Abstract
Yersinia ruckeri, the etiological agent of the enteric red mouth disease (ERM) of salmonids, produces Yrp1, a serralysin metalloprotease involved in pathogenesis. We describe here the hydrolytic and immunogenic properties of Yrp1. The protease was able to hydrolyze different matrix and muscle proteins as laminin, fibrinogen, gelatine, actin, and myosin but not type II and IV collagens. In addition, the Yrp1 protein, when inactivated by heat and used as an immunogen, was able to elicit a strong protection against the development of ERM. The analysis of different Y. ruckeri strains with (Azo+) or without (Azo−) Yrp1 activity showed that all of them contained the yrp1 operon. By using yrp1::lacZ operon fusions, protease production analysis, and complementation studies, it was possible to show that an Azo− strain was blocked at the transcription level. The transcriptional study of the yrp1 operon under different environmental conditions showed that it was regulated by osmolarity and temperature, without pH influence. Finally, when β-galactosidase activity was used as a probe in vivo, the progression of the disease in the fish could be visualized, and the tropism of the bacterium and affected organs could be defined. This system opens a vast field of study not only with regard to fish disease progression but also in pathogen interactions, temporal gene expression, carrier stages, antibiotic resistance selection, etc.
Yersiniosis or enteric red mouth disease (ERM) is a serious infectious disease in salmonids that causes important economic losses in many countries. The etiological agent is the gram-negative bacterium Yersinia ruckeri, which eventually produces hemorrhagic zones around the mouth as a characteristic symptom during the infection process. Once infected, fish grow and survive during weeks or even months without disease symptoms, thus remaining in a carrier stage. Under stress conditions outbreaks occur. Fish that survive may exhibit bacterial shedding of the intestine over long periods (12, 37). In addition, Y. ruckeri can remain infective in the aquatic environment (38), and it also has a biofilm-forming capacity (7). Despite the importance and extensive knowledge of Yersinia species in pathogenesis of mammals, there are few studies about Y. ruckeri, and the precise mechanisms of virulence are practically unknown. Iron-regulated outer membrane proteins (39), iron availability (10), or the presence of a thermolabile factor (16, 17) have been suggested to be involved in pathogenesis. Other authors have described the importance of extracellular products in the virulence of this bacterium (40-42). Based on this, Secades and Guijarro (45) purified the extracellular serralysin metalloprotease Yrp1 and two groups of strains, named Azo+ and Azo−, were defined according to the presence or absence of the Yrp1 proteolytic activity, respectively. More recently, Fernandez et al. (15) showed that the gene encoding Yrp1 is part of an operon containing a type I ABC transporter involved in protein secretion, encoded by three genes (yrpD, yrpE, and yrpF), together with gene inh, that encodes a protease inhibitor. Using a trout model, it was possible to show that inactivation of either yrp1 or yrpE by insertional mutagenesis resulted in a significant increase in the 50% lethal dose after inoculation by intraperitoneal injection, indicating the participation of the protease in pathogenesis (15). Thus, although this extracellular protease is a clear virulence factor in Y. ruckeri, there is no description in the literature of the involvement of this kind of enzyme in pathogenesis by other pathogenic Yersinia spp. (Y. enterocolitica, Y. pestis, and Y. pseudotuberculosis), with the exception of the recently described HreP protease in Y. enterocolitica (20).Virulence in these species has been related to the presence of a 70-kb plasmid (9), and chromosomal encoded virulence genes have also been described for different Yersinia species (for a review, see reference 36).
In many cases, virulence factors from a particular pathogen are under the control of environmental conditions. The ivi genes induced exclusively during the infection process are a clear example (29, 30). There are specific host induction factors and others related to environmental conditions, such as temperature, pH, iron availability, osmotic pressure, etc., that influence the expression of virulence genes (for reviews, see references 18, 22, and 31). In Yersinia species, there is an important group of virulence genes that are up- and downregulated by temperature (48). In fish pathogenic bacteria, temperature regulation is particularly important because the production of a specific protein may stop at a temperature corresponding to the upper limit of pathogenicity of the bacteria, which is below the optimal growth temperature (5, 45, 46). However, the level at which this regulation takes place is unknown. Other factors with regulatory effects on gene expression on fish pathogens, such as pH and osmolarity, have not been genetically studied. An additional way to study gene expression is in vivo analysis, a powerful technique that enables monitoring biological process through different approaches by coupling the gene of interest or its promoter to a reporter gene (6, 49). Despite the ever-increasing work on bacterial fish pathogens, only an in vivo study with a green fluorescent protein has been carried out in Edwardsiella tarda (25, 26).
On the other hand, good levels of protection against ERM disease have been reached by the use of preventive commercial vaccines made of dead bacterial cells (47). However, fish farm outbreaks that are probably due to the carrier stage mentioned above or to the existence of different serotypes do occur from time to time. For that reason, new approaches based on subunit or DNA vaccines could be used as an additional way to eliminate or minimize these outbreaks. Several proteins from fish bacterial pathogens have been shown to elicit an immune response against the respective infections (13, 23, 28), and DNA vaccines have been mainly studied for fish viral pathogens (14, 19, 27). Thus, a future form of prevention of infectious diseases in aquaculture could be a polyspecific vaccine based on the use of a mixture of antigens or DNA-encoding antigens from different pathogens that would protect against several diseases.
