Abstract
Background
p300 impacts the transcription of several genes involved in key pathways critical to PCa progression. Therefore, we evaluated the prognostic value of p300 expression and its correlation with nuclear alterations seen in tumor cells in men with long term follow-up after radical prostatectomy (RP).
Methods
NCI Cooperative Prostate Cancer Tissue Resource tissue microarray cores of 92 RP cases (56 non-recurrences and 36 PSA recurrences) were utilized for the study. p300 expression was assessed by quantitative immunohistochemistry and nuclear alterations in Feulgen-stained nuclei were evaluated by digital image analysis using the AutoCyte™ Pathology Workstation. Cox proportional hazards regression, Spearman’s rank correlation, and Kaplan-Meier plots were employed to analyze the data.
Results
p300 expression significantly correlated with nuclear alterations seen in tumor cells; specifically with circular form factor (p=0.012) and minimum feret (p=0.048). p300 expression in high grade tumors (Gleason score ≥7) was significantly higher compared to low grade tumors (Gleason score <7) [17.7% vs. 13.7%, respectively, p=0.03]. TNM stage, Gleason score, and p300 expression were univariately significant in the prediction of PCa biochemical recurrence free survival (p≤0.05). p300 expression remained significant in the multivariate model (p=0.03) while Gleason score showed a trend toward significance (p=0.06). Patients with a Gleason score ≥7 and p300 expression >24% showed the highest risk for PCa biochemical recurrence (p=0.002).
Conclusions
p300 expression correlates with nuclear alterations seen in tumor cells and has prognostic value in predicting long-term PCa biochemical recurrence free survival.
INTRODUCTION
Prostate cancer (PCa) is the second leading cause of cancer death among men in the United States, with an anticipated 218,890 newly diagnosed cases and nearly 27,000 deaths in 2007 (1). In a series of nearly 2,000 patients treated with radical prostatectomy at Johns Hopkins Hospital, 304 men developed PSA recurrence (15%) and were monitored without hormone therapy until demonstration of metastasis (2). Of these men, 34% developed distant metastases over a median period of 8 years from the time of the first postoperative PSA elevation (2). Han et al. (3) updated this study cohort, reporting 360 recurrences (17%) in 2,091 men with PCa. They used three preoperative or postoperative variables to create nomograms to assess biochemical recurrence-free survival probabilities. This study demonstrated the overall actuarial PSA-free survival probabilities at 5, 10, and 15 years to be 84%, 72% and 61%, respectively.
Clearly, the accumulation of repeated insults to the prostate over time through diet, infection, inflammation and aging results in a cascade of biological and molecular events which can result in malignancy. Therefore, PCa is a heterogeneous malignant disease where its’ development and progression depends upon the biology of inflammation of the prostate as well as hereditary (genetic susceptibility), epigenetic and somatic gene defects. Many of these alterations are permanent and reflect transition to malignancy and progression to metastasis.
In the search for new molecular biomarkers to predict biochemical recurrence free survival in men with PCa, several potential serologic and histological biomarkers have been evaluated (4-8). At the tissue level, Gleason score and pathological stage are significant predictors of biochemical recurrence and metastasis (9,10). Further, investigators have used nuclear structure alterations i.e. change in nuclear size, shape, DNA content and chromatin structure, to predict stage, biochemical recurrence and metastasis in men with PCa (11-15). Recently, Seligson et al. (16) showed that the levels of acetylated histones correlate with increasing tumor grade and global histone modification pattern is able to identify disease subtypes with distinct risks of tumor recurrence in men with PCa.
There are numerous transcriptional coactivators involved in transcription and chromatin remodeling in androgen dependent and independent PCa. p300, a transcriptional coactivator that acetylates histones found in the nucleosome, has been shown to be differentially expressed in a number of tumors (17-19). Debes et al. (20) demonstrated that p300 is involved in the IL-6-mediated transactivation of the androgen receptor (AR) in the absence of androgens in PCa cells. Others have shown a similar role of p300 in the presence of androgens (21). In addition, Debes et al. (22) showed that p300 plays a key role in PCa epithelial cell proliferation.
The National Cancer Institute (NCI) engaged multiple institutions to prepare the Cooperative Prostate Cancer Tissue Resource (CPCTR) tissue microarrays (TMAs). We obtained TMAs from this resource that included tumor tissue from a unique patient cohort of 92 men with long term follow-up to assess biochemical recurrence after surgical treatment for PCa. Using the TMAs from this patient cohort, we recently demonstrated the ability of nuclear morphometry determined by digital image analysis to predict biochemical recurrence with an AUC-ROC of 80% compared to pathology with an AUC-ROC of 67% (23). Using the same patient cohort, we asked if expression levels of p300, which acetylates core histone residues, could predict biochemical recurrence free survival in men with PCa. We also evaluated the association between p300 expression, nuclear structure alterations, Gleason score and pathologic stage.
