SUMMARY
The microbiome is being characterized by large-scale sequencing efforts, yet it is not known whether it regulates host metabolism in a general versus tissue-specific manner or which bacterial metabolites are important. Here, we demonstrate that microbiota have a strong effect on energy homeostasis in the colon compared to other tissues. This tissue specificity is due to colonocytes utilizing bacterially-produced butyrate as their primary energy source. Colonocytes from germfree mice are in an energy-deprived state and exhibit decreased expression of enzymes that catalyze key steps in intermediary metabolism including the TCA cycle. Consequently, there is a marked decrease in NADH/NAD+, oxidative phosphorylation, and ATP levels, which results in AMPK activation, p27kip1 phosphorylation, and autophagy. When butyrate is added to germfree colonocytes, it rescues their deficit in mitochondrial respiration and prevents them from undergoing autophagy. The mechanism is due to butyrate acting as an energy source rather than as an HDAC inhibitor.
INTRODUCTION
Diverse microbial communities reside at various sites within the human body (Camp et al., 2009; Eckburg et al., 2005; Savage, 1977). These microbiota and their genomes, referred to collectively as the microbiome, are being characterized by metagenomic sequencing as part of the Human Microbiome Project (Gill et al., 2006; Hildebrandt et al., 2009; Kurokawa et al., 2007; Qin et al.; Turnbaugh et al., 2009; Turnbaugh et al., 2007). The vast majority of microbes are bacteria that reside in the gut and are estimated to number 100 trillion, which is 10-fold greater than all of somatic and germ cell in the human body (Savage, 1977; Turnbaugh and Gordon, 2009). Moreover, taking the genetic diversity of the microbiome into account, it is estimated to harbor at least 100-fold more genes than the human genome (Hooper and Gordon, 2001). Based on current 16S and metagenomic sequence data, the gut microbiome is highly enriched for genes involved in energy production and metabolism (Gill et al., 2006; Qin et al.; Turnbaugh et al., 2009). These findings suggest that microbiota help facilitate the host’s ability to extract calories from their diet, but sequence-based data must be validated by experiments that investigate function.
To investigate the effect of the microbiome in host metabolism, germfree (GF) animals lacking microbiota have been studied (Gordon and Pesti, 1971; Hooper and Gordon, 2001; Wostmann, 1981). For example, GF mice have been compared to genetically identical mice that were raised conventionally (CONV-R) with “normal”, albeit undefined, microbiota. These studies support the idea that microbes increase host metabolic efficiency (Backhed et al., 2004; Turnbaugh et al., 2008; Turnbaugh and Gordon, 2009; Turnbaugh et al., 2009; Turnbaugh et al., 2006). For example, GF mice must consume 10–30% more food to maintain the same body weight as CONV-R controls (Backhed et al., 2004; Gordon and Pesti, 1971). Despite this increased food intake, GF mice are leaner with a ~40% decrease in the size of their epididymal fat pads (Backhed et al., 2004). They have a similar decrease in liver glycogen levels. GF mice also have lower blood glucose and insulin levels and are resistant to obesity induced by a high-fat diet (Backhed et al., 2007). However, these findings are complicated by the observation that GF mice exhibit increased locomotor activity (Backhed et al., 2007). Therefore, the increased food consumption and decreased body fat of GF mice may simply be due to increased energy expenditure. To demonstrate that microbiota directly affect metabolism, individual tissues from GF and CONV-R mice must be assessed for differences in key metabolic parameters (Camp et al., 2009). This approach has the potential to reveal general mechanisms as to how microbiota regulate metabolism as well as to identify tissue-specific differences.
RESULTS
Microbiota Influence Energy Homeostasis in the Colon
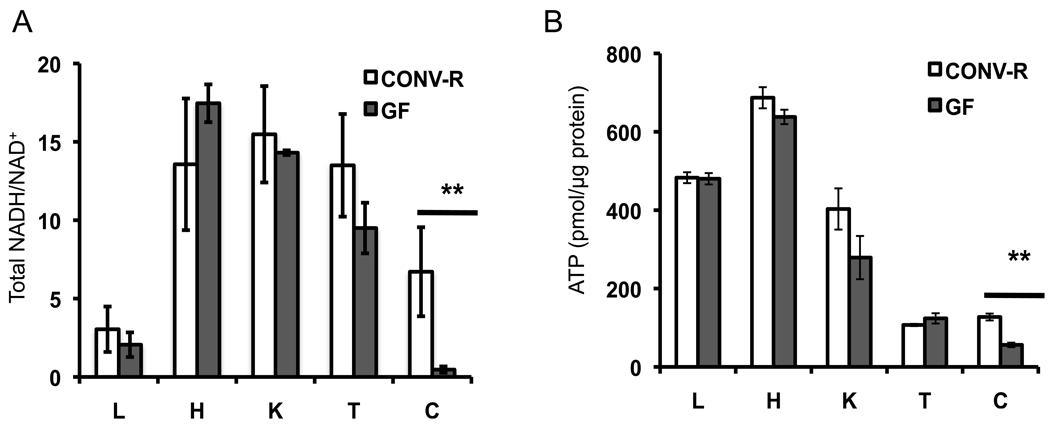
To investigate whether microbiota have tissue-specific effects on host metabolism, we analyzed two key biomarkers of energy homeostasis, NADH/NAD+ ratios and ATP levels, in several tissues from GF and CONV-R mice. We found no significant differences for either biomarker in liver, heart, kidney, or testis (Figure 1A, B). These results are consistent with previous studies from liver and heart of non-fasted GF and CONV-R mice (Backhed et al., 2007; Crawford et al., 2009). In stark contrast, NADH/NAD+ and ATP levels are significantly diminished in GF colon by 16 fold and 56%, respectively. These results indicate that microbiota have a particularly important role in regulating host metabolism in the colon.
Figure 1. Effects of Microbiota on Energy Homeostasis.
Total NADH/NAD+ ratios (A) and ATP levels (B) in various tissues from GF and CONV-R mice. L, liver; H, heart; K, kidney; T, testis; C, colon. 6 GF and 6 CONV-R mice were analyzed, and results are mean ± SEM. Significant differences are indicated (**p <0.01).
Microbiota Regulate the Abundance of mRNAs and Proteins Involved in Metabolism
To understand the effects of microbiota on colonocyte metabolism, we performed transcriptome profiling and quantitative mass spectrometry on freshly isolated colonocytes from GF and CONV-R mice. For the transcriptome profiling experiments, we identified 624 genes that are upregulated and 813 genes are downregulated in GF compared to CONV-R. All of these gene expression changes are statistically significant with false discovery rates below 0.01. The entire data set and statistical values are included as Table S1. Our transcriptome data includes genes previously shown to be regulated by microbiota in various tissues of zebrafish, mice, and piglets. For example, Angpt14/Fiaf is upregulated in GF compared to CONV while Gpx2, ApoB, Aqp4, and Pla2g4c are downregulated (Backhed et al., 2004; Chowdhury et al., 2007; Cresci et al.; Esworthy et al., 2003; Rawls et al., 2004). Several genes from our list were validated by RT-qPCR. In each case, the RT-qPCR data confirm the transcriptome profiling result (Figure S1A).
We also used quantitative mass spectrometry to assess proteomic differences between GF and CONV-R colonocytes. Using a two-dimensional shotgun mass spectrometry approach, we identified 2120 proteins per sample with no significant difference in the number of proteins identified between GF and CONV-R samples. Poisson regression analysis and normalization of individual spectral counts revealed that 136 proteins were upregulated and 80 proteins were downregulated in GF compared to CONV-R (p-values < 0.05). The entire protein list and statistical values are included as Table S2. A number of the differentially expressed proteins were confirmed by western blot analysis (Figure S1B).