We sought here to study some enzymatic properties, regulation, and in vivo expression of the Yrp1 protease from Y. ruckeri. Thus, by using purified Yrp1 protease we were able to determine its cleavage pattern over different matrix and muscle proteins. Experiments with Yrp1 toxoid were carried out in order to assess the induction of a protective immunity against Y. ruckeri disease. We show by PCR analysis that the presence of the yrp1 operon in all of the tested strains was independent from the Yrp1 phenotype. Complementation studies, together with yrp1::lacZ fusion analysis, showed that the yrp1 operon in an Azo− strain was blocked at the transcriptional level. Furthermore, similar studies showed that the yrp1 operon was regulated at the transcriptional level by osmotic and temperature conditions. Finally, the transcriptional fusion was used as a promoter probe to visualize gene expression in the fish. The yrp1 spatial expression was similar to the one found with other Y. ruckeri promoter fusions. This technique opens a new, wide, and varied way of investigating in vivo colonization, invasion, specific time gene expression, and different fish-pathogen interactions.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Y. ruckeri Azo− strains 146, 147, 3585, 955, and 956 and Azo+ strains 148, 149, 150, 4319, and 1386, as well as strain 150RI4, were described previously (45). Strains Al00 (Azo+) and Al02 (Azo+) (the present study) were isolated from Spanish fish farm outbreaks. Bacterial strains were routinely cultivated on nutrient broth (NB; Difco) or NB with 1.5% (wt/vol) agar at 18°C. Growth in liquid cultures was monitored by determining the absorbance at 600 nm at different times during incubation at 250 rpm.
Yrp1 substrate hydrolysis analysis.
Pure Yrp1 protein (0.8 μg), obtained as previously described by Secades and Guijarro (45), was incubated with different protein substrates of human origin (12-μg portions; Sigma Chemical Co.)—including fibrinogen, fibronectin, laminin, gelatine, collagen (types I, II, and IV), and the muscle proteins actin and myosin—at 18°C for 16 h in 25 mM Tris-HCl (pH 7.6) buffer containing 5 mM MgCl2. The reactions were terminated by adding 10 mM EDTA, and then samples were frozen, lyophilized, and resuspended in Laemmli sample buffer (24). Samples were loaded onto a sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis (SDS-10% PAGE) and, after electrophoresis, the gel was stained with Coomasie brilliant blue.
In vitro β-galactosidase determination in the yrp1::lacZ fusions and complementation studies.
Y. ruckeri 150RI4 (yrp1::lacZ fusion) was previously obtained by Fernandez et al. (15) by insertional mutagenesis with an internal fragment from the yrp1 gene and the suicide plasmid pIVET8 (30). Y. ruckeri 146RI1 (yrp1::lacZ fusion) (the present study) was obtained in a similar way. Portions (500 μl) of overnight cultures of Y. ruckeri 146RI1 and 150RI4 were used to inoculate 250-ml flasks containing 50 ml of NB and were then incubated at 18 or 28°C and 250 rpm. At different incubation times, cells were centrifuged at 12,000 × g for 5 min, and the β-galactosidase activity was assayed in cells by the Miller method (33). For the study of the influence of osmotic pressure, NB was prepared with a 100, 250, or 500 mM concentration of either NaCl, KCl, or d-xylose, and when the cultures reached an approximate optical density at 600 nm of 1.4, the samples were processed as previously described for the analysis of β-galactosidase activity. A similar method was used to assay the pH effect. Medium was buffered with 50 mM morpholine ethanesulfonate for pH 6 or 6.5 and with HEPES for pH 7, 7.5, and 8. In all cases, a standard NB was used as a control.
Complementation of the wild-type Azo− 146 strain was carried out by using plasmids pUK21B, pUK21C, or pUK21T containing yrp1 and inh genes, inh, yrpD, yrpE, and yrpF genes and all of the operon from Y. ruckeri 150, respectively (15). Y. ruckeri 146 was transformed by electroporation as described by Fernandez et al. (15).
Fish protection studies with Yrp1 toxoid.
Rainbow trouts (Oncorhynchus mykiss) weighing between 8 and 10 g were kept in 60-liter tanks at 18 ± 1°C in continually flowing dechlorinated water with feeding. Groups of 10 fish were injected intraperitoneally with 8 μg of heat-denatured (100°C for 2 min) Yrp1 protease in 0.1 ml of phosphate-buffered saline (PBS), obtained as described by Secades and Guijarro (45). Simultaneously, two control groups of 20 fish were injected. One of them was injected with a 0.1-ml portion of 102 heat-treated Y. ruckeri cells (100°C for 15 min) in PBS, and the other control group was injected with 0.1 ml of PBS. Fish were kept as described previously during 28 to 30 days, and then fish from each group were challenged with 0.1 ml of PBS containing 103 cells of Y. ruckeri 150, an inoculum that was previously found to kill >50% of the untreated fish 7 days after infection. Dead fish from each group were collected everyday and, after 10 days, the relative percent survival—defined as [1 − (% vaccinated mortality/% control mortality)] × 100—was determined.