MATERIAL AND METHODS
Prostate Tissue Specimens Dataset
The CPCTR-TMA is the result of a project funded by NCI RFA released in April, 2000 and four academic institutions [George Washington University Medical Center (Washington DC); Medical College of Wisconsin (Milwaukee, WI); New York University School of Medicine (New York, NY); and the University of Pittsburgh (Pittsburgh, PA)] were funded to form a national prostate cancer tissue resource, CPCTR. The resource is entirely funded by an individual Cooperative Agreement Grant from the NCI to each of the four participating sites (24,25). The CPCTR resource functions as a “virtual tissue bank” with a central database with all four participating sites working jointly with the NCI. Additionally, the methods for TMA construction employ a standardized protocol, a database containing standardized common data elements, and a supporting bioinformatics database with outcome results are also provided in a manuscript (26). Information about the NCI-CPCTR project and how to obtain these bioreagents can be found on the web at http://cpctr.cancer.gov.
NCI-CPCTR Patient Cohort
Pathological material from a total of 299 PCa chronologically consecutive radical prostatectomy patients were arrayed over four blocks with a single focus of tumor from each patient tumor represented in duplicate 0.6mm core spots. For determination of PSA recurrence, an algorithm was defined where the PSA values needed to increase >0.4 ng/dl (single value) or a PSA values >0.2 ng/dl with additional subsequent increasing values (27). The date of initial PSA rise (either the date of the single value >0.4 ng/ml or the date of the PSA value >0.2 ng/ml, before subsequent rising PSA values) was subtracted from the date of initial PSA nadir to determine the months to PSA recurrence. A total of 92 PCa cases (n = 56 non-recurrence and n = 36 recurrence) contained complete information for the study (Table 1).
Table 1.
Prostate Cancer Patients Demographics
| Variable Description | No Biochemical Recurrence (N = 56) | Biochemical Recurrence (N = 36) | p value |
|---|---|---|---|
| Median Age in Years (range) | 65.5 (47-76) | 64 (42-77) | 0.274a |
|
| |||
| Pathologic stage (%) | |||
| T2a | 9 (16.1) | 2 (5.6) | |
| T2b | 32 (57.1) | 17 (47.2) | 0.010b |
| T3a | 13 (23.2) | 9 (25.0) | 0.028c |
| T3b | 2 (3.6) | 8 (22.2) | |
|
| |||
| Gleason Score (%) | |||
| 5 | 6 (10.7) | 0 (0) | |
| 6 | 21 (37.5) | 10 (27.8) | 0.024b |
| 7 | 27 (48.2) | 23 (63.9) | 0.106c |
| 8 | 1 (1.8) | 2 (5.6) | |
| 9 | 1 (1.8) | 1 (2.8) | |
|
| |||
| Race (%) | |||
| White | 51 (91.1) | 32 (88.9) | 0.574b |
| Black | 1 (1.8) | 3 (8.3) | 0.505c |
| Others | 3 (5.3) | 1 (2.8) | |
| Unknown | 1 (1.8) | 0 (0) | |
Median test
Wilcoxon ranksum test
Fisher’s exact test
Measurement of p300 protein expression
Immunohistochemistry for p300 expression in PCa was performed on formalin-fixed paraffin biopsy sections using a DAKO AutoStainer. After dewaxing and dehydration, sections were placed in a rice steamer with citrate buffer (pH 6.0) for twenty minutes. The 6 micron sections were pretreated with 0.3% hydrogen peroxide for ten minutes, washed with deionized water and phosphate buffer (PBS, pH 7.4), and incubated with 0.5% Triton X-100 and 0.5% milk in PBS for 5 minutes at room temperature. The DAKO EnvisionPlus IHC kit was used for immunostaining. Briefly, the sections were blocked with 5% milk in PBS containing 0.1% Triton X-100 for 20 minutes and then incubated with the specific antibody for this protein (Santa Cruz Biotechnology, Santa Cruz, CA) at pre-determined dilutions with PBS containing 0.5% milk and 0.1% Triton X-100 at room temperature for one hour in a humidified chamber. After washing, the sections were sequentially incubated with biotinylated Envision secondary antibody, streptavidin-HRP, and freshly prepared DAB chromogen substrate. The p300 immunohistochemistry (IHC) stained tissues were counterstained with hematoxylin for one minute and mounted (supplementary figure 1).
The stained TMAs were scanned with a BLISS virtual slide scanner [Bacus Laboratories, Lombard, IL] at 40x magnification using the WebSlide® digital microscope slide format. This creates a database input file that lists information on every CPCTR-TMA core and provides an automatic link to the WebSlide® Net Viewer ActiveX Control (Bacus Labs, Lombard, IL) for a visual TMA core database. These BLISS virtual slide images were processed using a TMA score software program [Bacus Laboratories, Lombard, IL] that quantified p300 expression by measuring percentage of tumor area positive for the p300 antigen in each PCa case.
