The use of microorganisms to ferry cargo, previously described as microoxen by Weibel et al.,[1] provides an attractive route to propel and guide microstructures. In principle, a multitude of microorganisms could be utilized to enable motility and guidance in response to specific stimuli. Amongst these microorganisms, bacteria are small (micron sized) and especially relevant for ferrying micro and nanoscale cargo. In addition, bacteria are motile, capable of exhibiting different types of motion such as run-and-tumble, swarming, gliding and twitching.[2–5] These modes play an important role in guiding the response of bacteria to external stimuli - in phenomena such as chemotaxis, phototaxis and magnetotaxis.[6–8] There has been considerable interest in harnessing the motion of bacteria in the creation of biological motors or bioactuators on account of their small sizes, ability to respond to environmental cues, ability to convert chemical energy to motion (obviating need for external power supply) and higher swimming speed as compared to several microscopic artificially engineered devices.[1, 9] These advantages have recently been realized and there has been growing interest in utilizing the motion of intact microorganisms in propulsion.[1, 7, 10–13] However, previous use of bacterial propulsion has focused mainly on the utilization of large numbers of bacteria to propel 10–100 μm scale cargo. It is noteworthy, that in these studies, conjugation of bacteria to cargo was effected by blotting.[14]
We focus on the use of Escherichia coli (E. coli) bacteria to ferry smaller sub-micron scale cargo. These bacteria are well studied,[15, 16] are able to grow in vivo and have small sizes to enable passage in hard-to-reach spaces. As compared to previous methods for attachment of cargo to bacteria (primarily blotting based), we conjugate the bacteria to the cargo at a patterned silicon substrate using the specificity of antibody based interactions. This approach is motivated by the fact that many micro and nanostructures are fabricated using lithographic approaches that facilitate their creation on silicon substrates.[17, 18] Moreover, at the sub-micron scale it is often necessary to fabricate the structures using electron beam (e-beam) lithography which is a serial process yielding only small numbers of structures.[19, 20] If these structures are released from the surface prior to attachment to bacteria, as is required in solution based attachment schemes, the small concentrations of fabricated structures relative to the bacteria make the probability of attachment and subsequent tracking challenging. In principle, it may also be easier to geometrically orient the cargo relative to the bacterium at a surface as compared to in solution. Additionally, surface attachment provides control with respect to when and where the cargo-carrying bacteria could be released (on-demand release).
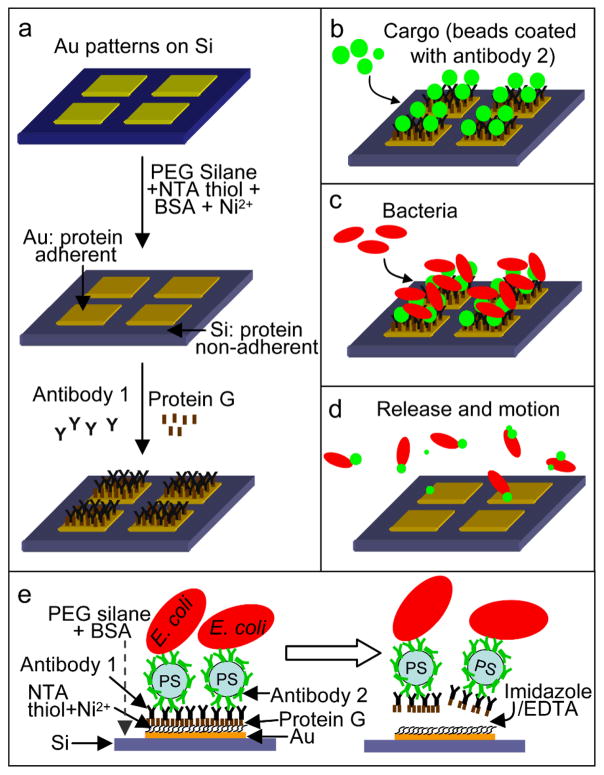
We developed a two antibody based method to attach and release motile cargo-carrying bacteria from patterned surfaces. Antibody based attachment facilitates selectivity for a particular bacterial species as well as specific locations (receptors) on the bacterium. Our surface attachment methodology enables surfaces to be coated with bacteria-cargo conjugates and these conjugates can be released on-demand and in response to specific chemical triggers. Figure 1 describes the schematic of our approach.