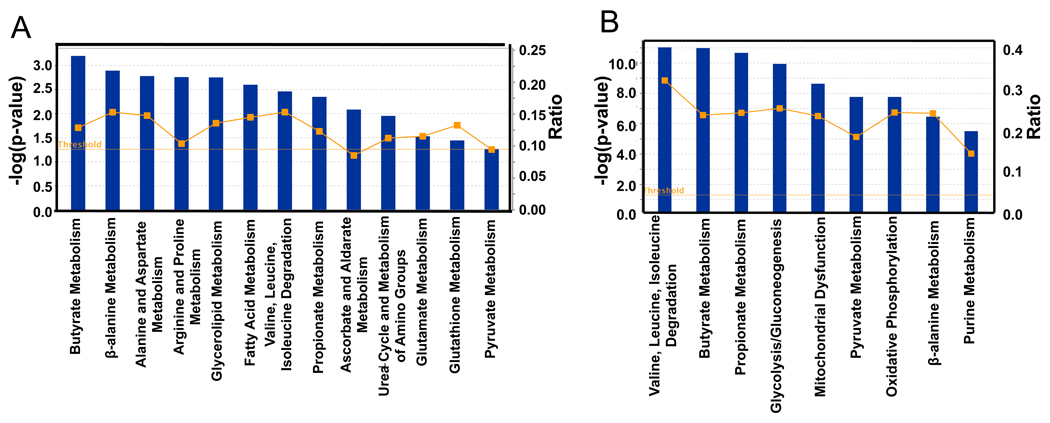
To identify biological pathways that are regulated by microbiota, we used Ingenuity Pathway Analysis (IPA) to analyze differences in gene expression. For both the transcriptome and proteome data, all of the top KEGG (Kyoto Encyclopedia of Genes and Genomes) categories have metabolic functions (Figure 2). There was significant concordance with the majority of the KEGG categories at the proteome level (Figure 2B) also being represented at the transcriptome level (Figure 2A). This overlap includes pathways that metabolize butyrate, which is a short-chain fatty acid (SCFAs) produced by microbes in the lumen of the colon at high (mM) levels. Butyrate is noteworthy because it is a histone deacetylase (HDAC) inhibitor that regulates gene expression and also serves as the primary energy source for the colonic epithelium (Boffa et al., 1978; Candido et al., 1978; Dashwood et al., 2006; Garcea and Alberts, 1980; Roediger, 1980, 1982). Considering that butyrate is a fatty acid oxidized in the mitochondria, it is reasonable that genes involved in fatty acid metabolism, glycerolipid metabolism, the tricarboxylic acid (TCA) cycle, oxidative phosphorylation, and mitochondrial dysfunction are also among the top KEGG categories (Figure 2).
Figure 2. Analysis of Microbiome Regulated Gene Products and Metabolites.
Top KEGG pathways of transcriptome (A) and proteome (B) identified as differences between GF and CONV-R. Blue bars correspond to −log (p-values) shown on y-axis at left. Straight yellow lines represent threshold of significance for these −log (p-values). Yellow boxes represent the ratio, shown on y-axis at right, and indicate the number of genes or proteins affected divided by total number within category. 6 GF and 6 CONV-R mice were used for the transcriptome analysis. For the proteome analysis, 3 GF and 3 CONV-R mice were used.
To provide additional insight into the role of microbiota in metabolism, we performed 1H NMR-based metabolomics on GF and CONV-R colonocytes. A principal components analysis scores plot revealed significant differences between GF and CONV-R samples (Figure S2). A number of metabolites differ between GF and CONV-R including glucose, lactate, creatine, glutamate, various amino acids, and 3-hydroxybutyrate (Figure S3). A number of amino acids, including alanine, glutamine, and branched chain amino acids such as isoleucine and valine, are elevated in GF colonocytes. These changes support the transcriptome and proteome data sets showing multiple KEGG categories related to amino acid metabolism. Additionally, 3–6 hydroxybutyrate (3-HB), a metabolic product of fatty acid and butyrate metabolism (two of the top KEGG categories), was significantly decreased in GF compared to CONV-R colonocytes.
Microbial Regulation of Oxidative Metabolism
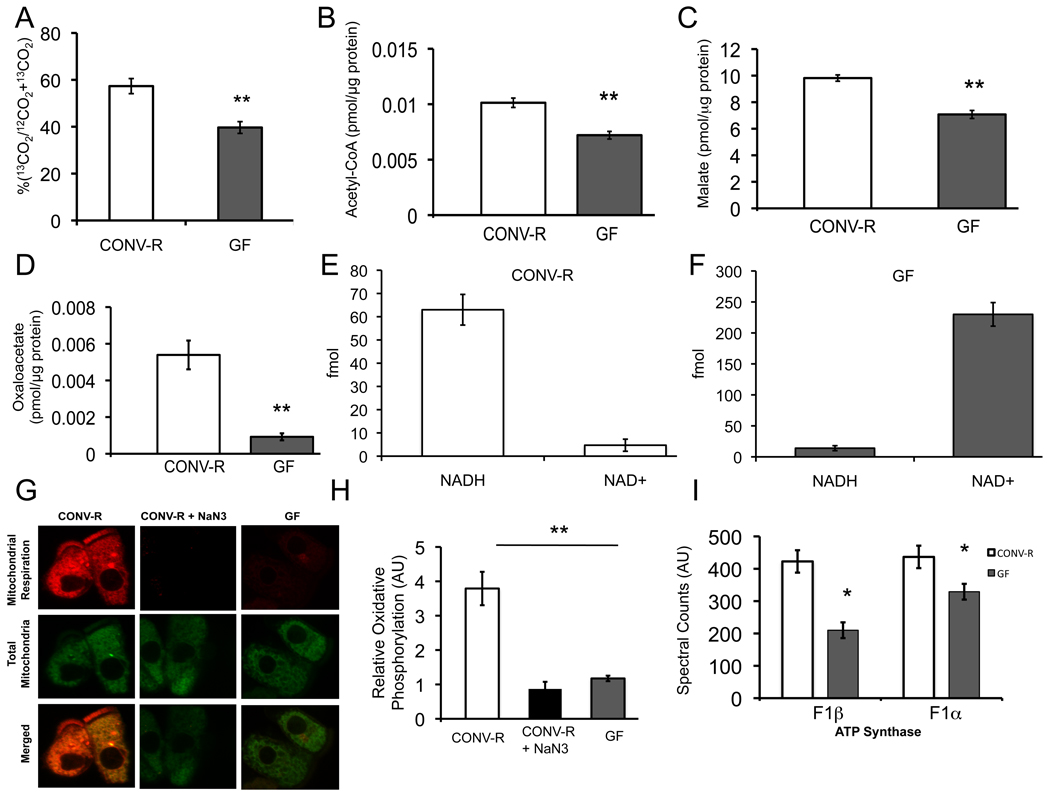
Based on our transcriptome and proteome data, microbiota regulate enzymes that catalyze reactions in intermediary metabolism. For example, 3 out of 6 enzymes involved in β-oxidation of fatty acids are downregulated including the acyl CoA dehydrogenase specific for SCFAs such as butyrate. To confirm that fatty-acid oxidation is diminished, we incubated freshly isolated colonocytes from CONV-R and GF mice with 13C-butyrate and measured conversion to labeled CO2. These experiments indicate GF colonocytes have a 31% decrease in labeled CO2 compared to CONV-R (Figure. 3A).
