, Wang G, Singh A, Huang D, Denmark T. A non-canonical MEK/ERK signaling pathway regulates autophagy via regulating Beclin 1. J Biol Chem. 2009;284:21412–21424. doi: 10.1074/jbc.M109.026013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webber JL, Tooze SA. Coordinated regulation of autophagy by p38alpha MAPK through mAtg9 and p38IP. EMBO J. 2010;29:27–40. doi: 10.1038/emboj.2009.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Pattingre S, Sinha S, Bassik M, Levine B. JNK1-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagy. Mol Cell. 2008;30:678–688. doi: 10.1016/j.molcel.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Li P, Fu S, Calay ES, Hotamisligil GS. Defective hepatic autophagy in obesity promotes ER stress and causes insulin resistance. Cell Metab. 2010;11:467–478. doi: 10.1016/j.cmet.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Goldman S, Baerga R, Zhao Y, Komatsu M, Jin S. Adipose-specific deletion of autophagy-related gene 7 (atg7) in mice reveals a role in adipogenesis. Proc Natl Acad Sci U S A. 2009;106:19860–19865. doi: 10.1073/pnas.0906048106. [DOI] [PMC free article] [PubMed] [Google Scholar]