Figure 1.
Schematic of the selective attachment and release of cargo-carrying bacteria from patterned surfaces. (a) Au patterned Si substrate. The substrate is treated so that Au become his-tag protein adherent (via thiolation with NTA thiol and treatment with Ni2+ ions) while Si is protein non-adherent (via PEG silanization and blocking with BSA). The Au surface is further functionalized with protein G and antibody 1 (a cargo specific antibody; goat anti-rabbit IgG). (b) Cargo coated with antibody 2 (rabbit anti-E. coli IgG) are then introduced causing them to bind to the antibodies on the Au patterns (c) Bacteria (E. coli) then specifically attach to the beads and (d) motile bacteria-cargo conjugates can be released on-demand using either imidazole or EDTA. (e) Schematic showing bound and released cargo-carrying bacteria.
First, we fabricated silicon (Si) substrates with gold (Au) patterns (an array of 80 μm × 80 μm Au square patterns spaced 40 μm apart) via photolithography and lift-off metallization[21] (see Supporting Information (SI) for details). The Si substrates were silanized with polyethylene glycol (PEG) silane to make them protein non-adherent.[22] Next, the Au surfaces were functionalized with nitrilotriacetic acid (NTA) thiol to enable subsequent binding of polyhistidine tagged (his-tag) proteins via Ni2+ ion chelation.[23] To further prevent non-specific protein adsorption on Si, the substrates were soaked overnight in bovine serum albumin (BSA). The substrates were then treated with nickel ions (Ni2+) by immersion in an aqueous nickel sulfate solution to allow the NTA end group on the thiolated Au patterns to chelate with Ni2+ ions. This step was followed by dipping the substrates in an aqueous his-tag protein G solution. The his-tag protein G selectively binds to the Ni2+-NTA complex, and only on the Au patterns (Figure 1a). We utilized 0.4 – 0.6 μm sized commercial polystyrene (PS) beads as the cargo. The size of cargo was selected to enable predominant attachment of a single bacterium to a single bead, while enabling tracking of bacteria-bead motion using optical microscopy. In principle, smaller cargo could be attached, but it would be challenging to clearly discern the cargo during motion. To sequentially attach the cargo and the bacteria to the surface, we utilized two antibodies with complementary binding sites. Antibody 1 was goat anti-rabbit IgG which enabled attachment of antibody 2 coated cargo to the protein G coated Au patterns [24, 25] (Figure 1b). The beads were coated with rabbit anti-E. coli (antibody 2) by adsorption prior to use. The Au patterns with attached antibody 2 coated beads were then utilized for E. coli attachment (via antibody 2; Figure 1c). Bacteria-bead conjugates were released from the Au patterns (Figure 1d), on-demand, using either imidazole or ethylenediaminetetraacetic acid (EDTA) which are well known for their strong affinity to metal ions. These chemicals preferentially bind (imidazole) or chelate (EDTA) Ni2+ as compared to the his-tagged protein G. This competitive binding advantage releases bacteria-bead conjugates from the substrate. This two-antibody based surface attachment methodology is versatile in that it allows both the type of cargo and the species of bacteria to be varied. Additionally, the attachment and release does not disable motility in bacteria and is chemically benign in that it is compatible with a range of cargo including those composed of metals, semiconductors, and polymers.