Figure 3. Regulation of Colonocyte Oxidative Metabolism.
(A) Percentage of 13C-butyrate metabolized to 13CO2 in CONV-R and GF colonocytes. (B–D) Measurement of intermediary metabolites. Levels of acetyl-CoA (B), malate (C), and oxaloacetate (D) in CONV-R and GF. (E, F) Levels of mitochondrial NADH and NAD+ in CONV-R (E) and GF (F) colonocytes. The different scales reflect different amounts of material loaded and detected by HPLC-LC-MS. (G) Oxidative metabolism indicated by MitoTracker Red CM-H2XRos (red fluorescence; top panel) and MitoTracker Green FM (green fluorescence; middle panel) in colonocytes from CONV-R, CONV-R with NaN3 (sodium azide, negative control), or GF. (H) Quantification of oxidative metabolism in different experimental groups. (I) Spectral counts from quantitative mass spectrometry of ATP synthase F1α and β. Results in A–D, G, and H are displayed as mean ± SE and significant differences are identified (*p <0.05, **p <0.01). For panels A–D, 4 GF and 4 CONV-R mice were used. In panels E–H, 3 mice were used per condition.
In addition to fatty-acid oxidation, the pyruvate dehydrogenase (PDH) complex is the other major pathway that gives rise to acetyl-CoA, and it is also downregulated in GF colonocytes. Therefore, we measured acetyl-CoA levels and observed a 30% decrease in GF compared to CONV-R (Figure 3B). Acetyl-CoA enters the TCA cycle, and 5 out of 8 TCA cycle enzymes are downregulated at the mRNA and/or protein level. To test whether diminished substrate and enzymes levels result in decreased TCA cycle function, we measured two TCA cycle intermediates. As expected, both malate and oxaloacetate are significantly diminished in GF (Figure 3C, D).
A primary objective of the TCA cycle is to reduce NAD+ to NADH, which enters the electron transport chain where oxidative phosphorylation culminates in ATP production. Diminished TCA cycle activity in GF colonocytes should result in a diminished NADH/NAD+ ratio in the mitochondria. To test this hypothesis, we isolated mitochondria from GF and CONV-R colonocytes and measured NADH and NAD+ by HPLC/LC-MS (Figure S4). Quantification of these data demonstrated that the mitochondrial NADH/NAD+ ratio was 0.061 ± 0.022 in GF compared to 13.56 ± 1.2 for CONV-R (Figure 3E, F).
To test whether the altered redox state results in diminished oxidative phosphorylation, we isolated colonocytes from GF and CONV-R mice and incubated them ex vivo with two mitotracker probes simultaneously. One probe fluoresced red upon being oxidized and provided a measure of oxidative phosphorylation (i.e., mitochondrial respiration), while the other probe fluoresced green regardless of mitochondrial respiration and therefore served as an internal control. As shown in Figure 3G, GF colonocytes exhibit a marked decrease in mitochondrial respiration (red in top row) but no difference in total mitochondria (green in middle row). The extent to which oxidative respiration is decreased in GF colonocytes is surprising because it more closely resembles sodium azide (NaN3)-treated negative controls than the CONV-R samples. Quantification of differences between these groups revealed a 70% drop in the rate of oxidative phosphorylation in GF compared to CONV-R (Figure 3H). These results are consistent with a significantly diminished mitochondrial NADH/NAD+ ratio (Figure 3E, F) and the reduction in ATP levels (Figure 1B) previously identified. Decreased ATP levels in GF colonocytes are also compatible with decreased protein expression of both ΰ and β catalytic subunits of F1-ATP synthase (Figure 3I, Figure S1B).
Energy Deprivation in GF Colonocytes Leads to Autophagy
Considering that GF colonocytes have ATP levels that are less than half of CONV-R controls, one might expect them to exhibit signs of energetic stress. AMPK (5’-adenosine monophosphate-activated protein kinase) is a metabolic sensor activated by falling ATP and/or rising AMP levels (Shaw et al., 2004). As expected, GF colonocytes have increased AMPK activation based on increased Thr172 phosphorylation (Figure 4A). In addition, we discovered that total AMPK levels are elevated in GF colonocytes (Figure 4A and Figure S5).
Figure 4. Microbial Regulation of Energy Metabolism and Autophagy.
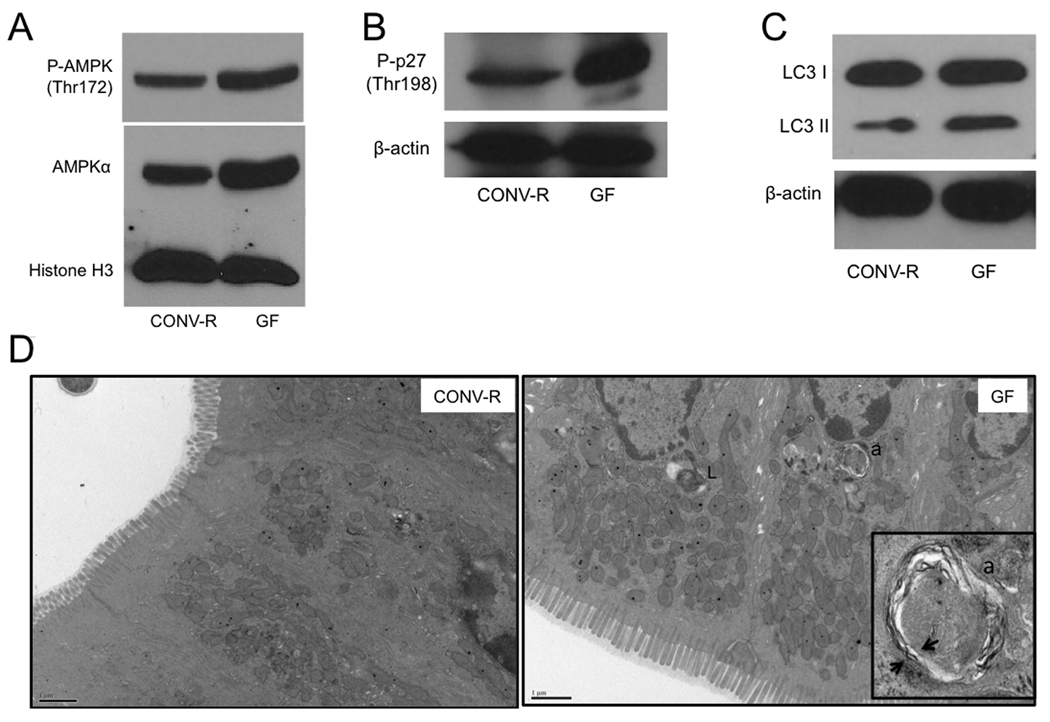
(A–C) Western blot analysis of phosho-AMPK and AMPK (A), phospho-p27 (B) and LC3-I and -II (C) with histone H3 or β-actin as loading controls in CONV-R and GF. (D) Transmission electron micrographs of CONV-R and GF colonic epithelium. Lysosomes (L) and autophagosomes (a) are indicated, and arrows show double membrane of autophagasome in inset. For A–C, western blots are representative of 4 experiments using 2 CONV-R and 2 GF mice with 2 technical replicates each.