Measurement of Nuclear Alterations
Using ~5μm sections prepared from the TMA blocks, Feulgen DNA-staining was performed per the manufacturer’s instructions (TriPath Imaging Inc, Burlington, NC). Next, a minimum of 125 intact, Feulgen-stained cancer nuclei were captured from the 0.6mm spots for each case using an AutoCyte Pathology Workstation (APW) [TriPath Imaging, Inc., Burlington, NC] and the QUIC-DNA software (11,12,28). The QUIC-DNA software calculated a total of 40 nuclear alterations [listed in Ref. (28)], including nuclear size, shape, DNA content and chromatin texture features (at a step size of one pixel), for each nuclei captured. For each case, the variance of each nuclear alteration was determined, thereby reducing the complexity of the nuclear alteration database to a single set of 40 variables for each case.
Statistical Methods
All data were analyzed using Stata™ v10.0 statistical analysis software (Stata Corporation, College Station, TX). A non-parametric k-sample chi-squared test for equality of medians was used to evaluate differences in the non-normally distributed ages. Wilcoxon’s ranksum test was used to test for distribution differences and Fisher’s exact test was used to test for differences in proportions between patients with and without biochemical recurrence. Correlations of p300 expression with Gleason score, pathologic stage and nuclear alterations were evaluated using Spearman’s rank correlation coefficients. Univariate Cox proportional hazards regression was used to identify significant prognostic factors for PCa biochemical recurrence. Ties were handled by the Breslow method, and the proportional hazard assumption was verified by examination of residual plots. We determined optimal cut-point for dichotomized p300 expression data using classification and regression tree analysis. Kaplan-Meier survival plots were created to demonstrate the ability of the p300 expression, pathologic stage and Gleason score to predict PSA recurrence free survival. Univariately significant variables were further considered in multivariate model. Statistical significance in this study was set as p ≤ 0.050.
RESULTS
The demographic and pathologic information for the biochemical (PSA) recurrence and non-recurrence groups of PCa patients are shown in Table 1. This table shows that patients with biochemical recurrence tended to have higher Gleason scores and higher pathologic stages. The mean p300 expression levels (% area positive for p300 immunostaining) in the biochemical (PSA) recurrence and non-recurrence groups of men were 18.69% ± 9.03% and 14.40% ± 6.53%, respectively (p = 0.009).
The p300 protein expression was significantly higher in high grade tumors (Gleason score ≥7: 17.70% ± 7.50%) compared to low grade tumors (Gleason score <7: 13.67% ± 7.83%) (p = 0.03). The mean p300 expression in pathologic stage T2 and T3 patients was 15.48% ± 7.16% and 17.20% ± 9.01%, respectively (p = 0.43). We observed significant associations between p300 protein expression and nuclear alterations seen in tumor cells in these CPCTR-TMA radical prostatectomy tissue samples. Of particular interest, it was noted that the circular form factor (rho = -0.26; p = 0.012) and minimum feret (rho = 0.21; p = 0.048) exhibited statistically significant correlations with p300 protein expression. An assessment of other nuclear features, such as area (rho = 0.16; p = 0.12), excess of gray value (rho = 0.17; p = 0.10) and standard deviation of gray value (rho = -0.17; p = 0.10), showed a trend toward statistical significance for correlation with p300 expression levels.
Gleason score showed significant correlation with several nuclear alterations seen in the tumor cells, including skewness of OD (rho = 0.24; p = 0.0212), excess of OD (rho = 0.22; p = 0.0337), DNA ploidy (rho = 0.25; p = 0.0155), variance (rho = -0.20; 0.049), sum average-AC (rho = -0.23; p = 0.0280), sum variance-AC (rho = -0.31; p = 0.0023), cluster shade (rho = -0.28; p = 0.0064) and second diagonal moment (rho = -0.27; p = 0.0084). The pathologic stage also showed significant correlation with several nuclear alterations seen in tumor cells including skewness of gray value (rho = 0.22; p = 0.0323), DNA ploidy (rho = 0.28; p = 0.0078), variance (rho = -0.22; p = 0.0397), cluster shade (rho = -0.23; p = 0.0249), and second diagonal moment (rho = -0.21; p = 0.0440).
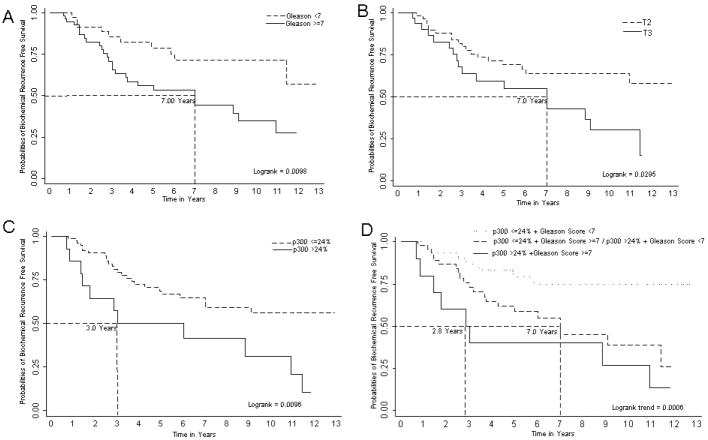
Upon univariate analyses, p300 expression as a continuous variable was a significant prognosticator (p = 0.021) for PCa biochemical recurrence. A dichotomized population for p300 expression was then defined with an optimal cutoff of 24% (85th percentile), specifically patients were categorized as having either low (≤24.0%) or high (>24.0%) p300 expression. Dichotomized pathologic stage, Gleason score and p300 expression were univariately significant (Table 2) for prediction of biochemical recurrence. However, when these three variables were considered together in a multivariate Cox proportional hazards model, only p300 expression was significant (Table 2). Figures 1A, 1B, and 1C show Kaplan-Meier survival curves for prediction of PCa biochemical recurrence free survival using pathologic stage, Gleason score, and p300 expression, respectively.
