Abstract
AIMS
To test prospectively the impact of VEGF-A gene polymorphisms on the pharmacodynamics of bevacizumab-chemotherapy in breast cancer patients.
METHODS
As part of the single-arm MO19391 trial, 137 women with locally recurrent or metastatic breast cancer receiving first-line bevacizumab-containing therapy were analysed. Patients received bevacizumab associated (76%) or not (24%) with taxane-based chemotherapy. Clinical evaluation included clinical response, time to progression (TTP) and a toxicity score corresponding to the sum of each maximum observed toxicity grade (hypertension, haemorrhage, arterial and venous thrombo-embolism). Functional VEGF-A polymorphisms at position −2578 C > A, −1498 T > C, −1154 G > A, −634 G > C and 936 C > T were analysed by PCR-RFLP (blood DNA).
RESULTS
Overall response rate (complete response (CR) + partial response (PR)) was 61%. Median TTP was 11 months. None of the VEGF-A polymorphisms was significantly linked to clinical response. Analysis of the 936C > T polymorphism revealed that the 96 patients homozygous for the 936C allele exhibited a marked tendency for a shorter TTP (median 9.7 months) than the 32 patients bearing the 936T allele (median 11.5 months, P = 0.022) of which 30 were CT and two were homozygous TT. Other polymorphisms did not influence TTP. VEGF-A−634 G > C was significantly related to the toxicity score with 39%, 49% and 81% of patients with score >1 in GG, GC and CC patients, respectively (P = 0.01).
CONCLUSIONS
The role for VEGF-A 936C > T polymorphism as a potential marker of TTP in breast cancer patients receiving bevacizumab-containing therapy concords with the known impact of VEGF-A 936C > T polymorphism on VEGF-A expression.
Keywords: bevacizumab, breast cancer, gene polymorphisms, pharmacogenetics, VEGF-A
WHAT IS ALREADY KNOWN ABOUT THIS SUBJECT
Functional polymorphisms on the VEGF-A gene, known to be linked to cancer risk or to VEGF-A plasma concentrations, have been identified. So far, limited knowledge has been published on the relationships between toxicity/efficacy of bevacizumab-based therapy and VEGF-A polymorphisms (tumoral DNA). We therefore prospectively tested the impact of these five gene polymorphisms (blood DNA) on the pharmacodynamics of bevacizumab-based treatment administered in metastatic breast cancer patients.
WHAT THIS STUDY ADDS
Present data obtained from a prospective study suggest a role for VEGF-A 936C > T polymorphism as a potential predictor of time to progression in breast cancer patients receiving bevacizumab-containing therapy. Also, the VEGF-A−634G > C polymorphism was linked to bevacizumab-related toxicity.
Introduction
Vascular endothelial growth factor A (VEGF-A) is a key angiogenic factor which promotes endothelial cell growth and tumour neovascularization. VEGF-A expression is a marker of invasiveness and tumour progression in various cancers, including breast cancer [1, 2]. Bevacizumab is a recombinant humanized monoclonal antibody that binds to VEGF-A and blocks VEGF binding to its receptors [3]. In a randomized phase III trial, the addition of bevacizumab to paclitaxel, administered as first-line treatment for metastatic breast cancer, significantly improved response rate and progression-free survival, along with minimal impact on toxicity [4].
In breast cancer, as well as in other cancer localizations, most studies have failed to identify biomarkers for predicting efficacy and/or toxicity of angiogenic agents, including bevacizumab [5–7]. One possible easy-to-perform approach for predicting inter-patient variability of drug effects is to study germinal gene polymorphisms potentially linked to drug pharmacodynamics and/or pharmacokinetics. Among genes potentially linked to inter-patient variability of bevacizumab pharmacodynamics is the VEGF-A gene which is highly polymorphic, with multiple common single nucleotide polymorphisms (SNPs) in the promoter, 5'untranslated and 3' untranslated regions [8]. Among them, five functional polymorphisms at positions −2578 C > A, −1498 T > C, −1154 G > A, −634 G > C and 936 C > T (Figure 1) have been associated with serum VEGF-A [9, 10] or cancer risk [11–13], including breast cancer risk or aggressiveness [10, 14, 15]. In a recent study conducted in patients receiving or not bevacizumab, it has been suggested that four out of these five polymorphisms may influence bevacizumab pharmacodynamics [16].
Figure 1.