In response to nutrient deprivation, AMPK activation induces autophagy, which is a catabolic process where the cell degrades its own cellular components in an attempt to maintain energy homeostasis (He and Klionsky, 2009; Yang and Klionsky, 2009). AMPK can induce autophagy by phosphorylating (Thr198) and stabilizing the cyclin-dependent kinase inhibitor 1B (p27kip1) (Liang et al., 2007). Therefore, we analyzed GF colonocytes and observed increased Thr198 phosphorylation compared to CONV-R controls (Figure 4B). Since AMPK and p27kip1 activation are higher in GF colonocytes, we expected that autophagy would also be increased. In support of this hypothesis, we detected increased levels of the autophagosome marker LC3-II in GF compared to CONV-R colonocytes (Figure 4C). In addition, we performed transmission electron microscopy (TEM) on colonic tissue to detect double-membrane autophagosomes. TEM ultramicrographs showed one or two autophagosomes per GF colonoycte compared to only one autophagosome for every ~30 cells in CONV-R colons (Figure. 4D). These findings indicate that microbiota decrease autophagy in the colon.
Butyrate-Producing Microbes Rescue GF Energetics and Autophagy
To demonstrate that the energy-deprived state and increased autophagy observed in GF colonocytes are a direct result of absence of microbes, we colonized GF mice with microbiota derived from CONV-R mice thus converting GF mice into CONV-D (conventionally-derived) mice. In addition to colonizing GF mice with a complete microbiota, we also colonized GF mice with a butyrate-producing bacterial strain called Butyrivibrio fibrisolvens (Diez-Gonzalez et al., 1999; Rumney et al., 1995). We then tested whether a complete colonization or colonization with B. fibrisolvens for three weeks would rescue oxidative phosphorylation, ATP levels, and autophagy. Mitotracker experiments clearly showed that CONV-D and B. fibrisolvens colonocytes exhibit the same degree of oxidative phosphorylation as CONV-R (Figure 5A, B). We also found similar ATP levels in CONV-D, B. fibrisolvens, and CONV-R colonocytes (Figure S6A). Finally, consistent with the ability of complete microbiota and B. fibrisolvens to restore energy homeostasis, these bacteria also restore autophagy to normal levels based on the LC3-II marker (Figure S6B).
Figure 5. Colonization and Butyrate Rescue Diminished Oxidative Metabolism.
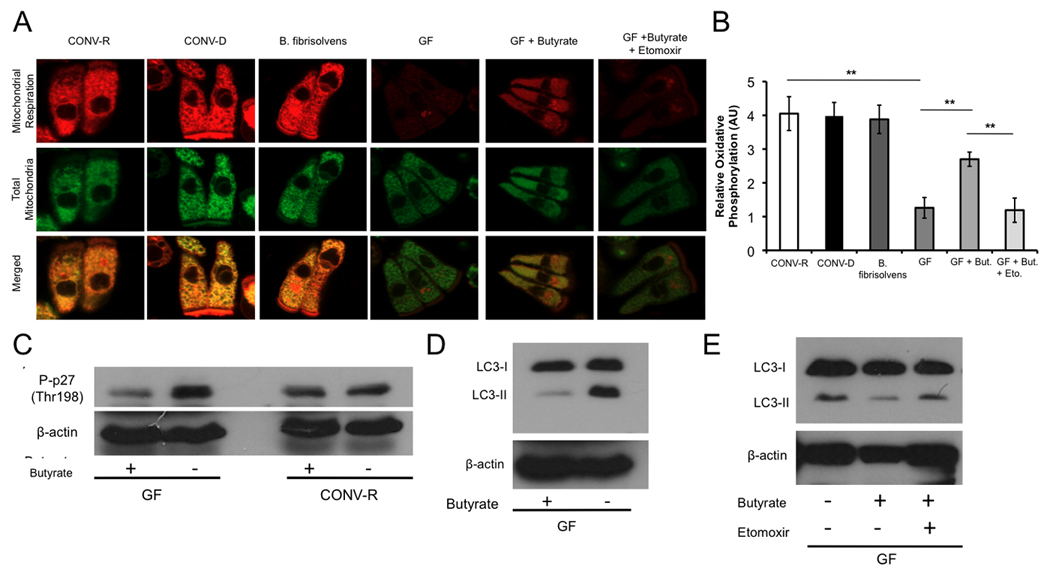
(A) Mitochondrial respiration indicated by MitoTracker Red CM-H2XRos (red fluorescence; top panel) and MitoTracker Green FM (green fluorescence; middle panel) in colonocytes from CONV-R, CONV-D, B. fibrisolvens, GF, GF with 10 mM butyrate, and GF with both 10 mM butyrate and 500 µM etomoxir. (B) Quantification of mitochondrial respiration in different experimental groups. (C–D) Western blot analysis of phospho-p27 (C) and LC3-I and -II (D) with and without 10 mM butyrate in CONV-R and GF with β-actin as loading controls. (E) Western blot analysis of LC3-I and –II of GF, GF with 10 mM butyrate, and GF with 10 mM butyrate and 500 µM etomoxir. For panels A and B, a total of 3 mice per condition were used, and results are displayed as mean ± SE, with significant differences indicated (**p <0.01). For panels C–E, western blots are representative of 4 experiments using 2 CONV-R and 2 GF mice with 2 technical replicates each.
Butyrate Functions as an Energy Source Rather than an HDAC Inhibitor
To provide direct evidence that butyrate is a causal factor, we added butyrate to GF colonocytes while they were being isolated over a 30–60 minute period. Butyrate had a dramatic effect ex vivo, increasing oxidative phosphorylation in GF from 30% to 70% of CONV-R levels (Figure 5A, B). We also tested whether butyrate inhibits autophagy in GF colonocytes. As expected, it decreases p27 phosphorylation and LC3-II compared to untreated GF colonocytes (Figure 5C, D).
Since butyrate is a SCFA and an HDAC inhibitor, it could rescue the GF defects by acting either as an energy source or epigenetic factor (Boffa et al., 1978; Candido et al., 1978; Dashwood et al., 2006; Garcea and Alberts, 1980; Roediger, 1980, 1982). To distinguish between these two possible mechanisms, we repeated the butyrate rescue experiments but added a compound that blocks fatty-acid oxidation. This pharmacological inhibitor, etomoxir, prevents butyrate from increasing oxidative phosphorylation in mitotracker experiments (Figure 5A, B). It also prevents butyrate from suppressing autophagy in GF colonocytes (Figure 5E).
The data presented above demonstrate that butyrate maintains energy homoeostasis and prevents autophagy by acting as an energy source rather than as an HDAC inhibitor. To provide additional support for this mechanism, we performed mitotracker experiments with three molecules with defined actions similar to butyrate. Propionate is a SCFA that enters the TCA through succinyl-CoA functions as an energy source and an HDAC inhibitor (Hinnebusch et al., 2002; Waldecker et al., 2008). In contrast, palmitate is a long-chain fatty acid (LCFA) that enters the TCA cycle as acetyl-CoA and only functions as an energy source, whereas Trichostatin A (TSA) only functions as an HDAC inhibitor. Consistent with the data presented above, propionate and palmitate stimulate mitochondrial respiration while TSA has no effect (Figure S7).