PCR detection of the yrp1 and yrpDE F genes in several Y. ruckeri strains.
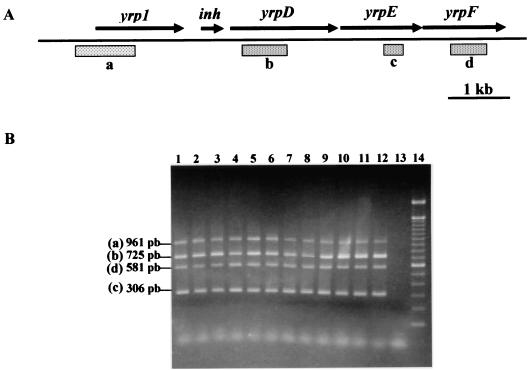
The specific primers designed for the present study from the respective gene sequences (EMBL accession no. AJ318052 and AJ421517) and used for PCR amplification in Y. ruckeri strains were as follows: for yrp1, XA2RP, nucleotides (nt) 289 to 272 upstream from the putative ATG start codon (5′-TATTCAACTGAAAGTGTA-3′), and XAC01, nt 672 to 656 (5′-ATAGCTCATAATACTGA-3′), generate a 961-bp PCR product; for yrpD, XA10RP, nt 157 to 173 (5′-GCATCAGGTAATGAAAT-3′), and XB5RP, nt 881 to 864 (5′-CAATCAGTTGATCAATA-3′), generate a 725-pb PCR product; for yrpE, XA2RP2, nt 672 to 688 (5′-ACGGTATGCCGAACTTA-3′), and XA2CO2, nt 978 to 962 (5′-CGGTACGATTTCCATTA-3′), generate a 306-pb PCR product; and for yrpF, XA2RP5, nt 433 to 450 (5′-GCCTTACTGGCTCAGGA-3′), and XA2CO5, nt 1013 to 997 (5′-TCCGCCTGCGACTGCTG-3′), generate a 581-pb PCR product. As a positive control, Y. ruckeri 150 was used. The different strains were grown in nutrient broth at 18°C, and 1 ml of stationary-phase cultures were centrifuged for 5 min at 12,000 × g, the pellet was resuspended in 100 μl of water, and the cells were lysed by boiling them for 10 min. Cell debris was then precipitated by centrifugation for 30 s, and 5-μl aliquots were used as a template DNA in the PCR assays. All PCR components (DNA polymerase, reaction buffer, and deoxynucleoside triphosphates) were provided by Biotools. The amplification reactions (25 cycles) were performed in two groups (yrp1-yrpD and yrpE-yrpF) in a Perkin-Elmer thermal cycler with 94°C for a 5-min initial denaturation, followed by denaturation at 94°C for 30 s, annealing at 40°C for 30 s for yrp1 and yrpD and at 45°C for 30 s for yrpE and yrpF, extension at 72°C for 1 min for yrp1 and yrpD and at 72°C for 30 s for yrpE and yrpF, and a final extension at 72°C for 7 min. The reaction products of the two groups were mixed, and 1.5% (wt/vol) agarose gel electrophoresis was used to separate the generated PCR amplicons.
In vivo β-galactosidase assay.
Rainbow trouts weighing 8 to 10 g were intraperitoneally injected with 103 cells of the Y. ruckeri 150RI4 (yrp1::lacZ fusion) strain or the Y. ruckeri 150 strain as a negative control. The fish were kept at 18°C as described above and, once dead, they were dissected and fixed in 4% paraformaldehyde in PBS for 20 min to 1 h. The fish were washed several times with PBS, followed by the addition of 5 ml of BetaBlue staining kit solution (Novagen/CN Biosciences, Inc.) to each fish, which were then incubated at 37°C until a blue color was apparent. Color progression was stopped by washing the samples five times with 10 ml of PBS, and the fish were finally stored at 4°C in 15% glycerol (vol/vol) in PBS. Photographs were taken with a digital Kodak DC 290 camera. Microscopic examinations of gill and intestine tissue samples were carried out by removing them from the fish and observing them with an Olympus BH12 microscope equipped with an Olympus DP12 digital camera. During the in vivo β-galactosidase assay, it was observed that some batches of fish showed a low β-galactosidase background activity in the intestine. Thus, batches of fish had to be checked before being used in the experiments.
Other determinations.
Proteolytic activity was assayed by using azocasein (Sigma) as a substrate, according to the method described by Secades and Guijarro (45). After an analysis of variance test, P values of <0.05 were considered significant.
RESULTS
Yrp1 protease has a wide range of substrate proteins.
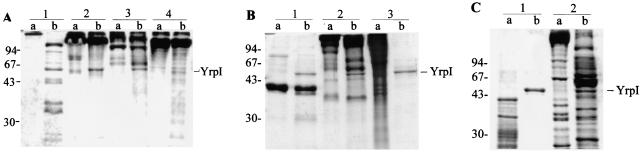
In order to define the role in virulence of the Yrp1 protease, different matrix and muscle proteins were used as possible substrates of the enzyme. As shown in Fig. 1A, lane 1, fibronectin was completely degraded, whereas type I collagen suffered a small hydrolysis (lane 4). In contrast, type II (lane 2) and IV (lane 3) collagens were refractory to hydrolysis. Other proteins (Fig. 1B), such as fibrinogen (lane 1), laminin (lane 2), and gelatine (lane 3), were hydrolyzed to different degrees, whereas gelatine was completely degraded. The muscle proteins actin and myosin (Fig. 1C, lanes 1 and 2, resepectively) were extensively degraded by Yrp1.