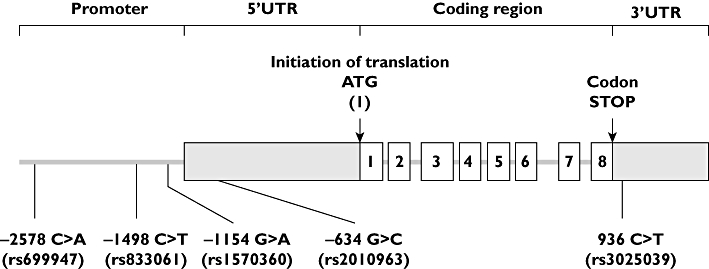
VEGF-A gene and analysed polymorphisms. UTR untranslated region
The aim of this prospective study was to test the impact of these five major functional VEGF-A gene polymorphisms on the efficacy of bevacizumab chemotherapy and on bevacizumab-related toxicity, in breast cancer patients treated as part of the large observational international MO 19391 (ATHENA) trial.
Methods
Patients and treatment
The single-arm, multinational, safety study MO19391 (ATHENA) EUDRACT # 2006-002529-21 was conducted in the context of general oncology practice in 2251 patients receiving bevacizumab in combination with taxane-based chemotherapy, for HER2 negative, locally recurrent or metastatic breast cancer. The study started in September 2006 and ended in August 2009. A companion study was conducted in 27 French centres to evaluate VEGF-A polymorphisms as potential predictors of treatment efficacy and safety. A total of 137 women were enrolled in this pharmacogenetic study, carried out with ethics committee approval. All patients gave informed consent. Patients received bevacizumab (10 mg kg−1 every 2 weeks or 15 mg kg−1 every 3 weeks), combined or not with a taxane-based standard chemotherapy, until disease progression, unacceptable toxicity or withdrawal. Adverse events related to bevacizumab were assessed according to the NCI CTCAE v3.0 criteria. Clinical response was assessed according to modified RECIST v1.0 criteria.
Pharmacogenetic analyses
VEGF-A gene polymorphisms were analysed by PCR-RFLP on DNA extracted from a 9 ml blood sample taken at baseline (Paxgene Blood DNA kit, Prenalytics), as previously described [8]. The positions of the analysed genotypes, given relative to the initiation of translation (Figure 1), were the following: −2578 C > A (promoter region), −1498 T > C (promoter region), −1154 G > A (promoter region), −634 G > C (5'UTR) and 936 C > T (3'UTR).
Statistics
Possible relationships between patient characteristics (age below vs. above the median value, Eastern Cooperative Oncology Groug (ECOG) performance status (PS) status 0 vs. 1–2, oestrogen receptor (ER) status, progesterone receptor (PR) status, number of metastatic lesions, associated chemotherapy taxane-containing regimen vs. non-taxane regimen vs. switched therapy) and clinical end-points were tested by means of chi-square or Fischer exact test. Time to progression (TTP) and overall survival (OS) were computed from the start of bevacizumab therapy. For OS, death due to any cause was considered. TTP and OS were analysed using the Kaplan-Meier method and the influence of the different polymorphisms was evaluated using a log-rank test. Survival curves for each polymorphism were also performed. Tumour response rate was analysed according to each polymorphism using a chi-square or Fischer exact test. To assess the possible relationships between VEGF-A gene polymorphisms and bevacizumab-related toxicities, we computed a toxicity score corresponding to the sum of each maximum observed toxicity grade in order to minimize the number of statistical tests. Thus, we did not report P values for each toxicity pattern but compared the genotype distribution of the five analysed polymorphisms between the group of patients having developed a toxicity score ≤ the median value and the group of patients exhibiting a toxicity score > the median value, by means of the Fischer exact test. Due to the number of analysed genotypes (five comparisons for each end-point), we applied the Bonferroni correction [17]: for a global risk α at 0.05, the P value to be applied in order to achieve a conclusive significant difference was thus 0.01. The above statistics were performed using SAS® 9.1.3. software. In addition, r2 values of linkage disequilibria between the five analysed VEGF-A gene polymorphisms, along with P values (Fischer's Exact test), were computed using the SHEsis software platform dedicated to genetic analyses (http:analysis.bio-x.cn) [18].
Results
Patient population and genotypes
Baseline characteristics of the 137 enrolled patients are shown in Table 1. Median duration of bevacizumab treatment was 7 months. The genotype distribution of the five analysed VEGF-A polymorphisms agreed with those predicted by the Hardy–Weinberg equilibrium. Strong linkage disequilibria were observed between VEGF-A gene polymorphisms located in the promoter and in the 5'UTR region (Table 2).
Table 1.