DISCUSSION
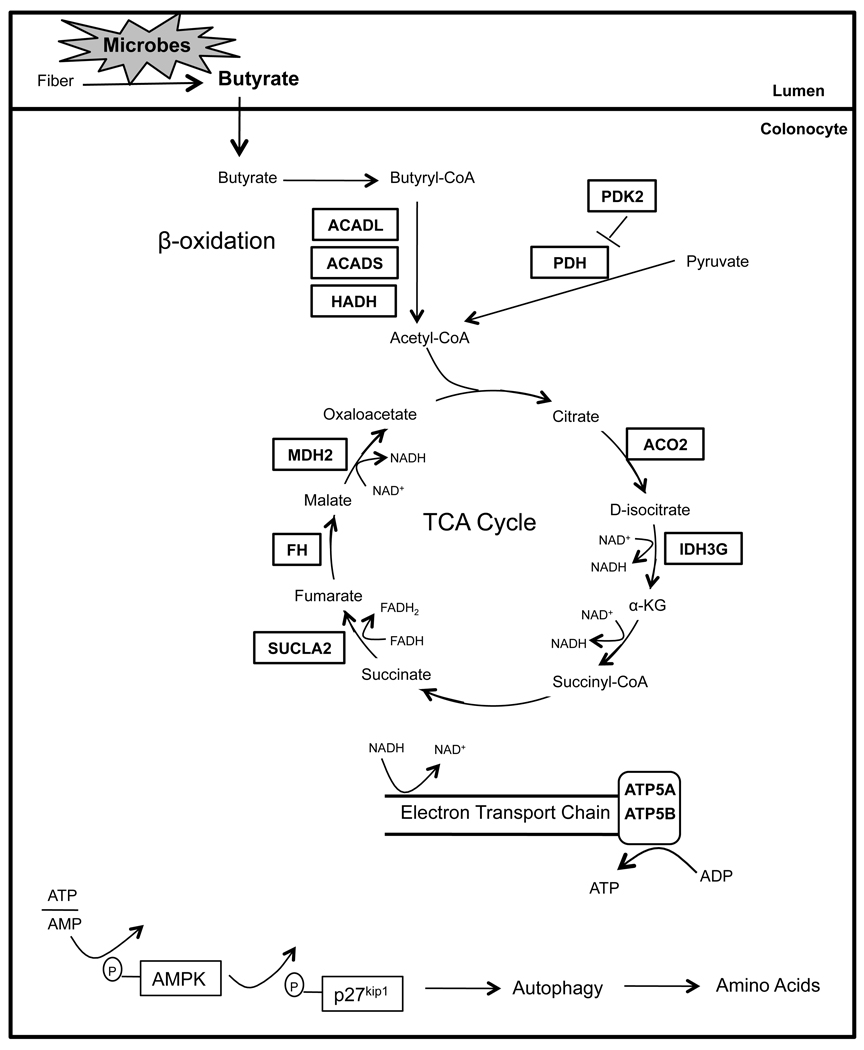
Many fundamental questions about how the microbiome regulates host metabolism have yet to be addressed. For example, does it have general or tissue-specific effects? Also, which bacterial metabolites have important biological activities? In our study, we demonstrate that microbiota have a strong effect maintaining NADH/NAD+ ratios and ATP levels in the colon but not liver, heart, kidney, or testis. We attribute this tissue specificity to the fact that colonocytes utilize bacterially-produced butyrate as their primary energy source, whereas most other cell types utilize glucose. The lumen of the colon has the highest density of microbes in the body, and specific bacteria belonging to genera such as Clostridium, Eubacterium, and Butyrivibrio ferment dietary fiber into butyrate at very high (mM) concentrations (Barcenilla et al., 2000; Pryde et al., 2002). As shown in Figure 6, butyrate is a SCFA transported into colonocytes, enters the mitochondria, and undergoes β oxidation to acetyl-CoA, which enters the TCA cycle resulting in the reduction of NAD+ to NADH. NADH enters the electron transport chain culminating in ATP production with CO2 as a byproduct.
Figure 6. Microbial Regulation of Colonocyte Metabolism.
Schematic showing that dietary fiber is fermented by microbes into butyrate in the lumen of the colon, which is transported into the colonocyte. In the colonocyte, butyrate promotes oxidative metabolism and inhibits autophagy. Based on transcriptome and proteome experiments, enzymes regulated by microbes are boxed. In all cases, boxed enzymes that function in β-oxidation and the TCA cycle are downregulated in GF colonocytes, revealing that microbes positively regulate their expression. Diminished ATP results in phosphorylation of AMPK and p27, which culminates in autophagy.
Our transcriptome and proteome experiments identified butyrate metabolism as the most affected pathway in GF colonocytes followed by related oxidative metabolic pathways. In GF colonocytes, the absence of microbial butyrate results in each one of the above metabolic processes (e.g., β oxidation, TCA cycle, oxidative phosphorylation) being diminished, which is correlated with key enzymes being downregulated (Figure 6). To demonstrate the importance of butyrate relative to other microbial metabolites, we rescued each of these processes by colonizing GF mice with butyrate-producing bacteria as well as by adding exogenous butyrate to GF colonocytes.
Considering that GF mice have colonocytes with less than half of normal ATP levels, one might expect them to exhibit signs of impaired colonic function. In fact, GF mice drink more water than CONV-R controls and exhibit mild diarrhea, both of which are indicative of perturbed water re-absorption (Gordon and Pesti, 1971). However, colonocyte function must be partially retained because GF mice do not die prematurely due to diarrhea-induced dehydration. Our results suggest this is due to GF colonocytes adapting to their suboptimal energetic state by undergoing autophagy. Reduced ATP levels activate AMPK, which, in turn, activates p27 to prevent apoptosis and induce autophagy in these cells (Figure 6). Autophagy involves protein catabolism and attenuated translation, giving rise to amino acids that can then be used for energetic purposes. Our NMR experiments found increased levels of amino acids, including branched chain amino acids, in GF compared to CONV-R colonocytes. The branched chain amino acids are the major nitrogen source for glutamine and alanine, which are also clearly elevated in the GF colonocytes. With this in mind, it is clear that microbes regulate energy homeostasis in the colon, and in the absence of microbiota, autophagy is used as a mechanism to derive amino acids that can be diverted toward energy production.
Dietary factors known as prebiotics promote the growth of certain bacteria at the expense of others and have implications for human health and disease. As our diets have shifted away from fiber and other complex carbohydrates toward processed, simple carbohydrates, the incidence of colorectal cancer (CRC) and inflammatory bowel diseases (IBD) such as ulcerative colitis and Crohn’s Disease have increased (Garcea and Alberts, 1980; Hamer et al., 2008; Latella and Caprilli, 1991). It is possible that increasing butyrate levels in the lumen and in colonocytes could help reverse this trend. In fact, butyrate enema therapy has been shown to ameliorate the inflammation associated with colitis in mouse models and in human clinical trials (Hamer et al., 2008).
EXPERIMENTAL PROCEDURES
Animals
Eight- to fourteen-week-old male C57BL/6 mice were used in all experiments. Conventionally-raised mice were reared in a specific pathogen-free (SPF) facility, while germfree mice were maintained following standard operating procedures at the National Gnotobiotic Resource Center at UNC. Detailed information is available from their website (www.med.unc.edu/ngrrc/about-us/) and upon request. All experiments were performed using protocols approved by the Institutional Animal Care and Use Committees (IACUC) of UNC and in accordance with federal guidelines.
Isolation of Colonocytes
Colonic epithelial cells were isolated as previously described (Roediger and Truelove, 1979) in order to exclude cell types other than colonocytes such as immune cells, smooth muscle, and enteric neurons. Briefly, animals were euthanized by CO2 asphyxiation followed by cervical dislocation. Proximal colons were dissected, flushed with sterile phosphate-buffered saline (PBS), and splayed open longitudinally along Whatmann paper. The colon was then washed 3 more times in PBS, pealed from paper, and placed in PBS solution containing 5mM EDTA. Submerged colon was then incubated on a rotator for ~30 minutes at 37 °C. Colonic tissue was removed leaving isolated colonocytes, which were pelleted, washed twice with PBS, and used for experiments.