FIG. 1.
SDS-PAGE profile analysis of degradation of different matrix and muscle proteins by Yrp1. Each protein (12 μg) was incubated in the absence (lanes a) or presence (lanes b) of the Yrp1 protease (0.8 μg) at 18°C during 16 h. Samples were loaded onto a SDS-10% PAGE gel and then stained with Coomasie brilliant blue. (A) Lanes: 1, fibronectin; 2, type II collagen; 3, type IV collagen; 4, type I collagen. (B) Lanes: 1, fibrinogen; 2, laminin; 3, gelatine. (C) Lanes: 1, actin; 2, myosin. The position of the molecular mass markers is indicated on the left in kilodaltons. The position of the Yrp1 is indicated on the right.
Yrp1 toxoid confers protective immunity against Y. ruckeri.
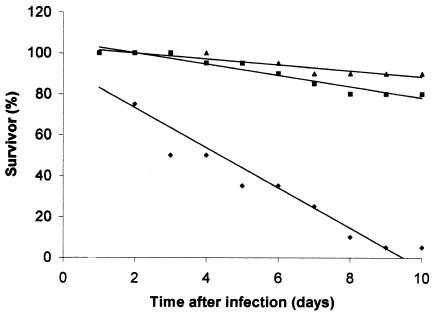
In order to determine the possible protective immunity properties of the Yrp1 protease on rainbow trout, a toxoid was prepared by inactivation of the protein by heating. The toxoid was then injected intramuscularly into fish. The animals were maintained for 28 to 30 days with feeding, and then they were challenged with intraperitoneal injections of 103 cells of the wild-type strain. The PBS-treated fish used as a control group showed 95% mortality after 10 days, whereas fish treated with toxoid or heat-killed cells showed 20 or 10% mortality, respectively (Fig. 2). Thus, in our experimental conditions, the relative percent survival value of the Yrp1 toxoid-treated fish was 79%, a value closer to that obtained with heat-killed cells (n = 90).
FIG. 2.
Vaccine challenge assay. Fish from each group were challenged with 103 Y. ruckeri 150 cells at 28 to 30 days after a 0.1-ml injection of 8 μg of heat-inactivated Yrp1 protease (▪), 102 heat-killed cells (▴), or control with PBS (♦). Dead fish were collected daily. These results represent the average of two independent experiments.
Detection of the yrp1 operon in several Y. ruckeri strains and regulation by temperature, osmolarity, and pH.
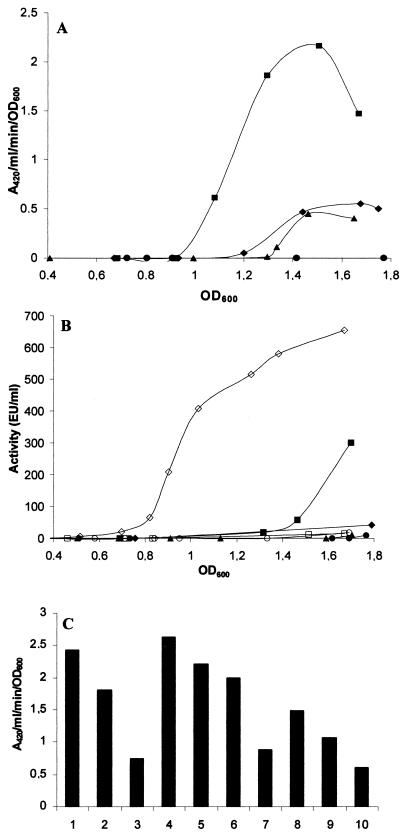
In a previous study (44), Y. ruckeri strains were defined in two groups, Azo+ and Azo−, according to their azocasein degradation capacity, which correlated with the presence and absence of the Yrp1 extracellular protease, respectively.By PCR analysis with specific oligonucleotides designed from yrp1 or yrpD, yrpE, and yrpF gene sequences from the yrp1 operon, it was possible to show, as can be observed in Fig. 3B, that all four genes were present in all of the analyzed strains, even those described as Azo−. In order to determine the reason for the absence of proteolytic activity in the Azo− strains containing the yrp1 operon, a yrp1::lacZ transcriptional gene fusion Azo− strain was constructed (146RI1). As shown in Fig. 4A, strain 146RI1 grown at 18°C presents a low level of β-galactosidase activity during growth. This low transcription level corresponds to an undetectable azocasein hydrolytic activity (Fig. 4B). In contrast, β-galactosidase and proteolytic activity production showed a continuous increase throughout growth in the Azo+ 150RI4 strain (Fig. 4A and B). Complementation studies with Azo− wild-type strain 146 as a recipient showed that only the strain carrying the yrp1 operon was able to hydrolyze azocasein. However, when the complementation was carried out with the yrp1 gene or the yrpDEF genes, there was no production of proteolytic activity.