The specificity of attachment of bacteria-bead conjugates was investigated by utilizing fluorescently labeled PS beads and bacteria (Figure 2). The beads were coated by adsorption with fluorescein isothiocyanate (FITC) labeled antibody 2 (rabbit anti-E. coli) so that they appeared green on fluorescence excitation. The bacteria were labeled with the nucleic acid stain Syto 61 so that they appeared red on fluorescence excitation. Fluorescent beads were first added to the substrate (0.1 mg/mL). After sufficient contact, the substrates were rinsed to remove unbound beads and a suspension of E. coli was added to the substrates (OD600 ~ 1). The substrates were then rinsed to remove unbound bacteria and observed under an upright, fluorescence microscope. Beads (Figure 2a) and bacteria (Figure 2b) specifically attached to the Au patterns following the functionalization scheme outlined above. From the fluorescence line intensity profiles it was evident that bead and bacteria binding were relatively uniform on the Au patterns and that non-specific adsorption on the Si substrate was minimal (Figures 2c–d). We performed several control experiments that suggest that if any of the molecular functionalization steps was skipped, beads and bacteria did not bind specifically to the Au patterns (SI for details). In addition to fluorescence microscopy, samples were visualized using scanning electron microscopy (SEM). For SEM, the samples were fixed with formaldehyde and glutaraldehyde, post-fixed with osmium tetroxide, dehydrated by placing in solutions of ethanol of increasing ethanol concentrations, transferred to solutions containing hexamethyldisilazane in increasing concentrations, air dried by leaving uncovered in the hood and sputter-coated with platinum (See SI for details).[26] SEM images confirmed the selective attachment of beads and bacteria on the Au patterns (Figure 2e) and to each other (Figure 2f).
Figure 2.
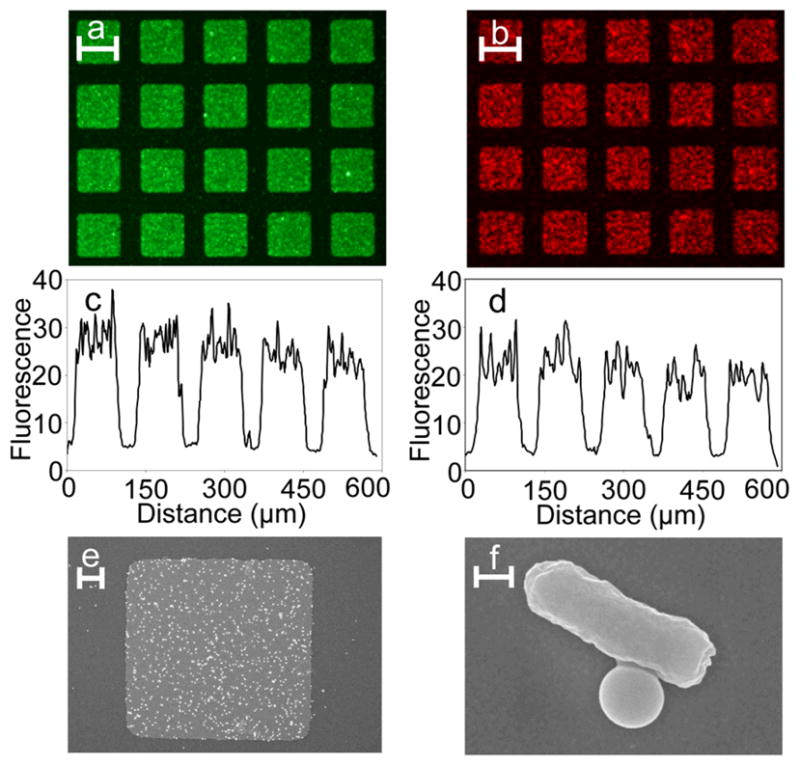
Verification of the specificity of attachment. (a–b) Fluorescence images and (c–d) line intensity plots of the selective attachment of beads (green; a and c) and E. coli bacteria (red; b and d) to Au patterns on Si substrates. (e) SEM image of a single square Au pattern with attached bacteria-bead conjugates and (f) zoom-in of a single bacteria-bead conjugate. Scale bars in panels a, b are 80 μm, panel e is 10 μm and panel f is 0.5 μm long.