Patient characteristics (n = 137)
| Age (years) | |
| Median | 56 |
| Range | 24–79 |
| Ethnicity | |
| Caucasian | 136 (99.3%) |
| Other | 1 (0.7%) |
| ECOG performance status | |
| 0 | 87 (63.5%) |
| 1 | 45 (32.8%) |
| 2 | 5 (3.6%) |
| Hormonal receptor status (primary disease) | |
| Oestrogen receptor (positive/negative) | 99/36 (72.3/26.3%) |
| Progesterone receptor (positive/negative) | 81/52 (59.1/38%) |
| Number of metastatic lesions | |
| ≤3 | 39 (29.1%) |
| >3 | 95 (70.9%) |
| Not documented | 3 |
| Associated chemotherapy | |
| Paclitaxel | 32 (24.2%) |
| Docetaxel | 55 (42%) |
| Navelbine | 3 (2.3%) |
| Taxane combination | 12 (9.2%) |
| Non-taxane combination | 3 (2.3%) |
| Switched chemotherapy | 26 (19.8%) |
| Not documented | 6 |
| Number of cycles with bevacizumab | |
| Median | 12 |
| Range | 3–36 |
| 3 cycles | 6 (4.6%) |
| 4 to 6 cycles | 26 (19.8%) |
| 7 to 9 cycles | 21 (16%) |
| >9 cycles | 78 (59.5%) |
| Not documented | 6 |
Table 2.
Linkage disequilibria between VEGF-A gene polymorphisms (r2 and p computed on SHEsis software)
| −2578C > A | −1498T > C | −1154G > A | −634G > C | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CC | CA | AA | TT | TC | CC | GG | GA | AA | GG | GC | CC | ||
| −1498T > C | TT | 39 | 4 | 0 | |||||||||
| TC | 3 | 55 | 2 | ||||||||||
| CC | 0 | 1 | 2 | ||||||||||
| r2= 0.816 P < 0.00001 | |||||||||||||
| −1154G > A | GG | 41 | 16 | 3 | 41 | 17 | 2 | ||||||
| GA | 1 | 44 | 11 | 2 | 44 | 11 | |||||||
| AA | 0 | 0 | 17 | 0 | 0 | 17 | |||||||
| r2= 0.572 P < 0.00001 | r2= 0.566 P < 0.00001 | ||||||||||||
| −634G > C | GG | 7 | 20 | 29 | 6 | 20 | 30 | 14 | 25 | 17 | |||
| GC | 20 | 40 | 2 | 23 | 40 | 0 | 31 | 32 | 0 | ||||
| CC | 15 | 0 | 0 | 14 | 1 | 0 | 15 | 0 | 0 | ||||
| r2= 0.381 P < 0.00001 | r2= 0.415 P < 0.00001 | r2= 0.272 P < 0.00001 | |||||||||||
| 936C > T | CC | 34 | 41 | 24 | 33 | 44 | 23 | 47 | 39 | 14 | 45 | 43 | 12 |
| CT | 8 | 17 | 6 | 10 | 15 | 6 | 13 | 15 | 3 | 10 | 18 | 3 | |
| TT | 0 | 2 | 1 | 0 | 2 | 1 | 0 | 3 | 0 | 1 | 2 | 0 | |
| r2= 0.007 P = 0.17 | r2= 0.001 P = 0.63 | r2= 0.002 P = 0.48 | r2= 0.004 P = 0.29 | ||||||||||
Impact of gene polymorphisms on treatment efficacy
Clinical response, assessable in 127 patients, showed 10 complete responses (CR), 67 partial responses (PR), 41 stable diseases (SD) and nine patients with progressive disease (PD), accounting for an overall response rate (CR + PR) of 60.6%. None of the analysed VEGF-A polymorphisms was significantly linked to clinical response (Table 3).
Table 3.
Links between VEGF-A genotypes and treatment efficacy*
| Genotype | CR + PR | SD + PD | P† | Median TTP (month) | P‡ |
|---|---|---|---|---|---|
| −2578 | |||||
| CC | 25 (64.1%) | 14 (35.9%) | 9.0 | ||
| CA | 29 (51.8%) | 27 (48.2%) | 0.20 | 10.7 | 0.71 |
| AA | 20 (71.4%) | 8 (28.6%) | 11.5 | ||
| −1498 | |||||
| TT | 27 (64.3%) | 15 (35.7%) | 8.9 | ||
| TC | 29 (50.9%) | 28 (49.1%) | 0.14 | 11.1 | 0.54 |
| CC | 19 (73.1%) | 7 (26.9%) | 11.5 | ||
| −1154 | |||||
| GG | 34 (60.7%) | 22 (39.3%) | 8.9 | ||
| GA | 31 (58.5%) | 22 (41.5%) | 0.87 | 12.0 | 0.36 |
| AA | 10 (66.7%) | 5 (33.3%) | 10.4 | ||
| −634 | |||||
| GG | 34 (66.7%) | 17 (33.3%) | 11.1 | ||
| GC | 33 (55.9%) | 26 (44.1%) | 0.54 | 10.7 | 0.46 |
| CC | 9 (60%) | 6 (40%) | 6.9 | ||
| 936 | |||||
| CC | 53 (57%) | 40 (43%) | 9.7 | ||
| CT + TT | 22 (71%) | 9 (29%) | 0.21 | 11.5 | 0.022 |
127 and 137 patients were assessable for clinical response and TTP, respectively. Due to missing genotypes (poor quality DNA), relationships between clinical end-points and genotypes were performed on lower figures, depending on the analysed genotype.