FIG. 3.
PCR analysis of the yrp1 operon from Y. ruckeri strains. Y. ruckeri strains were grown in NB, and then two independent PCR (yrp1-yrpD and yrpE-yrpF) reactions were performed with specific primers. After the PCRs, the generated amplicons from each strain were mixed and separated in a 1.5% agarose gel. (A) Diagram of the yrp1 operon showing the genes and locations of the amplified fragments represented on the agarose gel. (B) The generated amplicons were yrp1 (961 bp), yrpD (725 bp), yrpE (581 bp), and yrpF (306 bp). Lanes: 1, 146−; 2, 147−; 3, 148+; 4, 149+; 5, 150+; 6, 1386+; 7, 3585−; 8, 4319+; 9, 955−; 10, 956−; 11, Al00+; 12, Al02+; 13, negative control; 14, molecular mass markers. The superscript symbols following the strain numbers (+ or −) indicate Azo+ or Azo− strains, respectively. The positions and sizes of the generated amplicons are indicated.
FIG. 4.
Analysis of yrp1::lacZ fusions and azocasein degradation of Y. ruckeri Azo + and Azo− strains during growth at different temperatures and osmolarities. (A) Activation of yrp1::lacZ fusion in strains 150RI4 (Azo+) and 146RI1 (Azo−). The expression of β-galactosidase was examined. Symbols: ▪, 150RI4 at 18°C; ♦, 150RI4 at 28°C; ▴, 146RI1 at 18°C; •, 146RI1 at 28°C. (B) Production of Yrp1 in strains 150RI4 (Azo+) and 146RI1 (Azo−). Caseinolytic activity was determined as described by Secades and Guijarro (44). Symbols: ▪, strain 150 at 18°C; ♦, strain 150 at 28°C; ▴, strain 146 at 18°C; •, strain 146 at 28°C. Open symbols indicate strain 146 at 18°C, complemented with the plasmids pUK21B (○), pUK21C (□), or pUK21T (⋄). (C) Activation of yrp1::lacZ fusion in strain 150RI4 at different NaCl, KCl, and d-xylose concentrations. Bars: 1, 2, and 3, NaCl at 100, 250, and 500 mM, respectively; 4, NB as a control; 5, 6, and 7, KCl at 100, 250, and 500 mM, respectively; 8, 9, and 10, d-xylose at 100, 250, and 500 mM, respectively.
Examination of the yrp1::lacZ fusions under different environmental conditions showed that, although bacterial growth was better at 28°C (45), the level of yrp1 expression was higher at 18°C than at 28°C (Fig. 4A). These data correspond with the proteolytic activity found at both temperatures (Fig. 4B). In addition, as the osmotic pressure of the medium increased, a slight decrease in transcription of the yrp1 operon was observed, with a significant inhibition of the transcription at either 500 mM NaCl (P = 0.004) or KCl (P = 0.001) (Fig. 4C).Incubation of the bacterium in the presence of the sugar d-xylose, which is not metabolized, caused greater repression (P = 0.002) (Fig. 4C). On the contrary, no major changes in transcription levels were observed at pH values between 6 and 8 (P = 0.669) (data not shown).
Visualization of yrp1 expression in fish by using yrp1::lacZ fusion.
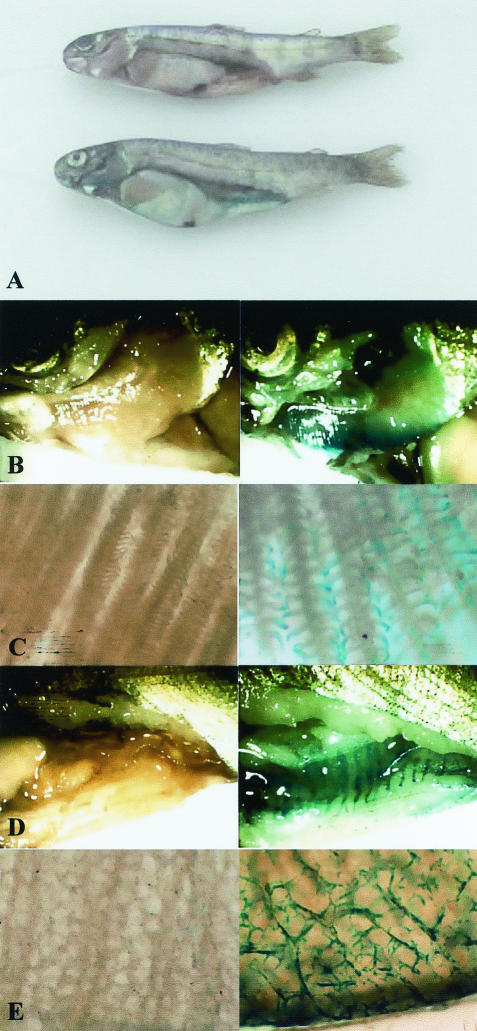
In studies with strain 150RI4, which contains the yrp1::lacZ transcriptional gene fusion (15), it was possible to monitor the in vivo expression of the yrp1 virulence gene promoter driving β-galactosidase activity. We found that fish that were intraperitoneally injected with the Y. ruckeri 150RI4 strain showed two clear macroscopic and defined β-galactosidase activity zones, corresponding with the gill and intestine tissues (Fig. 5A). A low and diffuse blue-green color could be visualized through the enlarged spleen and liver (Fig. 5A). In Fig. 5, a clear picture of the gills (Fig. 5B) and intestines (Fig. 5D) may be observed. A microscopic picture of the yrp1::lacZ fusion expression in gills can be seen in Fig. 5C, where the gill arches and filaments seem to be completely covered by the bacterium. In addition, a similar picture is evident in the intestine tissue, which shows an intense color along the capillary system (Fig. 5E).