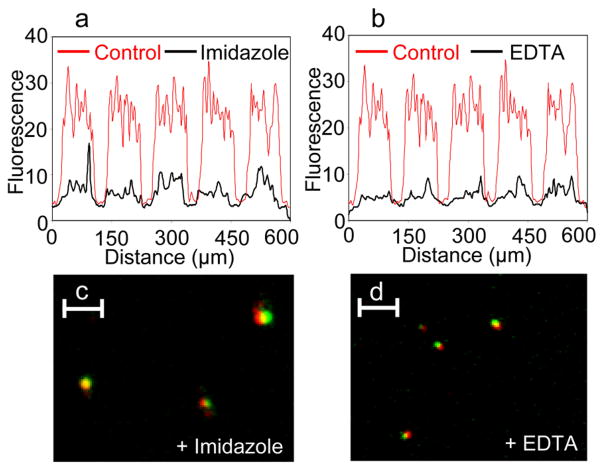
The release of the bacteria-bead conjugates was monitored by measuring fluorescence on the Au patterns after treatment with either imidazole (300 mM imidazole in motility buffer) or EDTA (50 mM EDTA in motility buffer) for 30 minutes at room temperature using an upright, fluorescence microscope. The motility buffer consisted of 10 mM PO43−, 0.1 mM EDTA, 1 μM methionine and 10 mM lactic acid (pH 7.3).[27] Additionally, the released bacteria-cargo conjugates in the supernatant solution were fixed with formaldehyde and glutaraldehyde and visualized using a dual-fluorescence microscope (SI for details). Figures 3a and b shows fluorescence intensity line scans over Au patterns after treatment with imidazole and EDTA respectively. The control, treated with PBS, is included in red as a reference. A significant drop in fluorescence intensity was observed after treatment with imidazole or EDTA as compared to treatment with PBS indicating relatively high release of bacteria-bead conjugates on-demand. Using dual-fluorescence microscopy, we were also able to visualize bacteria-bead conjugates in the supernatants released via imidazole (Figure 3c) or EDTA (Figure 3d). The co-localization of the red fluorescence of the bacteria with the green fluorescence of the bead indicates release of the intact bacteria-bead conjugates. The observed relative size difference of bacteria and beads between Figures 3c and 3d can be attributed to the fact that these conjugates were at different focal planes during imaging.
Figure 3.
Release of bacteria-bead (E. coli-bead) conjugates from the Au patterned substrates. (a–b) Fluorescence line intensity plots across Au patterned substrates treated with (a) imidiazole or (b) EDTA (shown in black) as compared to control substrates treated with PBS (shown in red in panels a and b). Also shown are released bacteria-bead conjugates observed in the supernatant solution after release via (c) imidazole or (d) EDTA. These conjugates appear as co-localized red (E. coli)-green (bead) spots. Scale bars in panels c and d are 5 μm long.
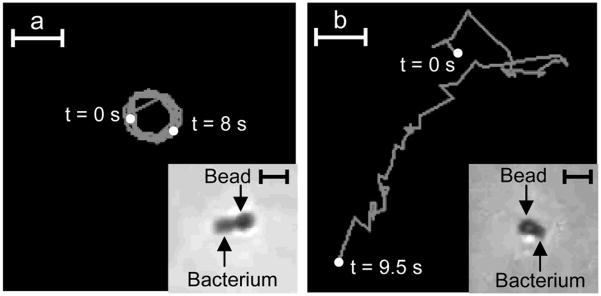
We tracked the motion of individual bacteria-cargo conjugates in solution after release from the surface by either imidazole or EDTA (Figure 4). Both imidazole and EDTA were made in the motility buffer as described above. The supernatants after release, containing the bacteria-cargo conjugates were imaged between two glass coverslips separated by a double sticky tape spacer 120 μm in thickness using an inverted microscope, 40 × DIC objective and video imaging (see SI for details). After release, we occasionally observed that the conjugates would get stuck to the coverslip, likely due to van der Waal forces. In those instances, bacteria moved in circular trajectories (Figure 4a); this observation is similar to previous studies where bacteria were tethered to surfaces via their flagella.[28, 29] The mean speed of rotation of the conjugate in Figure 4a was about 10 μm/s. In contrast, the ‘substrate-free’ bacteria-bead conjugates were observed to move in trajectories as shown in Figure 4b with mean speeds of about 18 μm/s (see SI for details). These trajectories and mean speeds are consistent with those observed for typical bacterial motility and motion in the literature. Additionally, these trajectories are distinctively different from Brownian motion based bead trajectories.[16, 30]
Figure 4.