Fischer Exact Test
Log Rank Test. CR complete response, PR partial response, SD stable disease, TPP time to progression.
Median follow-up was 16 months. All patients (n = 137) were assessable for TTP and OS. At time of analysis, 82 patients had developed progressive disease (60%) and 26 patients had died (19%), all from breast cancer. Median TTP was 11 months. Interestingly, analysis of the VEGF-A 936C > T polymorphism revealed that patients bearing the 936T allele (936CT, 30 patients and 936TT, two patients) exhibited a marked trend towards a longer TTP as compared with patients homozygous for the 936C allele (n = 96, Log Rank P = 0.022, Table 3, Figure 2). Median TTP was 11.5 months (95% CI 10.2, 25.8) in 936CT or 936TT patients vs. 9.7 months (95% CI 7.8, 12) in 936 CC patients. Other polymorphisms did not influence TTP (Table 3).
Figure 2.
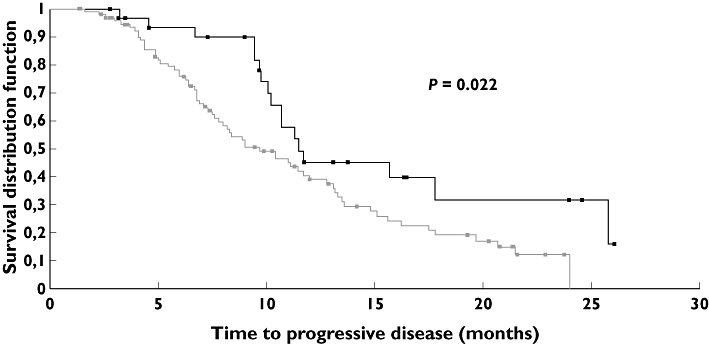
Time to progression (TTP) according to VEGF- 936C > T polymorphism. Median TTP was 9.7 months in 936CC patients vs. 11.5 months in patients bearing the 936T allele (Log Rank, P = 0.022). CC (n = 96) : 65 events ( ); CT (n = 30) + TT (n = 2) : 17 events (
); CT (n = 30) + TT (n = 2) : 17 events ( )
)
The influence of the VEGF-A gene polymorphisms on OS did not reveal any significant relationship, but was limited by the few number of events (n = 26).
None of the patient characteristics was associated with clinical response, TTP, or OS.
Impact of gene polymorphisms on bevacizumab-related toxicity
The analysed bevacizumab-related toxicities were hypertension, haemorrhage, arterial and venous thrombo-embolism. Globally, these toxicities were mild (Table 4). The toxicity score ranged between 0 and 6 (mean 1.8, median 1): 70 patients had a score 0–1 and 67 patients had a score 2 to 6. The toxicity score was not related to any of the patient characteristics. Table 5 shows that the VEGF-A−634 G > C polymorphism was significantly related to the toxicity score, with 39%, 49% and 81% of patients with score >1 in GG, GC and CC patients, respectively (P = 0.01). Close examination of each toxicity pattern included in this score revealed that patients with the −634C allele were prone to develop hypertension and thrombo-embolism.
Table 4.
Description of the bevacizumab-related toxicity*
| Grade 0 n (%) | Grade 1 n (%) | Grade 2 n (%) | Grade 3 n (%) | Grade 4 n (%) | |
|---|---|---|---|---|---|
| Hypertension | 81 (59.1%) | 23 (16.8%) | 20 (14.6%) | 13 (9.5%) | 0 |
| Haemorrhage | 37 (27.0%) | 85 (62.0%) | 15 (10.9%) | 0 | 0 |
| Arterial thrombo-embolism | 135 (98.5%) | 0 | 1 (0.7%) | 0 | 1 (0.7%) |
| Venous thrombo-embolism | 128 (93.4%) | 1 (0.7%) | 4 (2.9%) | 3 (2.2%) | 1 (0.7%) |
Maximum observed toxicity grade (NCI-CTCAE criteria).
Table 5.