FIG. 5.
Macroscopic and microscopic observation of yrp1::lacZ gene expression in fish. Fish were infected by intraperitoneal injection with 103 cells of Y. ruckeri 150RI4 (yrp1::lacZ) and, after death, they were treated with a X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) solution as described in Materials and Methods. Control fish were injected with the Y. ruckeri wild-type strain 150. (A) Dissected rainbow trout showing the internal organs. The control, 150 strain fish is shown above the strain 150 RI4 fish. Panels B, C, D, and E (left column) show the 150 wild-type strain (control fish) in the left and the 150RI4 strain (yrp1::lacZ) in the right column. (B and C) Macroscopic and microscopic details of gills, respectively. (D and E) Macroscopic and microscopic details of intestine, respectively.
In order to determine whether the pattern of yrp1 expression in vivo was specific for this particular gene, different Y. ruckeri promoters were cloned by shotgun cloning in plasmid pIVET8, and the respective insertional mutant strains were generated as described by Fernandez et al. (15). All of them showed an expression similar to that of yrp1, both in intensity as well as in distribution in fish tissues (data not shown).
DISCUSSION
Degradation studies with Yrp1 protease showed that the protein digests a wide variety of matrix and muscle proteins. This behavior shares some similarities with the degradation pattern obtained with Fpp1 protease from the fish pathogenic bacterium Flavobacterium psychrophilum (46) and other metalloproteases related to tissue damage or invasion (for reviews, see reference 34). Invasive processes involve the degradation of extracellular matrix and basement membranes. It is particularly interesting the fact that laminin, a major component of basement membranes, was digested by this protease. Although the proteins used in these experiments were not of fish origin, all of them have conserved sequences among vertebrates. This, together with the clear preference for the hydrolysis of laminin in relation to the other assayed proteins, suggests that fish laminin could be one of the major natural substrates of this protease. Thus, this degradation may be the cause of membrane alterations leading to erosion and pores in capillary vessels, which results in the leakage of blood through microhemorrhages in particular areas such as the mouth and intestine, which is characteristic of this disease. The fact that a yrp1-deficient mutant seems to produce attenuated symptoms supports this hypothesis (data not shown). These results, together with the fact that the Yrp1 protease is a virulence factor not essential for disease development and growth of the bacterium (15), allowed us to speculate that this protein could be involved in the invasion of different tissues during progression of the infection and may also have a nutritional role.
According to the extracellular location of the Yrp1 protease and its involvement in virulence, it was reasonable to expect some protection against the ERM disease when a toxoid of the protein was used as an immunogen. Thus, the protection against ERM was made possible by using active immunization with the Yrp1 toxoid through intramuscular injections. The efficacy of protection was found to be high, confirming the role of the Yrp1 toxoid as a subunit immunogen. The level of protection obtained with dead Y. ruckeri cells as a vaccine is similar to the one described by Altinok et al. (2), with 95% mortality in unvaccinated fish. Several proteins from bacterial fish pathogens have been shown to elicit protective immunity against the respective disease. Such are the cases of the OspA lipoprotein antigen of Piscirickettsia salmonis (23), a porin of Aeromonas salmonicida (28), and an adhesin of Aeromonas hydrophila (13). However, until now there has never been a description of subunit vaccine made of a toxoid protease in fish. According to this result, it would be interesting to study the application of Yrp1 toxoid through an immersion bath in order to facilitate its utilization together with the commercial vaccine. At the same time, a putative DNA vaccine using a part of the yrp1 gene encoding an immunogenic nontoxic peptide could be assayed.
The Azo− phenotype seems to be a widespread phenomenon in Y. ruckeri (45). The fact that all of the analyzed Azo− strains had the yrp1 operon was surprising. Based on this finding it can be concluded that this operon is present in Y. ruckeri as a genotypic characteristic. In spite of this, a basal level of expression of this operon occurs in Azo− strains. When one of the Azo− strains was analyzed by transcriptional fusion, it was observed that there was a low level of transcription of the yrp1operon, which apparently is not enough for the detection of proteolytic activity, indicating that a transcriptional blockage was the cause of the Azo− phenotype. Complementation studies showed that only when the whole operon, carrying either its own promoter or the lacZ promoter from the plasmid, was introduced in the Azo− strain was there azocasein hydrolysis. All of these results indicate that the whole yrp1 operon is transcriptionally inactive or with very low expression levels in this Azo− strain, and this in turn suggests that transcriptional level is a major regulation mechanism of the yrp1 operon. Although this result could not be extrapolated to all of the Azo− strains, it does suggest, in accordance with the presence of the operon in all of them, that transcriptional regulation is the basis for the protease-negative phenotype. The phenotypic lack of some virulence factors in the collection of strains is a common characteristic of pathogenic bacteria. Although the strains used in the present study were isolated from different outbreaks, laboratory subculture could give place to the Azo− phenotype.