Table 2.
Cox proportional hazards regression
| Variable | N | Univariate
|
Multivariate
|
Stratification based upon Gleason Score & p300 status
|
|||
|---|---|---|---|---|---|---|---|
| HR (95% CI) | P value | HR (95% CI) | P value | HR (95% CI) | P value | ||
| TNM Stage | |||||||
| T2 | 60 | 1.00 | 0.034 | 1.00 | 0.170 | ||
| T3 | 32 | 2.03 (1.06 - 3.92) | 1.60 (0.82 - 3.15) | ||||
|
| |||||||
| Gleason Score | |||||||
| <7 | 37 | 1.00 | 0.013 | 1.00 | 0.060 | ||
| ≥7 | 55 | 2.54 (1.22 - 5.30) | 2.07 (0.97 - 4.43) | ||||
|
| |||||||
| p300 | |||||||
| <=24% | 78 | 1.00 | 0.013 | 1.00 | 0.032 | ||
| >24% | 14 | 2.47 (1.21 - 5.03) | 2.19 (1.07 - 4.47) | ||||
|
| |||||||
| Gleasonp300 | |||||||
| 1¶ | 33 | 1.00 | |||||
| 2§ | 49 | 2.81 (1.19 - 6.66) | 0.018 | ||||
| 3* | 10 | 5.18 (1.88 - 14.30) | 0.002 | ||||
Gleason Score <7 and p300 ≤24%
Gleason Score <7 and p300 >24% / Gleason Score ≥7 and p300 ≤24%
Gleason Score ≥7 and p300 >24%
Figure 1.
Kaplan-Meier plots showing ability of Gleason score (A), pathological stage (B), p300 expression (C), and Gleason score & p300 combined (D) to predict biochemical recurrence free survival. Logrank test and Logrank trend test were used to test equality of survivor functions across two groups and three ordered groups respectively.
Additionally, we stratified the NCI-CPCTR patients based upon Gleason score and p300 expression status. Table 2 and Figure 1D show the ability of Gleason score and p300 expression status combined to predict PCa biochemical recurrence free survival. Because there were only 4 patients with a Gleason score <7 & high p300 expression, this subcategory was merged with cases having Gleason score ≥7 & low p300 expression for these analyses. Patients with Gleason score ≥7 & high p300 protein expression had a significantly higher risk of PCa biochemical recurrence (p = 0.002) (Table 2 & Figure 1D).
DISCUSSION
The nucleosome, i.e. the fundamental unit of chromatin organization, is composed of 146 base pairs of DNA wrapped in 1.65 turns around an octamer of the four core histones, H2A, H2B, H3, and H4 (29). Chromatin remodeling directly influences the activity of DNA as it relates to transcription, replication, and recombination and is regulated by two highly conserved mechanisms, post-translational modifications of histone residues (e.g. acetylation, methylation) and ATP-dependent nucleosome position reorganization.
Seligson et al. (16) showed that PCa cells have global level modifications in individual histones and that altered patterns of these modifications are predictive of clinical outcome. Polycomb group protein EZH2 causes methylation of histone H3 lysine 9 and histone H3 lysine 27 and its overexpression is associated with poor prognosis (30-32). The p300/CBP histone acetyltransferase (HAT) causes acetylation of all four core histone residues of the nucleosome. Hence, modifications of the nucleosome’s net charge by neutralizing the positive charge of lysine ɛ-amino group alters DNA-histone interactions (cross-talk), which then modify transcriptional activity of the cell (33). Also, other nucleosome assembly proteins functionally interact and augment the activity of p300/CBP, and the presence of core histones appears to regulate the interaction between p300 and key nucleosome assembly proteins that establish various chromatin organization states, impacting nuclear structure (nuclear importins and Lamins A & C) and functions (i.e. cell proliferation, DNA repair etc.) (34).