Links between VEGF-A genotypes and bevacizumab-related toxicity*
| Polymorphism | n | % of patients with grade 1-2-3-4 hypertension | % of patients with grade 1-2-3-4 haemorrhage | % of patients with grade 1-2-3-4 arterial thromboembolism | % of patients with grade 1-2-3-4 venous thromboembolism | % of patients with toxicity score > 1 |
|---|---|---|---|---|---|---|
| −2578C > A | ||||||
| CC | 42 | 45.2% | 66.7% | 4.8% | 4.8% | 57.1% |
| CA | 60 | 41.7% | 71.7% | 0% | 6.7% | 48.3%, P = 0.18 |
| AA | 31 | 29.0% | 83.9% | 0% | 6.5% | 35.5% |
| −1498T > C | ||||||
| TT | 44 | 43.2% | 65.9% | 4.5% | 4.5% | 52.3% |
| TC | 61 | 44.3% | 72.1% | 0% | 8.2% | 50.8%, P = 0.39 |
| CC | 30 | 26.7% | 83.3% | 0% | 6.7% | 36.7% |
| −1154G > A | ||||||
| GG | 60 | 41.7% | 66.7% | 3.3% | 5.0% | 53.3% |
| GA | 57 | 40.4% | 77.2% | 0% | 10.5% | 47.4%, P = 0.43 |
| AA | 17 | 29.4% | 82.4% | 0% | 0% | 35.3% |
| −634G > C | ||||||
| GG | 56 | 30.4% | 82.1% | 0% | 5.4% | 39.3% |
| GC | 63 | 44.4% | 68.3% | 1.6% | 6.3% | 49.2%, P = 0.012 |
| CC | 16 | 56.3% | 62.5% | 6.3% | 12.5% | 81.3% |
| 936C > T | ||||||
| CC | 100 | 41.0% | 75.0% | 1.0% | 9.0% | 49.0% |
| CT | 31 | 35.5% | 67.7% | 3.2% | 0% | 48.4%, P = 1.00 |
| TT | 3 | 33.3% | 66.7% | 0% | 0% | 33.3% |
P values were not computed for each toxicity pattern in order to minimize the number of statistical tests. Fischer Exact tests were performed for the toxicity score.
Discussion
Elevated tumoral expression of VEGF-A, usually referred to as VEGF, is a very strong marker of cancer progression in node-negative breast carcinoma not receiving any adjuvant treatment [1, 2], and a strong predictor of poor efficacy of tamoxifen and chemotherapy in metastatic breast cancer patients [19]. This may suggest that breast tumours overexpressing VEGF-A may be good candidates for anti-VEGF target therapies, even though such a link has not been demonstrated so far [20]. Among the numerous studies aimed at evaluating the predictive value of VEGF-A expression on bevacizumab efficacy, one study conducted on 56 advanced breast cancer patients reported that low baseline circulating VEGF was significantly linked to longer TTP [21]. Interpretation of biomarker studies such as VEGF-A showed the difficulty to discriminate between its predictive value and its prognostic value. In this regard, the randomized trial by Dowlati et al. [22] conducted on 163 non-small-cell-lung cancer patients receiving carboplatin-paclitaxel ± bevacizumab is very informative. Patients with high baseline VEGF plasma concentrations had an increased probability of response with the addition of bevacizumab (33% vs. 7.7% with and without bevacizumab, respectively).
VEGF-A expression is controlled by complex mechanisms involving VEGF-A mRNA splicing that generates multiple VEGF-A isoforms including a pro-angiogenic isoform family and an anti-angiogenic isoform family [23] and various controls at the gene level itself, particularly at the 5' and 3' untranslated regions that contain key regulatory elements. Importantly, functional gene polymorphisms have been described in these regulatory regions, as well as in the promoter region (Figure 1). Since analysis of germinal VEGF-A polymorphisms from a blood sample is much more reliable and easy than analysis of tumoral or plasma VEGF-A (ex vivo release of VEGF has been reported in blood samples), the aim of the present study was to analyse prospectively the possible relationships between the five major functional germinal VEGF-A polymorphisms and pharmacodynamics of first-line bevacizumab-based therapy in metastatic breast cancer patients included in the single-arm safety MO19391 trial. This pharmacogenetic companion study was conducted on 137 women, whose characteristics were not significantly different from that of the entire cohort of patients (data not shown). Patient characteristics were not related to any of the analysed clinical end-points, thus justifying the univariate analyses performed to test the possible relationships between genotypes and toxicity score, TTP or OS.
The presently analysed pharmacodynamic–pharmacogenetic relationships were specific to bevacizumab for the toxicity since we strictly considered the bevacizumab-related toxicity, i.e. hypertension, haemorrhage and thrombo-embolism (Table 4) [24]. Patients bearing the −634 C allele presented a significant higher risk to develop toxicity (Table 5). This result contrasts with data from Schneider et al. [16] focused on hypertension, who reported on 177 advanced breast cancer patients receiving bevacizumab-paclitaxel that VEGF-A−634CC genotype was associated with less grade 3–4 hypertension. This polymorphism, located in the 5' untranslated region, has been shown to affect the protein translation efficiency and the −634C allele has been associated with lower VEGF-A expression [25]. However, the basis of the possible link between VEGF-A expression and bevacizumab-related systemic toxicity is clearly unknown.