The production of the Yrp1 protease by Y. ruckeri is temperature dependent (45). The activation of the yrp1 promoter occurred at the end of the growth phase, and it was repressed when bacteria were grown at 28°C. Thus, Yrp1 production is regulated, in part, in response to environmental signals such as temperature. Maximal Yrp1 production was consistent with the temperature at which the bacterium causes the disease, a usual temperature in many phases of the salmonid's production cycle in aquaculture. Yrp1 production stopped abruptly at the temperature optimal for bacterial growth, which is lethal for the salmonid's life. Thus, Yrp1 induction may be an environmental adaptation to the optimal temperature conditions for efficient infection and colonization. This regulatory behavior was already phenotypically observed for siderophore synthesis in Vibrio salmonicida (5), Fpp1 protease production in F. psychrophilum (45), lipopolysaccharide changes in A. hydrophila (1), and the expression of aprX protease (3) and lipA lipase genes (50) of Pseudomonas fluorescens. A well-studied system of adaptive gene expression to the environmental temperature for an efficient infection is the one developed by other Yersinia species (Y. enterocolitica, Y. pseudotuberculosis, and Y. pestis), wherein two types of regulatory effects influence the expression of genes encoding some proteins involved in infection. Thus, some genes are downregulated and others are upregulated at 37 and 26°C, respectively (8, 9, 48). In that sense, the Yrp1 protease from Y. ruckeri is a new example of the expression of a virulence gene that is regulated by specific environmental conditions, being highly expressed at temperatures found in the host and repressed at higher temperatures. A related case of regulation by temperature is the one described for yplA (44), yst (32), inv (35), and ure (11) virulence genes in Y. enterocolitica, among others, which are repressed at temperatures below that of the host.
The effect of osmolarity on virulence gene expression has only been studied to a limited degree. This is particularly important in the case of fish pathogens because osmolarity could define in some cases the range of fish species (saltwater or freshwater fish) that a pathogen such as Y. ruckeri could infect. The expression of yrp1 was maximal in NB without added NaCl, KCl,or xylose. The fact that a significant decrease of expression of the yrp1 operon took place at a low d-xylose concentration (100 mM), together with the low level of expression detected with 500 mM NaCl or KCl, indicates that the yrp1 operon is influenced by the osmotic pressure of the medium, and this probably limits the capacity to generate disease to saltwater fish. In A. hydrophila, virulence is related to osmolarity in an opposite way. Thus, it was more virulent for fish when it was grown at high osmolarity, increasing, at the same time, its caseinolytic and hemolytic extracellular activities (1). Osmolarity also affects the regulation of some Y. enterocolitica genes such as yst (32) and fleABC (21) or the type IV pilus gene cluster of Y. pseudotuberculosis (4).
The use of an operon fusion with β-galactosidase activity as a label was very useful in determining the in vivo expression of the yrp1 gene. This approach has been used in different organisms but, as far as we know, this is the first time that it has been applied to fish. Clearly, the yrp1 gene expression occurred mainly in the gill and intestine tissue. The fact that different bacterial genes were expressed in the same location strongly suggests that, more than a specific pattern of gene expression in tissues, this result shows the location of Y. ruckeri in fish. This result also shows that other tissues and organs are less invaded. It is not clear why Y. ruckeri has such a tropism, but it may be related to the presence in both tissues of an accessible superficial capillary system. More studies using immersion experiments should be done for defining the entry site, colonization, and invasion and to show the importance of the gastrointestinal tract as a portal of entry, as suggested by Ross et al. (43) for this bacterium and as has been shown for E. tarda (25). This technique could be a useful tool for studying temporal gene expression, as well as for performing experiments on strain competition, the development of infection and colonization, routes of infection, etc.
Taking theses results as a whole, we conclude that the Yrp1 protease, the first virulence factor defined in Y. ruckeri, presents similarities in its mode of regulation to other virulence genes from mammal pathogenic Yersinia and plays a nonessential, although relevant, role during infection and ERM disease development in salmonids.
Acknowledgments
This study was supported in part by the Spanish MCYT (grant AGL2000-0869). L.F. and A.M. were recipients of an FPI grant from the Spanish Ministerio de Ciencia y Tecnologia.
We thank A. del Cerro and B. Alvarez for help and advice and also A. F. Braña for assistance and critical reading of the manuscript.