Transcriptome Analysis
Colonocytes were prepared from GF and CONV-R mice (3 biological replicates each) and flash-frozen. Total RNA was prepared using Trizol reagent (Invitrogen, Carlsbad, CA), and integrity was assessed using the RNA 6000 Nano-LabChip kit followed by analysis using a Bioanalyzer (Agilent, Palo Alto, CA). Hybridizations to the MouseWG-6 v2.0 BeadChip arrays (Illumina, San Diego, CA), washing, and scanning was performed at the University of Tennessee at Memphis Core Facility. Data was analyzed using R/limma (Smyth, 2004).
Quantitative Mass Spectrometry
Whole cell proteins from colonocytes were extracted by sonication in a buffer of 8M urea, 25mM Tris pH7.6, and 100mM NaCl for 5 minutes. Proteins were subjected to overnight trypsin digestion followed by isoelectric focusing of peptides on a pH3.5–4.5 immobilized pH gradient strip. Peptides were then further separated by reverse-phase LC coupled inline with a linear ion-trap mass spectrometer. MS and MS/MS sans were acquired using data-dependent settings and searched against a NCBI mouse proteome database (ftp://ftp.ncbi.nih.gov/refseq/M_musculus/mRNA_Prot/mouse.protein.faa.gz). Results were filtered using an in house derived algorithm. Quantification was performed using redundant spectral counting where spectral counts (SC) for an individual protein are normalized to the overall spectra collected in each fraction for the individual and then by total spectral counts for all proteins(Carvalho et al., 2008a; Carvalho et al., 2008b; Kislinger et al., 2006). More details of digestion conditions, acquisition and search parameters, and data filtering algorithms and quantification statistics are described in supplemental methods.
NMR Metabolomics
Colonocytes were processed using the methanol-chloroform-water method (Beckonert et al., 2007), and the polar phase was lyophilized, stored at −80°C, thawed and suspended in 550 ml of a 1.0 mM solution of 2,2-dimethyl-silapentane-5-sulfonate sodium salt (DSS) containing 0.01% (w/v) sodium azide as a bacteriocide. The solutions were transferred to 5mm NMR tubes. NMR spectra were acquired on a Varian Inova 600 MHz using a 5 mm pulsed field gradient, inverse detection probe (Varian, Inc., Palo Alto, CA). The spectra were acquired at 25°C with 2048 transients using a sweep width of 6900 Hz and digitized with 32K points. The pulse sequence included a one sec solvent pre-saturation period and a 3.4 sec acquisition time. The data were processed using the ACD NMR Manager software version 12 (Advanced Chemistry Development, Toronto, Canada). A 0.3 Hz exponential line broadening was applied to the data. The spectra were phased, baseline corrected, and integrated using high-resolution binning with bin widths of 0.001ppm. The regions from 0.5 to 4.7 ppm and 4.9 to 9.0 were included in the integration. The regions below 0.5 and above 9.0 contained only noise and the region from 4.7 to 4.9 contained the residual solvent peak. The data were normalized by setting the sum of all integrals equal to 1000 for each spectrum. Metabolite identification was facilitated by the Chenomx NMR metabolite database, version 6 (Chenomx Inc, Edmonton, Canada). The statistical analyses including principal components analysis (PCA) and partial least squares (PLS) were performed using SimcaP+ version 12 (Umetrics, Umea, Sweden). Pareto scaling was applied to the binned NMR data prior to statistical analysis.
Butyrate Oxidation to CO2
Colonocytes were incubated in Exetainer breath storage tubes with a 4% 13C-butyrate and 96% unlabeled butyrate for 1hr at 37 °C and stopped with sodium azide treatment. The amount of 13CO2 generated from the colonocytes was measured with a 20/20 gas isotope ratio mass spectrometer (Europa Scientific, Cincinnati, OH) at Metabolic Solutions (Nashua, NH). Dissolved CO2 in solution was liberated to the tube headspace by addition of 100 uL-saturated citric acid. The ratio of 13CO2 to 12CO2 (mass 45 to 44) was measured directly from the sample tube headspace. All samples were compared to an internal reference gas (5% CO2, balance 75% N2, 20% O2) that had been calibrated against the International standard PeeDeeBelmnite (PDB). The results were expressed as the delta (δ) value per mil (%) and then converted to units of atom % 13C and defined by: Atom % 13C = 13CO2 / (13CO2 + 12CO2) × 100%. Changes in 13CO2 enrichment before and after butyrate administration were calculated. CO2 standards at three different 13C enrichments were run before and after the run to check instrument performance. The analytical precision of the instrument is 0.0001 atom % 13C.
HPLC/LC-MS Analysis of NADH/NAD+
Mitochondria were isolated from CONV-R and GF colon in accordance to the manufacture’s instructions with the mitochondrial isolation kit from Biovision (Mountain View, CA). Isolated mitochondria were then resuspended in 0.1 % ammonium hydroxide and were analyzed for NAD+ and NADH content by HPLC separation with subsequent detection by a triple-quadrupole mass spectrometer operating in positive mode. Peak areas were calculated using ThermoXcaliber (San Jose, CA). Chromatographic peaks were integrated for samples and areas were compared to peak area for standards (5 pmols) for each compound. Further details concerning the methodology are included in the supplemental methods section.
Metabolic Assays
All metabolic assays were performed with Biovision (Mountain View, CA) metabolism kits. Specifically, acetyl-CoA (K317-100), malate (K637-100), oxaloacetate (K659-100), NAD/NADH (K337-100), and ATP (K354-100) were used in accordance to manufacturer specifications. For all assays, flash-frozen tissue was deproteinized with Perchloric acid kit (K808-200), which was also purchased from Biovision. All experiments were performed on 4–6 independent biological replicates and 2 technical replicates per experimental group.
Confocal Microscopy
The reagents MitoTracker Red CM-H2XRos, and MitoTracker Green FM were purchased from Invitrogen (Carlsbad, CA). Cells were viewed with Olympus IX81 confocal microscope equipped with FITC, Texas Red, Rhodamine, and DAPI filters. All fluorescent micrographs were acquired with the same exposure and focal plane. Image J (Bethesda, MD) was used to quantify integrated density (total fluorescence) per cell. Mitotracker dyes were added at 500 nM (MitoTracker Red CM-H2XRos), and 500 nM (MitoTracker Green FM) while isolating colonocytes. Image J was used to analyze mean fluorescence for both mitotracker dyes. The red-to-green fluorescence is an indication of oxidative metabolism in the colonocytes. All micrographs were taken with identical exposures. Z-stacks were acquired for each image and for each cell the individual z-stacks did not differ in red-to-green mean fluorescence between focal planes. 3 independent biological replicates were analyzed per experimental group.