The main objective of the study was to assess the influence of VEGF-A polymorphisms on the efficacy of a bevacizumab-based first-line therapy in advanced breast cancer patients. The majority of patients received concomitant taxane therapy. No relationship was observed between clinical response and VEGF-A genotypes. In contrast, patients bearing the VEGF-A 936T genotype presented a marked tendency towards a longer TTP than homozygous VEGF-A 936CC patients (Figure 2). The 936C > T polymorphism is located in a region of the gene corresponding to the 3'untranslated region of the VEGF mRNA and this mRNA domain is highly implicated in VEGF mRNA stability [26]. Three studies have reported that the VEGF-A 936T allele was significantly associated with decreased VEGF-A plasma concentrations [10, 27, 28]. Accordingly, in a series of 49 head and neck patients, our group reported that tumours harboring the VEGF-A 936T allele exhibited lower VEGF-A concentrations at the tumoral level [8]. To our knowledge, the opposite pattern concerning the impact of VEGF-A 936C > T genotype on VEGF-A expression has never been reported in the literature. It can thus be considered that the VEGF-A 936T allele likely governs VEGF-A expression. Thus, the longer TTP presently observed in patients bearing the 936T allele perfectly concords with the observations of Burstein et al. [21] showing a longer TTP in metastatic breast cancer patients with low plasma VEGF. In both studies, patients received bevacizumab-based therapy. It is however not clear-cut whether these observations reflect a prognostic or a predictive value. One can hypothesize that the 936T allele provides an advantage to bevacizumab-treated patients by two complementary mechanisms: a lower target level (at the systemic and tumoral levels), along with a less pro-angiogenic tumour environment reducing the risk of tumour growth.
Schneider et al. [16] investigated the same VEGF-A genotypes as ours, in tumour blocks of metastatic breast cancer patients receiving first-line paclitaxel or paclitaxel-bevacizumab therapy. They found that the VEGF-A−1154A and −2578A alleles were significantly associated with longer overall survival (but neither TTP nor responsiveness) only in patients included in the paclitaxel-bevacizumab arm. This may suggest a bevacizumab-dependent predictive value of VEGF-A−1154 and −2578 genotypes. The differences between the results of Schneider et al. and ours stand on the clinical end points, the nature of the VEGF-A genotypes and the nature of the analysed tissues. In our study, no linkage disequilibria were observed between 936C > T, on the one hand, and −2578C > A or −1154G > A on the other (Table 2). As opposed to 936C > T genotype for which the functional impact on VEGF-A expression has been consistently observed [8, 10, 27, 28], the influence of −2578C > A and −1154G > A genotypes on expression and/or cancer risk is controversial [13]. Schneider et al. [16] analysed the influence of −2578C > A and −1154G > A genotypes on tumoral VEGF-A expression and did not observe significant relationships.
In conclusion, present data point out the potential role of VEGF-A 936C > T functional polymorphism for predicting disease progression in metastatic breast cancer patients receiving bevacizumab-based therapy. Further studies aimed at improving the knowledge of the impact of the numerous VEGF-A polymorphisms on outcome of bevacizumab-treated patients are warranted.
Acknowledgments
We aknowledgement Roche for financial support.
Competing Interests
J-YP, ME and GM have received funds for research by Roche. XP received an honorarium from Roche SA. CV has been reimbursed by Roche and other pharmaceutical companies for attending several conferences. MP is a Roche employee. There are no other competing interests to declare.