The p300 HAT domain is essential for physiological processes of cell proliferation, differentiation and apoptosis (35-37). Mammals lacking p300 gene exhibit defects in neurulation, cell proliferation and heart development (38). In addition to histone modifications, p300/CBP can acetylate and modify activity of several non-histone proteins [reviewed in Ref.(39)] including p53 (40,41), HMG I(Y) (42), HMG14 (43), GATA-1 (44,45), c-Myb (46), E2F-1 (47), EKLF (48), ACTR, TIF2, SRC-1 (49), Tat (50,51), TCF (52), TFIIE and TFIIF (53). Further, p300/CBP depletion causes cyclin E down-regulation (17), which in association with CDK2, controls DNA replication, centrosome duplication and histone gene expression (54).
Additionally, p300/CBP is required for effective ligand-dependent gene activation by nuclear receptor (55). The p300 protein acetylates the androgen receptor (AR) at three lysine residues in its DNA binding domain (21). Point mutations in these AR acetylation sites selectively prevent androgen-induction of androgen responsive genes, hampers coactivation of the AR by SRC-1, p300, Tip60 and Ubc9, and results in a 10-fold increase in the binding of the co-repressor NCoR (56). High levels of AR are associated with aggressive clinicopathologic parameters and decreased PCa recurrence free survival (57). Furthermore, IL-6 cytokine mediated transactivation of AR-dependent genes in the absence of androgens requires p300 HAT activity, implicating p300 in PCa progression (20).
The role of p300 in PCa molecular pathogenesis is an important event that impacts transcription of several genes involved in key pathways critical to PCa recurrence and progression. Hence, our observation on the prognostic clinical value of p300 protein expression and its potential role in transcription and effects on chromatin organization provide confirmation of results from other laboratories (16,20-22,37,38,43,55,58) and extend our understanding of its role in PCa progression.
In conclusion, p300 expression in PCa tissue may be a useful biomarker for predicting progression and is one step in a series of finding additional tissue biomarkers that will improve early prognostic decisions on PCa patient management.
Supplementary Material
Acknowledgments
Funding was provided by Johns Hopkins University Prostate Cancer Specialized Programs of Research Excellence (SPORE) [Grant number: P50CA58236], the Patana Fund and the Early Detection Research Network (EDRN) of the National Cancer Institute [Grant number: CA086323-06].
References
- 1.Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ. Cancer statistics, 2007. CA: a cancer journal for clinicians. 2007;57(1):43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- 2.Pound CR, Partin AW, Eisenberger MA, Chan DW, Pearson JD, Walsh PC. Natural history of progression after PSA elevation following radical prostatectomy. Jama. 1999;281(17):1591–1597. doi: 10.1001/jama.281.17.1591. [DOI] [PubMed] [Google Scholar]
- 3.Han M, Partin AW, Zahurak M, Piantadosi S, Epstein JI, Walsh PC. Biochemical (prostate specific antigen) recurrence probability following radical prostatectomy for clinically localized prostate cancer. J Urol. 2003;169(2):517–523. doi: 10.1097/01.ju.0000045749.90353.c7. [DOI] [PubMed] [Google Scholar]
- 4.DeMarzo AM, Nelson WG, Isaacs WB, Epstein JI. Pathological and molecular aspects of prostate cancer. Lancet. 2003;361(9361):955–964. doi: 10.1016/S0140-6736(03)12779-1. [DOI] [PubMed] [Google Scholar]
- 5.Nelson WG, De Marzo AM, Isaacs WB. Prostate cancer. The New England journal of medicine. 2003;349(4):366–381. doi: 10.1056/NEJMra021562. [DOI] [PubMed] [Google Scholar]
- 6.Rhodes DR, Sanda MG, Otte AP, Chinnaiyan AM, Rubin MA. Multiplex biomarker approach for determining risk of prostate-specific antigen-defined recurrence of prostate cancer. Journal of the National Cancer Institute. 2003;95(9):661–668. doi: 10.1093/jnci/95.9.661. [DOI] [PubMed] [Google Scholar]
- 7.Tricoli JV, Schoenfeldt M, Conley BA. Detection of prostate cancer and predicting progression: current and future diagnostic markers. Clin Cancer Res. 2004;10(12 Pt 1):3943–3953. doi: 10.1158/1078-0432.CCR-03-0200. [DOI] [PubMed] [Google Scholar]
- 8.Veltri R. Molecular biology of serum biomarkers of prostate cancer. In: Kirby R, Partin A, Feneley M, Parsons J, editors. Prostate cancer: Principles and Practice. London & New York: Taylor & Francis; 2006. pp. 269–284. [Google Scholar]