Western Blot Analyses
Colonocytes or tissues were prepared as described above, except in butyrate rescue experiments, where 10 mM butyrate was added during the isolation of colonocytes. Cells or tissues were flash-frozen, and homogenized or agitated in 8.3 M urea lysis buffer (Reyes et al., 1997) with NaF and Na orthovanadate and incubated on wet ice for 30 minutes. Samples were then centrifuged at 14,000 rpm for 20 minutes at 4 °C. Protein supernatant was transferred to a new 1.5 mL eppendorf tube and protein concentration was determined by Bradford assay. Protein homogenates were separated on 10, 12, and 4–20% gradient SDS-polyacrylamide gels and transferred onto PVDF membrane. Membranes were then incubated on rocker in 3% BSA or 5% non-fat dry milk with NaF for 1 hr at RT. Blocked membranes were incubated in 1° Ab overnight on rotator at 4°C. Antibodies were used at 1:250 to 1:2000 and included pyruvate dehydrogenase cocktail (Mitosciences, Eugene, OR; MSP02), ATP synthase F1β (Invitrogen, Carlsbad, CA; A-21351), phospho-AMPK (Cell Signaling), AMPKα (Cell Signaling, Danvers, MA), LC3 (Cell Signaling, Danvers, MA), phospho-p27 (R&D Systems; YXH02), β-actin (Abcam; ab8226), and total Histone H3 (Upstate; 05-928). Blots were washed 3X for 10 min/wash in 1X TBST or 1X PBST (for non-phospho Westerns) at RT and incubated with horseradish peroxidase-conjugated secondary antibodies (1:2000 to 1:4000) for 1 hr and 30 min on rocker at room temperature. Membranes were then washed 3 additional times in 1XTBST or 1XPBST (for non-phospho Westerns), treated with ECL reagent, and developed for analysis.
Transmission Electron Microscopy
Tissues were dissected, rinsed with PBS, and fixed in 0.15M sodium phosphate buffer, pH 7.4 containing 2% paraformaldehyde and 2.5% glutaraldehyde overnight. After buffer washes, the tissue was postfixed for 1 hour with 1% osmium tetroxide/0.15M sodium phosphate buffer. Samples were dehydrated using increasing concentrations of ethanol, infiltrated and embedded in Polybed 812 epoxy resin (Polysciences, Inc., Warrington, PA). 1-micrometer sections were prepared to select representative areas by light microscopy and 70nm ultrathin sections were then cut using a diamond knife. Sections were mounted on 200 mesh copper grids and staining with 4% aqueous uranyl acetate and Reynolds’ lead citrate. Sections were observed with a LEO EM910 transmission electron microscope operating at 80kV (LEO Electron Microscopy, Thornwood, NY) and photographed using a Gatan Orius SC1000 CCD Digital Camera and Digital Micrograph 3.11.0 (Gatan, Inc., Pleasanton, CA).
Statistics
For all metabolic assays significant differences between experimental groups were determined by two-tail t-test (p<0.05). In experiments where more than two groups were compared such as the mitotracker experiments, we performed an ANOVA followed by a Tukey post-hoc test to determine significant differences. For gene expression, data were analyzed with R/limma (Smyth, 2004), and FDR and FDR-adjusted p-values were calculated. For quantitative mass spectrometry data, a Poisson regression analysis was performed to determine significance of differentially expressed proteins between samples. All data are expressed as mean ± SE.
Supplementary Material
ACKNOWLEDGEMENTS
We thank V. Madden, S. Ray, and B. Bagnell at the UNC Microscopy Services Laboratory for advice with confocal microscopy and performing transmission electron microscopy. We would also like to thank L. Collins at the UNC Biomarker Mass Spectrometry Facility for his technical assistance in performing the HPLC/LC-MS experiments. We acknowledge W. Pathmasiri and P. Sandusky for technical assistance with NMR, and the National Gnotobiotic Resource Center (P40RRO118603 and P30 DK 34987) for maintaining germfree mice. We also thank J. Rawls, G. Camp, and R. Chandler for critical comments on this manuscript. This work was supported by the National Institutes of Health (CA125237 to S.J.B.), American Institute of Cancer Research, and The Prevent Cancer Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no conflicts of interest.
REFERENCES
- Backhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, Semenkovich CF, Gordon JI. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci U S A. 2004;101:15718–15723. doi: 10.1073/pnas.0407076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backhed F, Manchester JK, Semenkovich CF, Gordon JI. Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proc Natl Acad Sci U S A. 2007;104:979–984. doi: 10.1073/pnas.0605374104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barcenilla A, Pryde SE, Martin JC, Duncan SH, Stewart CS, Henderson C, Flint HJ. Phylogenetic relationships of butyrate-producing bacteria from the human gut. Appl Environ Microbiol. 2000;66:1654–1661. doi: 10.1128/aem.66.4.1654-1661.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckonert O, Keun HC, Ebbels TM, Bundy J, Holmes E, Lindon JC, Nicholson JK. Metabolic profiling, metabolomic and metabonomic procedures for NMR spectroscopy of urine, plasma, serum and tissue extracts. Nat Protoc. 2007;2:2692–2703. doi: 10.1038/nprot.2007.376. [DOI] [PubMed] [Google Scholar]
- Boffa LC, Vidali G, Mann RS, Allfrey VG. Suppression of histone deacetylation in vivo and in vitro by sodium butyrate. J Biol Chem. 1978;253:3364–3366. [PubMed] [Google Scholar]
- Camp JG, Kanther M, Semova I, Rawls JF. Patterns and scales in gastrointestinal microbial ecology. Gastroenterology. 2009;136:1989–2002. doi: 10.1053/j.gastro.2009.02.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candido EP, Reeves R, Davie JR. Sodium butyrate inhibits histone deacetylation in cultured cells. Cell. 1978;14:105–113. doi: 10.1016/0092-8674(78)90305-7. [DOI] [PubMed] [Google Scholar]
- Carvalho PC, Fischer JS, Chen EI, Yates JR, 3rd, Barbosa VC. PatternLab for proteomics: a tool for differential shotgun proteomics. BMC Bioinformatics. 2008a;9:316. doi: 10.1186/1471-2105-9-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho PC, Hewel J, Barbosa VC, Yates JR., 3rd Identifying differences in protein expression levels by spectral counting and feature selection. Genet Mol Res. 2008b;7:342–356. doi: 10.4238/vol7-2gmr426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury SR, King DE, Willing BP, Band MR, Beever JE, Lane AB, Loor JJ, Marini JC, Rund LA, Schook LB, Van Kessel AG, Gaskins HR. Transcriptome profiling of the small intestinal epithelium in germfree versus conventional piglets. BMC Genomics. 2007;8:215. doi: 10.1186/1471-2164-8-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford PA, Crowley JR, Sambandam N, Muegge BD, Costello EK, Hamady M, Knight R, Gordon JI. Regulation of myocardial ketone body metabolism by the gut microbiota during nutrient deprivation. Proc Natl Acad Sci U S A. 2009;106:11276–11281. doi: 10.1073/pnas.0902366106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cresci GA, Thangaraju M, Mellinger JD, Liu K, Ganapathy V. Colonic gene expression in conventional and germ-free mice with a focus on the butyrate receptor GPR109A and the butyrate transporter SLC5A8. J Gastrointest Surg. 14:449–461. doi: 10.1007/s11605-009-1045-x. [DOI] [PubMed] [Google Scholar]