REFERENCES
- 1.Gasparini G, Toi M, Gion M, Verderio P, Dittadi R, Hanatani M, Matsubara I, Vinante O, Bonoldi E, Boracchi P, Gatti C, Suzuki H, Tominaga T. Prognostic significance of vascular endothelial growth factor protein in node-negative breast carcinoma. J Natl Cancer Inst. 1997;89:139–47. doi: 10.1093/jnci/89.2.139. [DOI] [PubMed] [Google Scholar]
- 2.Linderholm B, Tavelin B, Grankvist K, Henriksson R. Vascular endothelial growth factor is of high prognostic value in node-negative breast carcinoma. J Clin Oncol. 1998;16:3121–8. doi: 10.1200/JCO.1998.16.9.3121. [DOI] [PubMed] [Google Scholar]
- 3.Presta LG, Chen H, O'Connor SJ, Chisholm V, Meng YG, Krummen L, Winkler M, Ferrara N. Humanization of an anti-vascular endothelial growth factor monoclonal antibody for the therapy of solid tumors and other disorders. Cancer Res. 1997;57:4593–9. [PubMed] [Google Scholar]
- 4.Miller K, Wang M, Gralow J, Dickler M, Cobleigh M, Perez EA, Shenkier T, Cella D, Davidson NE. Paclitaxel plus bevacizumab versus paclitaxel alone for metastatic breast cancer. N Engl J Med. 2007;357:2666–76. doi: 10.1056/NEJMoa072113. [DOI] [PubMed] [Google Scholar]
- 5.Jubb AM, Hurwitz HI, Bai W, Holmgren EB, Tobin P, Guerrero AS, Kabbinavar F, Holden SN, Novotny WF, Frantz GD, Hillan KJ, Koeppen H. Impact of vascular endothelial growth factor-A expression, thrombospondin-2 expression, and microvessel density on the treatment effect of bevacizumab in metastatic colorectal cancer. J Clin Oncol. 2006;24:217–27. doi: 10.1200/JCO.2005.01.5388. [DOI] [PubMed] [Google Scholar]
- 6.Miller K, Trigo JM, Wheeler C, Barge A, Rowbottom J, Sledge G, Baselga J. A multicenter phase II trial of ZD6474, a vascular endothelial growth factor receptor-2 and epidermal growth factor receptor tyrosine kinase inhibitor, in patients with previously treated metastatic breast cancer. Clin Cancer Res. 2005;11:3369–76. doi: 10.1158/1078-0432.CCR-04-1923. [DOI] [PubMed] [Google Scholar]
- 7.Bidard FC, Mathiot C, Degeorges A, Etienne-Grimaldi MC, Delva R, Pivot X, Veyret C, Bergougnoux L, de Cremoux P, Milano G, Pierga JY. Clinical value of circulating endothelial cells and circulating tumor cells in metastatic breast cancer patients treated first line with bevacizumab and chemotherapy. Ann Oncol. 2010;21:1765–71. doi: 10.1093/annonc/mdq052. [DOI] [PubMed] [Google Scholar]
- 8.Formento JL, Etienne-Grimaldi MC, Francoual M, Pagès G, Onesto C, Formento P, Chamorey E, Dassonville O, Poissonnet G, Milano G. Influence of the VEGF-A 936C > T germinal polymorphism on tumoral VEGF expression in head and neck cancer. Pharmacogenomics. 2009;10:1277–83. doi: 10.2217/pgs.09.54. [DOI] [PubMed] [Google Scholar]
- 9.Koukourakis MI, Papazoglou D, Giatromanolaki A, Bougioukas G, Maltezos E, Sivridis E. VEGF gene sequence variation defines VEGF gene expression status and angiogenic activity in non-small cell lung cancer. Lung Cancer. 2004;46:293–8. doi: 10.1016/j.lungcan.2004.04.037. [DOI] [PubMed] [Google Scholar]
- 10.Krippl P, Langsenlehner U, Renner W, Yazdani-Biuki B, Wolf G, Wascher TC, Paulweber B, Haas J, Samonigg H. A common 936 C/T gene polymorphism of vascular endothelial growth factor is associated with decreased breast cancer risk. Int J Cancer. 2003;106:468–71. doi: 10.1002/ijc.11238. [DOI] [PubMed] [Google Scholar]
- 11.Ku KT, Wan L, Peng HC, Tsai MH, Tsai CH, Tsai FJ. Vascular endothelial growth factor gene-460 C/T polymorphism is a biomarker for oral cancer. Oral Oncol. 2005;41:497–502. doi: 10.1016/j.oraloncology.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 12.Lee SJ, Lee SY, Jeon HS, Park SH, Jang JS, Lee GY, Son JW, Kim CH, Lee WK, Kam S, Park RW, Park TI, Kang YM, Kim IS, Jung TH, Park JY. Vascular endothelial growth factor gene polymorphisms and risk of primary lung cancer. Cancer Epidemiol Biomarkers Prev. 2005;14:571–5. doi: 10.1158/1055-9965.EPI-04-0472. [DOI] [PubMed] [Google Scholar]
- 13.Jain L, Vargo CA, Danesi R, Sissung TM, Price DK, Venzon D, Venitz J, Figg WD. The role of vascular endothelial growth factor SNPs as predictive and prognostic markers for major solid tumors. Mol Cancer Ther. 2009;8:2496–508. doi: 10.1158/1535-7163.MCT-09-0302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu H, Shu XO, Cui Y, Kataoka N, Wen W, Cai Q, Ruan ZX, Gao YT, Zheng W. Association of genetic polymorphisms in the VEGF gene with breast cancer survival. Cancer Res. 2005;65:5015–9. doi: 10.1158/0008-5472.CAN-04-2786. [DOI] [PubMed] [Google Scholar]