- 9.Epstein JI, Allsbrook WC, Jr, Amin MB, Egevad LL. The 2005 International Society of Urological Pathology (ISUP) Consensus Conference on Gleason Grading of Prostatic Carcinoma. The American journal of surgical pathology. 2005;29(9):1228–1242. doi: 10.1097/01.pas.0000173646.99337.b1. [DOI] [PubMed] [Google Scholar]
- 10.Gleason DF. Histologic grading of prostate cancer: a perspective. Human pathology. 1992;23(3):273–279. doi: 10.1016/0046-8177(92)90108-f. [DOI] [PubMed] [Google Scholar]
- 11.Veltri RW, Partin AW, Epstein JE, Marley GM, Miller CM, Singer DS, Patton KP, Criley SR, Coffey DS. Quantitative nuclear morphometry, Markovian texture descriptors, and DNA content captured on a CAS-200 Image analysis system, combined with PCNA and HER-2/neu immunohistochemistry for prediction of prostate cancer progression. Journal of cellular biochemistry. 1994;19:249–258. [PubMed] [Google Scholar]
- 12.Veltri RW, Partin AW, Miller MC. Quantitative nuclear grade (QNG): a new image analysis-based biomarker of clinically relevant nuclear structure alterations. Journal of cellular biochemistry. 2000;(Suppl 35):151–157. doi: 10.1002/1097-4644(2000)79:35+<151::aid-jcb1139>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 13.Veltri RW, O’Dowd GJ, Orozco R, Miller MC. The role of biopsy pathology, quantitative nuclear morphometry, and biomarkers in the preoperative prediction of prostate cancer staging and prognosis. Seminars in urologic oncology. 1998;16(3):106–117. [PubMed] [Google Scholar]
- 14.Mohler JL, Figlesthaler WM, Zhang XZ, Partin AW, Maygarden SJ. Nuclear shape analysis for the assessment of local invasion and metastases in clinically localized prostate carcinoma. Cancer. 1994;74(11):2996–3001. doi: 10.1002/1097-0142(19941201)74:11<2996::aid-cncr2820741117>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 15.Khan MA, Walsh PC, Miller MC, Bales WD, Epstein JI, Mangold LA, Partin AW, Veltri RW. Quantitative alterations in nuclear structure predict prostate carcinoma distant metastasis and death in men with biochemical recurrence after radical prostatectomy. Cancer. 2003;98(12):2583–2591. doi: 10.1002/cncr.11852. [DOI] [PubMed] [Google Scholar]
- 16.Seligson DB, Horvath S, Shi T, Yu H, Tze S, Grunstein M, Kurdistani SK. Global histone modification patterns predict risk of prostate cancer recurrence. Nature. 2005;435(7046):1262–1266. doi: 10.1038/nature03672. [DOI] [PubMed] [Google Scholar]
- 17.Bandyopadhyay D, Okan NA, Bales E, Nascimento L, Cole PA, Medrano EE. Down-regulation of p300/CBP histone acetyltransferase activates a senescence checkpoint in human melanocytes. Cancer research. 2002;62(21):6231–6239. [PubMed] [Google Scholar]
- 18.Muraoka M, Konishi M, Kikuchi-Yanoshita R, Tanaka K, Shitara N, Chong JM, Iwama T, Miyaki M. p300 gene alterations in colorectal and gastric carcinomas. Oncogene. 1996;12(7):1565–1569. [PubMed] [Google Scholar]
- 19.Fan S, Ma YX, Wang C, Yuan RQ, Meng Q, Wang JA, Erdos M, Goldberg ID, Webb P, Kushner PJ, Pestell RG, Rosen EM. p300 Modulates the BRCA1 inhibition of estrogen receptor activity. Cancer research. 2002;62(1):141–151. [PubMed] [Google Scholar]
- 20.Debes JD, Schmidt LJ, Huang H, Tindall DJ. p300 mediates androgen-independent transactivation of the androgen receptor by interleukin 6. Cancer research. 2002;62(20):5632–5636. [PubMed] [Google Scholar]
- 21.Fu M, Wang C, Reutens AT, Wang J, Angeletti RH, Siconolfi-Baez L, Ogryzko V, Avantaggiati ML, Pestell RG. p300 and p300/cAMP-response element-binding protein-associated factor acetylate the androgen receptor at sites governing hormone-dependent transactivation. The Journal of biological chemistry. 2000;275(27):20853–20860. doi: 10.1074/jbc.M000660200. [DOI] [PubMed] [Google Scholar]
- 22.Debes JD, Sebo TJ, Lohse CM, Murphy LM, Haugen DA, Tindall DJ. p300 in prostate cancer proliferation and progression. Cancer research. 2003;63(22):7638–7640. [PubMed] [Google Scholar]
- 23.Veltri RW, Miller MC, Isharwal S, Marlow C, Makarov DV, Partin AW. Prediction of Prostate-Specific Antigen Recurrence in Men with Long-term Follow-up Postprostatectomy Using Quantitative Nuclear Morphometry. Cancer Epidemiol Biomarkers Prev. 2008;17(1):102–110. doi: 10.1158/1055-9965.EPI-07-0175. [DOI] [PubMed] [Google Scholar]