- Dashwood RH, Myzak MC, Ho E. Dietary HDAC inhibitors: time to rethink weak ligands in cancer chemoprevention? Carcinogenesis. 2006;27:344–349. doi: 10.1093/carcin/bgi253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diez-Gonzalez F, Bond DR, Jennings E, Russell JB. Alternative schemes of butyrate production in Butyrivibrio fibrisolvens and their relationship to acetate utilization, lactate production, and phylogeny. Arch Microbiol. 1999;171:324–330. doi: 10.1007/s002030050717. [DOI] [PubMed] [Google Scholar]
- Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esworthy RS, Binder SW, Doroshow JH, Chu FF. Microflora trigger colitis in mice deficient in selenium-dependent glutathione peroxidase and induce Gpx2 gene expression. Biol Chem. 2003;384:597–607. doi: 10.1515/BC.2003.067. [DOI] [PubMed] [Google Scholar]
- Garcea RL, Alberts BM. Comparative studies of histone acetylation in nucleosomes, nuclei, and intact cells. Evidence for special factors which modify acetylase action. J Biol Chem. 1980;255:11454–11463. [PubMed] [Google Scholar]
- Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, Gordon JI, Relman DA, Fraser-Liggett CM, Nelson KE. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon HA, Pesti L. The gnotobiotic animal as a tool in the study of host microbial relationships. Bacteriol Rev. 1971;35:390–429. doi: 10.1128/br.35.4.390-429.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamer HM, Jonkers D, Venema K, Vanhoutvin S, Troost FJ, Brummer RJ. Review article: the role of butyrate on colonic function. Aliment Pharmacol Ther. 2008;27:104–119. doi: 10.1111/j.1365-2036.2007.03562.x. [DOI] [PubMed] [Google Scholar]
- He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrandt MA, Hoffmann C, Sherrill-Mix SA, Keilbaugh SA, Hamady M, Chen YY, Knight R, Ahima RS, Bushman F, Wu GD. High-fat diet determines the composition of the murine gut microbiome independently of obesity. Gastroenterology. 2009;137:1716–1724. e1711–e1712. doi: 10.1053/j.gastro.2009.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch BF, Meng S, Wu JT, Archer SY, Hodin RA. The effects of short-chain fatty acids on human colon cancer cell phenotype are associated with histone hyperacetylation. J Nutr. 2002;132:1012–1017. doi: 10.1093/jn/132.5.1012. [DOI] [PubMed] [Google Scholar]
- Hooper LV, Gordon JI. Commensal host-bacterial relationships in the gut. Science. 2001;292:1115–1118. doi: 10.1126/science.1058709. [DOI] [PubMed] [Google Scholar]
- Kislinger T, Cox B, Kannan A, Chung C, Hu P, Ignatchenko A, Scott MS, Gramolini AO, Morris Q, Hallett MT, Rossant J, Hughes TR, Frey B, Emili A. Global survey of organ and organelle protein expression in mouse: combined proteomic and transcriptomic profiling. Cell. 2006;125:173–186. doi: 10.1016/j.cell.2006.01.044. [DOI] [PubMed] [Google Scholar]
- Kurokawa K, Itoh T, Kuwahara T, Oshima K, Toh H, Toyoda A, Takami H, Morita H, Sharma VK, Srivastava TP, Taylor TD, Noguchi H, Mori H, Ogura Y, Ehrlich DS, Itoh K, Takagi T, Sakaki Y, Hayashi T, Hattori M. Comparative metagenomics revealed commonly enriched gene sets in human gut microbiomes. DNA Res. 2007;14:169–181. doi: 10.1093/dnares/dsm018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latella G, Caprilli R. Metabolism of large bowel mucosa in health and disease. Int J Colorectal Dis. 1991;6:127–132. doi: 10.1007/BF00300209. [DOI] [PubMed] [Google Scholar]
- Liang J, Shao SH, Xu ZX, Hennessy B, Ding Z, Larrea M, Kondo S, Dumont DJ, Gutterman JU, Walker CL, Slingerland JM, Mills GB. The energy sensing LKB1-AMPK pathway regulates p27(kip1) phosphorylation mediating the decision to enter autophagy or apoptosis. Nat Cell Biol. 2007;9:218–224. doi: 10.1038/ncb1537. [DOI] [PubMed] [Google Scholar]
- Pryde SE, Duncan SH, Hold GL, Stewart CS, Flint HJ. The microbiology of butyrate formation in the human colon. FEMS Microbiol Lett. 2002;217:133–139. doi: 10.1111/j.1574-6968.2002.tb11467.x. [DOI] [PubMed] [Google Scholar]
- Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, Mende DR, Li J, Xu J, Li S, Li D, Cao J, Wang B, Liang H, Zheng H, Xie Y, Tap J, Lepage P, Bertalan M, Batto JM, Hansen T, Le Paslier D, Linneberg A, Nielsen HB, Pelletier E, Renault P, Sicheritz-Ponten T, Turner K, Zhu H, Yu C, Jian M, Zhou Y, Li Y, Zhang X, Qin N, Yang H, Wang J, Brunak S, Dore J, Guarner F, Kristiansen K, Pedersen O, Parkhill J, Weissenbach J, Bork P, Ehrlich SD. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawls JF, Samuel BS, Gordon JI. Gnotobiotic zebrafish reveal evolutionarily conserved responses to the gut microbiota. Proc Natl Acad Sci U S A. 2004;101:4596–4601. doi: 10.1073/pnas.0400706101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes JC, Muchardt C, Yaniv M. Components of the human SWI/SNF complex are enriched in active chromatin and are associated with the nuclear matrix. J Cell Biol. 1997;137:263–274. doi: 10.1083/jcb.137.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roediger WE. Role of anaerobic bacteria in the metabolic welfare of the colonic mucosa in man. Gut. 1980;21:793–798. doi: 10.1136/gut.21.9.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roediger WE. Utilization of nutrients by isolated epithelial cells of the rat colon. Gastroenterology. 1982;83:424–429. [PubMed] [Google Scholar]
- Roediger WE, Truelove SC. Method of preparing isolated colonic epithelial cells (colonocytes) for metabolic studies. Gut. 1979;20:484–488. doi: 10.1136/gut.20.6.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumney CJ, Duncan SH, Henderson C, Stewart CS. Isolation and characteristics of a wheatbran-degrading Butyrivibrio from human faeces. Lett Appl Microbiol. 1995;20:232–236. doi: 10.1111/j.1472-765x.1995.tb00435.x. [DOI] [PubMed] [Google Scholar]
- Savage DC. Microbial ecology of the gastrointestinal tract. Annu Rev Microbiol. 1977;31:107–133. doi: 10.1146/annurev.mi.31.100177.000543. [DOI] [PubMed] [Google Scholar]
- Shaw RJ, Kosmatka M, Bardeesy N, Hurley RL, Witters LA, DePinho RA, Cantley LC. The tumor suppressor LKB1 kinase directly activates AMP-activated kinase and regulates apoptosis in response to energy stress. Proc Natl Acad Sci U S A. 2004;101:3329–3335. doi: 10.1073/pnas.0308061100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Backhed F, Fulton L, Gordon JI. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe. 2008;3:213–223. doi: 10.1016/j.chom.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Gordon JI. The core gut microbiome, energy balance and obesity. J Physiol. 2009;587:4153–4158. doi: 10.1113/jphysiol.2009.174136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, Egholm M, Henrissat B, Heath AC, Knight R, Gordon JI. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. The human microbiome project. Nature. 2007;449:804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- Waldecker M, Kautenburger T, Daumann H, Busch C, Schrenk D. Inhibition of histone-deacetylase activity by short-chain fatty acids and some polyphenol metabolites formed in the colon. J Nutr Biochem. 2008;19:587–593. doi: 10.1016/j.jnutbio.2007.08.002. [DOI] [PubMed] [Google Scholar]
- Wostmann BS. The germfree animal in nutritional studies. Annu Rev Nutr. 1981;1:257–279. doi: 10.1146/annurev.nu.01.070181.001353. [DOI] [PubMed] [Google Scholar]
- Yang Z, Klionsky DJ. An overview of the molecular mechanism of autophagy. Curr Top Microbiol Immunol. 2009;335:1–32. doi: 10.1007/978-3-642-00302-8_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.