Tracking the motion of individual bacteria-bead conjugates (E. coli propelling sub-micron beads) upon release from the substrate. (a) Circular trajectory traced by a released bacteria-bead conjugate (inset) when the conjugate sticks to the surface of the coverslip after release (imaged for 8 s, mean speed was about 10 μm/s). (b) Trajectory traced by a bacteria-bead conjugate (inset) after release from the surface (imaged for 9.5 s, mean speed was about 18 μm/s). Scale bars in panels a and b are 5 μm long, inset scale bars in panels a and b are 2 μm long.
In conclusion, we have demonstrated a methodology for selective attachment and on-demand release of motile cargo-carrying bacteria from patterned substrates; the strategy can be extended to e-beam fabricated nanostructures with appropriate sacrificial layers for release. Our methodology involves integrating fabrication, self-assembly, antibody-based attachment and chemical release. We note that in the current study, we utilized a polyclonal antibody (antibody 2) for attachment of the bead to the bacterium. We believe that the specificity as well as efficiency of bead attachment to the bacteria can be improved by use of monoclonal antibodies to a specific receptor on the bacterial surface. Additionally, since E. coli are peritrichous, attachment yields would possibly be enhanced by de-flagellating them or by utilizing flagella specific antibodies.[31, 32] We anticipate use of this strategy in the development of micro and nanoscale autonomous devices, miniaturized medicine and lab-on-a-chip applications. For example, it is conceivable that micro and nanofabricated tools and devices for diagnostics, therapeutics and surgery can be attached to motile bacteria using this approach.[33] Additionally, genetic engineering tools can also be used to synthesize bacteria with localized receptors for directed conjugation of cargo as well as bacteria that respond and hence ferry cargo in reponse to different chemical stimuli.
Experimental Section
Fabrication and patterning of the substrates
An array of 80 μm × 80 μm Au squares spaced 40 μm apart was prepared on a Si substrate using photolithography and lift-off metallization (see SI for details).[21] The substrates were treated to make Au his-tag protein adherent and Si protein non-adherent. To achieve this, the substrates were cleaned with hot piranha and dipped in polyethylene glycol silane (PEG Silane; Gelest Inc.) to silanize the Si. Subsequently, the Au patterns on the substrate were thiolated with nitrilotriacetic acid thiol (NTA thiol, Prochimia).[23] To further block non-specific protein adsorption, the substrates were dipped in bovine serum albumin for 16 h. The substrates were then treated with nickel sulfate (to bind Ni2+ to NTA), his-tagged protein G (the his-tag binds to Ni2+-NTA; Abcam) and antibody 1 which binds to protein G via its Fc region (cargo capturing antibody anti-rabbit IgG; AbD Serotec). [24, 25]
Attachment of beads and bacteria on the Au patterns on the substrate
The cargo used in this study comprised of polystyrene (PS) beads (diameter 0.4 – 0.6 μm; Spherotech; SI for details) coated with FITC labeled bacteria capturing antibody (rabbit anti-E.coli IgG, AbD Serotec). The beads were spatially attached by immersing the substrates in a solution containing the beads. After rinsing the unbound beads, the substrates were imaged using a Nikon AZ100 multizoom microscope (B-2E/C filter set; Nikon Microscopy). Fluorescence intensity line plots were generated using ImageJ (NIH). The bacterial strain used in this study is Escherichia coli W3110 (ATCC; # 39936). The bacteria were attached to the beads by adding the substrates to a bacterial suspension. After rinsing off the unbound bacteria, the bacteria on the substrates were stained using Syto 61 (red fluorescent nucleic acid stain; Invitrogen) and imaged using the Nikon AZ100 multizoom microscope (G-2E/C filter set; Nikon Microscopy). Fluorescence intensity line plots were generated using ImageJ.