- 24.Berman JJ, Datta M, Kajdacsy-Balla A, Melamed J, Orenstein J, Dobbin K, Patel A, Dhir R, Becich MJ. The tissue microarray data exchange specification: implementation by the Cooperative Prostate Cancer Tissue Resource. BMC bioinformatics. 2004;5:19. doi: 10.1186/1471-2105-5-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Melamed J, Datta MW, Becich MJ, Orenstein JM, Dhir R, Silver S, Fidelia-Lambert M, Kadjacsy-Balla A, Macias V, Patel A, Walden PD, Bosland MC, Berman JJ. The cooperative prostate cancer tissue resource: a specimen and data resource for cancer researchers. Clin Cancer Res. 2004;10(14):4614–4621. doi: 10.1158/1078-0432.CCR-04-0240. [DOI] [PubMed] [Google Scholar]
- 26.Patel AA, Kajdacsy-Balla A, Berman JJ, Bosland M, Datta MW, Dhir R, Gilbertson J, Melamed J, Orenstein J, Tai KF, Becich MJ. The development of common data elements for a multi-institute prostate cancer tissue bank: the Cooperative Prostate Cancer Tissue Resource (CPCTR) experience. BMC cancer. 2005;5:108. doi: 10.1186/1471-2407-5-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liao Z, Datta MW. A simple computer program for calculating PSA recurrence in prostate cancer patients. BMC urology. 2004;4:8. doi: 10.1186/1471-2490-4-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Veltri RW, Marlow C, Khan MA, Miller MC, Epstein JI, Partin AW. Significant variations in nuclear structure occur between and within Gleason grading patterns 3, 4, and 5 determined by digital image analysis. The Prostate. 2007;67(11):1202–1210. doi: 10.1002/pros.20614. [DOI] [PubMed] [Google Scholar]
- 29.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389(6648):251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 30.Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Science. 5595. Vol. 298. New York, NY: 2002. Role of histone H3 lysine 27 methylation in Polycomb-group silencing; pp. 1039–1043. [DOI] [PubMed] [Google Scholar]
- 31.Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell. 2002;111(2):185–196. doi: 10.1016/s0092-8674(02)00975-3. [DOI] [PubMed] [Google Scholar]
- 32.Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, Ghosh D, Pienta KJ, Sewalt RG, Otte AP, Rubin MA, Chinnaiyan AM. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419(6907):624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 33.Vo N, Goodman RH. CREB-binding protein and p300 in transcriptional regulation. The Journal of biological chemistry. 2001;276(17):13505–13508. doi: 10.1074/jbc.R000025200. [DOI] [PubMed] [Google Scholar]
- 34.Shikama N, Chan HM, Krstic-Demonacos M, Smith L, Lee CW, Cairns W, La Thangue NB. Functional interaction between nucleosome assembly proteins and p300/CREB-binding protein family coactivators. Molecular and cellular biology. 2000;20(23):8933–8943. doi: 10.1128/mcb.20.23.8933-8943.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goodman RH, Smolik S. CBP/p300 in cell growth, transformation, and development. Genes & development. 2000;14(13):1553–1577. [PubMed] [Google Scholar]
- 36.Giordano A, Avantaggiati ML. p300 and CBP: partners for life and death. Journal of cellular physiology. 1999;181(2):218–230. doi: 10.1002/(SICI)1097-4652(199911)181:2<218::AID-JCP4>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 37.Ait-Si-Ali S, Polesskaya A, Filleur S, Ferreira R, Duquet A, Robin P, Vervish A, Trouche D, Cabon F, Harel-Bellan A. CBP/p300 histone acetyl-transferase activity is important for the G1/S transition. Oncogene. 2000;19(20):2430–2437. doi: 10.1038/sj.onc.1203562. [DOI] [PubMed] [Google Scholar]
- 38.Yao TP, Oh SP, Fuchs M, Zhou ND, Ch’ng LE, Newsome D, Bronson RT, Li E, Livingston DM, Eckner R. Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell. 1998;93(3):361–372. doi: 10.1016/s0092-8674(00)81165-4. [DOI] [PubMed] [Google Scholar]
- 39.Sterner DE, Berger SL. Acetylation of histones and transcription-related factors. Microbiol Mol Biol Rev. 2000;64(2):435–459. doi: 10.1128/mmbr.64.2.435-459.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu L, Scolnick DM, Trievel RC, Zhang HB, Marmorstein R, Halazonetis TD, Berger SL. p53 sites acetylated in vitro by PCAF and p300 are acetylated in vivo in response to DNA damage. Molecular and cellular biology. 1999;19(2):1202–1209. doi: 10.1128/mcb.19.2.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90(4):595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 42.Munshi N, Merika M, Yie J, Senger K, Chen G, Thanos D. Acetylation of HMG I(Y) by CBP turns off IFN beta expression by disrupting the enhanceosome. Molecular cell. 1998;2(4):457–467. doi: 10.1016/s1097-2765(00)80145-8. [DOI] [PubMed] [Google Scholar]