- 15.Jin Q, Hemminki K, Enquist K, Lenner P, Grzybowska E, Klaes R, Henriksson R, Chen B, Pamula J, Pekala W, Zientek H, Rogozinska-Szczepka J, Utracka-Hutka B, Hallmans G, Försti A. Vascular endothelial growth factor polymorphisms in relation to breast cancer development and prognosis. Clin Cancer Res. 2005;11:3647–53. doi: 10.1158/1078-0432.CCR-04-1803. [DOI] [PubMed] [Google Scholar]
- 16.Schneider BP, Wang M, Radovich M, Sledge GW, Badve S, Thor A, Flockhart DA, Hancock B, Davidson N, Gralow J, Dickler M, Perez EA, Cobleigh M, Shenkier T, Edgerton S, Miller KD. ECOG 2100. Association of vascular endothelial growth factor and vascular endothelial growth factor receptor-2 genetic polymorphisms with outcome in a trial of paclitaxel compared with paclitaxel plus bevacizumab in advanced breast cancer: ECOG 2100. J Clin Oncol. 2008;26:4672–8. doi: 10.1200/JCO.2008.16.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bland JM, Altman DG. Multiple significance tests: the Bonferroni method. BMJ. 1995;310:170. doi: 10.1136/bmj.310.6973.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shi YY, He L. SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Res. 2005;15:97–8. doi: 10.1038/sj.cr.7290272. [DOI] [PubMed] [Google Scholar]
- 19.Foekens JA, Peters HA, Grebenchtchikov N, Look MP, Meijer-van Gelder ME, Geurts-Moespot A, van der Kwast TH, Sweep CG, Klijn JG. High tumor levels of vascular endothelial growth factor predict poor response to systemic therapy in advanced breast cancer. Cancer Res. 2001;61:5407–14. [PubMed] [Google Scholar]
- 20.Murukesh N, Dive C, Jayson GC. Biomarkers of angiogenesis and their role in the development of VEGF inhibitors. Br J Cancer. 2010;102:8–18. doi: 10.1038/sj.bjc.6605483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burstein HJ, Chen YH, Parker LM, Savoie J, Younger J, Kuter I, Ryan PD, Garber JE, Chen H, Campos SM, Shulman LN, Harris LN, Gelman R, Winer EP. VEGF as a marker for outcome among advanced breast cancer patients receiving anti-VEGF therapy with bevacizumab and vinorelbine chemotherapy. Clin Cancer Res. 2008;14:7871–7. doi: 10.1158/1078-0432.CCR-08-0593. [DOI] [PubMed] [Google Scholar]
- 22.Dowlati A, Gray R, Sandler AB, Schiller JH, Johnson DH. Cell adhesion molecules, vascular endothelial growth factor, and basic fibroblast growth factor in patients with non-small cell lung cancer treated with chemotherapy with or without bevacizumab – an Eastern Cooperative Oncology Group Study. Clin Cancer Res. 2008;14:1407–12. doi: 10.1158/1078-0432.CCR-07-1154. [DOI] [PubMed] [Google Scholar]
- 23.Harper SJ, Bates DO. VEGF-A splicing: the key to anti-angiogenic therapeutics? Nat Rev Cancer. 2008;8:880–7. doi: 10.1038/nrc2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koutras AK, Fountzilas G, Makatsoris T, Peroukides S, Kalofonos HP. Bevacizumab in the treatment of breast cancer. Cancer Treat Rev. 2010;36:75–82. doi: 10.1016/j.ctrv.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 25.Watson CJ, Webb NJA, Bottomley MJ, Brenchley PEC. Identification of polymorphisms within the vascular endothelial growth factor (VEGF) gene: correlation with variation in VEGF protein production. Cytokine. 2000;12:1232–35. doi: 10.1006/cyto.2000.0692. [DOI] [PubMed] [Google Scholar]
- 26.Levy AP, Levy NS, Goldberg MA. Hypoxia-inducible protein binding to vascular endothelial growth factor mRNA and its modulation by the von Hippel-Lindau protein. J Biol Chem. 1996;271:25492–7. doi: 10.1074/jbc.271.41.25492. [DOI] [PubMed] [Google Scholar]
- 27.Renner W, Kotschan S, Hoffmann C, Obermayer-Pietsch B, Pilger E. A common 936 C/T mutation in the gene for vascular endothelial growth factor is associated with vascular endothelial growth factor plasma levels. J Vasc Res. 2000;37:443–8. doi: 10.1159/000054076. [DOI] [PubMed] [Google Scholar]
- 28.Zhai R, Gong MN, Zhou W, Thompson TB, Kraft P, Su L, Christiani DC. Genotypes and haplotypes of the VEGF gene are associated with higher mortality and lower VEGF plasma levels in patients with ARDS. Thorax. 2007;62:718–22. doi: 10.1136/thx.2006.069393. [DOI] [PMC free article] [PubMed] [Google Scholar]