Chemical release of bacteria–bead conjugates from the gold patterns
The bacteria–bead conjugates were released from the surface of the substrates by immesing in either imidazole or EDTA (SI for details). The substrates were imaged using the Nikon AZ100 multizoom microscope (G-2E/C filter set) and fluorescence intensity profiles were generated using ImageJ. The released bacteria–bead conjugates were fixed and imaged using a 3i Marianas microscope (Intelligent Imaging Innovations Inc.) under a GFP/RFP filter set to simultaneously image the red and green fluorescences of the bacteria and beads respectively.
Tracking motion of individual bacteria-bead conjugates
Individual bacteria-bead conjugates were imaged using a Nikon Eclipse TS100 inverted microscope under a 40 × DIC objective with video imaging (using Nikon’s Q Capture Pro software). To track the trajectories of the bacteria-bead conjugates, the video was converted to an image series (TIF series) using ImageJ. The trajectories were plotted and parameters such as the mean speed were calculated using ImageJ’s manual tracking plugin.
Supplementary Material
Footnotes
This work was supported by the NIH Director’s New Innovator Award Program, part of the NIH Roadmap for Medical Research, through grant number 1-DP2-OD004346-01. Information about the NIH Roadmap can be found at http://nihroadmap.nih.gov. The authors acknowledge support from the Camille-Dreyfus Foundation and access to microscopy facilities provided by the Integrated Imaging Center at Johns Hopkins University.
Supporting Information is available on the WWW under http://www.small-journal.com or from the author.
Contributor Information
Dr. Rohan Fernandes, Department of Chemical and Biomolecular Engineering, Johns Hopkins University, 3400 N Charles Street, Baltimore, MD 21218, USA
Mary Zuniga, Department of Chemical and Biomolecular Engineering, Johns Hopkins University, 3400 N Charles Street, Baltimore, MD 21218, USA.
Fritz R. Sassine, Department of Chemical and Biomolecular Engineering, Johns Hopkins University, 3400 N Charles Street, Baltimore, MD 21218, USA
Mert Karakoy, Department of Chemical and Biomolecular Engineering, Johns Hopkins University, 3400 N Charles Street, Baltimore, MD 21218, USA.
Prof. Dr. David H. Gracias, Department of Chemical and Biomolecular Engineering and Department of Chemistry, Johns Hopkins University, 3400 N Charles Street, Baltimore, MD 21218, USA.
References
- 1.Weibel DB, Garstecki P, Ryan D, DiLuzio WR, Mayer M, Seto JE, Whitesides GM. Proc Natl Acad Sci USA. 2005;102:11963. doi: 10.1073/pnas.0505481102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sokolov A, Apodaca MM, Grzybowski BA, Aranson IS. Proc Natl Acad Sci USA. 2010;107:969. doi: 10.1073/pnas.0913015107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harshey RM. Mol Microbiol. 1994;13:389. doi: 10.1111/j.1365-2958.1994.tb00433.x. [DOI] [PubMed] [Google Scholar]
- 4.Lapidus IR, Berg HC. J Bacteriol. 1982;151:384. doi: 10.1128/jb.151.1.384-398.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Skerker JM, Berg HC. Proc Natl Acad Sci USA. 2001;98:6901. doi: 10.1073/pnas.121171698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lux R, Shi W. Crit Rev Oral Biol Med. 2004;15:207. doi: 10.1177/154411130401500404. [DOI] [PubMed] [Google Scholar]