- 43.Bergel M, Herrera JE, Thatcher BJ, Prymakowska-Bosak M, Vassilev A, Nakatani Y, Martin B, Bustin M. Acetylation of novel sites in the nucleosomal binding domain of chromosomal protein HMG-14 by p300 alters its interaction with nucleosomes. The Journal of biological chemistry. 2000;275(15):11514–11520. doi: 10.1074/jbc.275.15.11514. [DOI] [PubMed] [Google Scholar]
- 44.Hung HL, Lau J, Kim AY, Weiss MJ, Blobel GA. CREB-Binding protein acetylates hematopoietic transcription factor GATA-1 at functionally important sites. Molecular and cellular biology. 1999;19(5):3496–3505. doi: 10.1128/mcb.19.5.3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boyes J, Byfield P, Nakatani Y, Ogryzko V. Regulation of activity of the transcription factor GATA-1 by acetylation. Nature. 1998;396(6711):594–598. doi: 10.1038/25166. [DOI] [PubMed] [Google Scholar]
- 46.Tomita A, Towatari M, Tsuzuki S, Hayakawa F, Kosugi H, Tamai K, Miyazaki T, Kinoshita T, Saito H. c-Myb acetylation at the carboxyl-terminal conserved domain by transcriptional co-activator p300. Oncogene. 2000;19(3):444–451. doi: 10.1038/sj.onc.1203329. [DOI] [PubMed] [Google Scholar]
- 47.Marzio G, Wagener C, Gutierrez MI, Cartwright P, Helin K, Giacca M. E2F family members are differentially regulated by reversible acetylation. The Journal of biological chemistry. 2000;275(15):10887–10892. doi: 10.1074/jbc.275.15.10887. [DOI] [PubMed] [Google Scholar]
- 48.Zhang W, Bieker JJ. Acetylation and modulation of erythroid Kruppel-like factor (EKLF) activity by interaction with histone acetyltransferases. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(17):9855–9860. doi: 10.1073/pnas.95.17.9855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen H, Lin RJ, Xie W, Wilpitz D, Evans RM. Regulation of hormone-induced histone hyperacetylation and gene activation via acetylation of an acetylase. Cell. 1999;98(5):675–686. doi: 10.1016/s0092-8674(00)80054-9. [DOI] [PubMed] [Google Scholar]
- 50.Kiernan RE, Vanhulle C, Schiltz L, Adam E, Xiao H, Maudoux F, Calomme C, Burny A, Nakatani Y, Jeang KT, Benkirane M, Van Lint C. HIV-1 tat transcriptional activity is regulated by acetylation. The EMBO journal. 1999;18(21):6106–6118. doi: 10.1093/emboj/18.21.6106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ott M, Schnolzer M, Garnica J, Fischle W, Emiliani S, Rackwitz HR, Verdin E. Acetylation of the HIV-1 Tat protein by p300 is important for its transcriptional activity. Curr Biol. 1999;9(24):1489–1492. doi: 10.1016/s0960-9822(00)80120-7. [DOI] [PubMed] [Google Scholar]
- 52.Waltzer L, Bienz M. Drosophila CBP represses the transcription factor TCF to antagonize Wingless signalling. Nature. 1998;395(6701):521–525. doi: 10.1038/26785. [DOI] [PubMed] [Google Scholar]
- 53.Imhof A, Yang XJ, Ogryzko VV, Nakatani Y, Wolffe AP, Ge H. Acetylation of general transcription factors by histone acetyltransferases. Curr Biol. 1997;7(9):689–692. doi: 10.1016/s0960-9822(06)00296-x. [DOI] [PubMed] [Google Scholar]
- 54.Ewen ME. Where the cell cycle and histones meet. Genes & development. 2000;14(18):2265–2270. doi: 10.1101/gad.842100. [DOI] [PubMed] [Google Scholar]
- 55.Chakravarti D, LaMorte VJ, Nelson MC, Nakajima T, Schulman IG, Juguilon H, Montminy M, Evans RM. Role of CBP/P300 in nuclear receptor signalling. Nature. 1996;383(6595):99–103. doi: 10.1038/383099a0. [DOI] [PubMed] [Google Scholar]
- 56.Fu M, Rao M, Wang C, Sakamaki T, Wang J, Di Vizio D, Zhang X, Albanese C, Balk S, Chang C, Fan S, Rosen E, Palvimo JJ, Janne OA, Muratoglu S, Avantaggiati ML, Pestell RG. Acetylation of androgen receptor enhances coactivator binding and promotes prostate cancer cell growth. Molecular and cellular biology. 2003;23(23):8563–8575. doi: 10.1128/MCB.23.23.8563-8575.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li R, Wheeler T, Dai H, Frolov A, Thompson T, Ayala G. High level of androgen receptor is associated with aggressive clinicopathologic features and decreased biochemical recurrence-free survival in prostate: cancer patients treated with radical prostatectomy. The American journal of surgical pathology. 2004;28(7):928–934. doi: 10.1097/00000478-200407000-00013. [DOI] [PubMed] [Google Scholar]
- 58.Yao TP, Ku G, Zhou N, Scully R, Livingston DM. The nuclear hormone receptor coactivator SRC-1 is a specific target of p300. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(20):10626–10631. doi: 10.1073/pnas.93.20.10626. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.