- 7.Martel S, Tremblay CC, Ngakeng S, Langlois L. Appl Phys Lett. 2006;89:233904. [Google Scholar]
- 8.Ragatz L, Jiang ZY, Bauer CE, Gest H. Arch Microbiol. 1995;163:1. doi: 10.1007/BF00262196. [DOI] [PubMed] [Google Scholar]
- 9.Sitti M. Nature. 2009;458:1121. doi: 10.1038/4581121a. [DOI] [PubMed] [Google Scholar]
- 10.Behkam B, Sitti M. Conf Proc IEEE Eng Med Biol Soc. 2006;1:2421. doi: 10.1109/IEMBS.2006.259841. [DOI] [PubMed] [Google Scholar]
- 11.Park SJ, Bae H, Kim J, Lim B, Park J, Park S. Lab Chip. 2010;10:1706. doi: 10.1039/c000463d. [DOI] [PubMed] [Google Scholar]
- 12.Steager E, Kim CB, Patel J, Bith S, Naik C, Reber L, Kim MJ. Appl Phys Lett. 2007;90:263901. [Google Scholar]
- 13.Steager EB, Patel JA, Kim CB, Yi DK, Lee W, Kim MJ. Microfluid Nanofluid. 2007:5. [Google Scholar]
- 14.Darnton N, Turner L, Breuer K, Berg HC. Biophys J. 2004;86:1863. doi: 10.1016/S0006-3495(04)74253-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Adler J, Templeton B. J Gen Microbiol. 1967;46:175. doi: 10.1099/00221287-46-2-175. [DOI] [PubMed] [Google Scholar]
- 16.Berg HC, Brown DA. Nature. 1972;239:500. doi: 10.1038/239500a0. [DOI] [PubMed] [Google Scholar]
- 17.Leong TG, Randall CL, Benson BR, Bassik N, Stern GM, Gracias DH. Proc Natl Acad Sci USA. 2009;106:703. doi: 10.1073/pnas.0807698106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cho JH, Azam A, Gracias DH. Langmuir. 2010;26:16534. doi: 10.1021/la1013889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cho JH, Gracias DH. Nano Lett. 2009;9:4049. doi: 10.1021/nl9022176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cho JH, James T, Gracias DH. Proc SPIE. 2010;7767:776704. [Google Scholar]
- 21.Veiseh M, Zhang Y, Hinkley K, Zhang M. Biomed Microdev. 2001;3:45. [Google Scholar]
- 22.Veiseh M, Zhang M. J Am Chem Soc. 2006;128:1197. doi: 10.1021/ja055473q. [DOI] [PubMed] [Google Scholar]
- 23.Sigal GB, Bamdad C, Barberis A, Strominger J, Whitesides GM. Anal Chem. 1996;68:490. doi: 10.1021/ac9504023. [DOI] [PubMed] [Google Scholar]
- 24.Bjorck L, Kronvall G. J Immunol. 1984;133:969. [PubMed] [Google Scholar]
- 25.Sjobring U, Bjorck L, Kastern W. J Biol Chem. 1991;266:399. [PubMed] [Google Scholar]
- 26.Perkins EM, McCaffery JM. Methods Mol Biol. 2007;372:467. doi: 10.1007/978-1-59745-365-3_33. [DOI] [PubMed] [Google Scholar]
- 27.Kalinin Y, Neumann S, Sourjik V, Wu M. J Bacteriol. 2010;192:1796. doi: 10.1128/JB.01507-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Manson MD, Tedesco PM, Berg HC. J Mol Biol. 1980;138:541. doi: 10.1016/s0022-2836(80)80017-9. [DOI] [PubMed] [Google Scholar]
- 29.Silverman M, Simon M. Nature. 1974;249:73. doi: 10.1038/249073a0. [DOI] [PubMed] [Google Scholar]
- 30.Behkam B, Sitti M. Appl Phys Lett. 2008;93:223901. [Google Scholar]
- 31.Stocker BAD, Campbell JC. J Gen Microbiol. 1959;20:670. doi: 10.1099/00221287-20-3-670. [DOI] [PubMed] [Google Scholar]
- 32.Novotny C, Carnahan J, Brinton CC., Jr J Bacteriol. 1969;98:1294. doi: 10.1128/jb.98.3.1294-1306.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fernandes R, Gracias DH. Mater Today. 2009;12:14. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.