REFERENCES
- 1.Aguilar, A., S. Merino, X. Rubires, and J. M. Tomas. 1997. Influence of osmolarity on lipopolysaccharides and virulence of Aeromonas hydrophila serotype O:34 strains grown at 37°C. Infect. Immun. 65:1245-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altinok, I., J. M. Grizzle, and Z. Liu. 2001. Detection of Yersinia ruckeri in rainbow trout blood by use of the polymerase chain reaction. Dis. Aquat. Org. 44:29-34. [DOI] [PubMed] [Google Scholar]
- 3.Burger, M., R. G. Woods, C. McCarthy, and I. R. Beachem. 2000. Temperature regulation of protease in Pseudomonas fluorescens LS107d2 by an ECF sigma factor and a transmembrane activator. Microbiology 146:3149-3155. [DOI] [PubMed] [Google Scholar]
- 4.Collyn, F., M. A. Lety, S. Nair, V. Escuyer, A. Ben Younes, M. Simonet., and M. Marceau. 2002. Yersinia pseudotuberculosis harbors a type IV pilus gene cluster that contributes to pathogenicity. Infect. Immun. 70:6196-6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Colquhoun, D. J., and H. Sorum. 2001. Temperature-dependent siderophore production in Vibrio salmonicida. Microbial Pathol. 31:213-219. [DOI] [PubMed] [Google Scholar]
- 6.Contag, C. H., and M. H. Bachmann. 2002. Advances in in vivo bioluminescence imaging of gene expression. Annu. Rev. Biomed. Eng. 4:235-260. [DOI] [PubMed] [Google Scholar]
- 7.Coquet, L., P. Cosette, L. Quillet, F. Petit, G.-A. Junter, and T. Jouenne. 2002. Occurrence and phenotypic characterization of Yersinia ruckeri strains with biofilm-forming capacity in a rainbow trout farm. Appl. Environ. Microbiol. 68:470-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cornelis, G. R. 1988. The Yersinia deadly kiss. J. Bacteriol. 180:5495-5504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cornelis, G. R., A. Boland, A. P. Boyd, C. Genijen, M. Iriarte, C. Neyt, M.-P. Sory, and I. Stanier. 1998. The virulence plasmid of Yersinia, an antihost genome. Microbiol. Mol. Biol. Rev. 62:1315-1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davies, R. L. 1991. Yersinia ruckeri produces four iron-regulated outer membranes proteins but does not produce detectable siderophores. J. Fish. Dis. 14:563-570. [Google Scholar]
- 11.de Koning-Ward, T. F., and R. M. Robins-Browne. 1997. A novel mechanism of urease regulation in Yersinia enterocolitica. FEMS Microbiol. Lett. 147:221-226. [DOI] [PubMed] [Google Scholar]
- 12.Everlyn, T. P. T. 1996. Infection and disease, p. 339-362. In G. Iwana and T. Nakanishi (ed.), The fish immune system: organism, pathogen and environment. Academic Press, Inc., San Diego, Calif.
- 13.Fangk, H. M., C. Lingr, and G. M. Sin. 2002. Enhancement of protective immunity in blue gourami, Trichogaster trichopterus (Pallas), against Aeromonas hydrophila and Vibrio anguillarum by A. hydrophila major adhesion. J. Fish Dis. 23:137-145. [Google Scholar]
- 14.Fernadez-Alonso, M., A. Rocha, and J. M. Coll. 2001. DNA vaccination by immersion and ultrasound to trout viral haemorrhagic septicaemia virus. Vaccine 19:3067-3075. [DOI] [PubMed] [Google Scholar]
- 15.Fernandez, L., P. Secades, J. R. Lopez, I. Marquez, and J. A. Guijarro. 2002. Isolation and análisis of a protease gene with an ABC transport system in the fish pathogen Yersinia ruckeri: insertional mutagenesis and involvement in virulence. Microbiology 148:2233-2243. [DOI] [PubMed] [Google Scholar]
- 16.Furones, M. D., M. L. Gilpin, D. J. Alderman, and C. B. Munn. 1990. Virulence of Yersinia ruckeri serotype 1 strains is associated with a heat sensitive factor (HSF) in cell extracts. FEMS Microbiol. Lett. 66:339-344. [DOI] [PubMed] [Google Scholar]
- 17.Furones, M. D., M. L. Gilpin, and C. B. Munn. 1993. Culture media for the differentiation of isolates of Yersinia ruckeri based on detection of a virulence factor. J. Appl. Bacteriol. 74:360-366. [DOI] [PubMed] [Google Scholar]
- 18.Griffin, E. 1991. Environmental regulation of bacterial virulence-implication for vaccine design and production. Trends Biotechnol. 9:309-315. [DOI] [PubMed] [Google Scholar]
- 19.Heppell, J., and H. L. Davis. 2000. Application of DNA vaccine technology to aquaculture. Adv. Drug Deliv. Rev. 43:29-43. [DOI] [PubMed] [Google Scholar]
- 20.Heusipp, G., G. M. Young, and V. L. Miller. 2001. HreP, an in vivo-expressed protease of Yersinia enterocolitica, is a new member of a family of subtilisin/kexin-like proteases. J. Bacteriol. 183:3556-3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kapatral, V., and S. A. Minnich. 1995. Co-ordinate, temperature-sensitive regulation of the three Yersinia enterocolitica. Mol. Microbiol. 9:49-56. [DOI] [PubMed] [Google Scholar]
- 22.Konkel, M. E., and K. Tilly. 2000. Temperature-regulated expression of bacterial virulence genes. Microb. Infect. 2:157-166. [DOI] [PubMed] [Google Scholar]
- 23.Kuzyk, M. A., J. Burian, D. Machander, D. Dolhaine, S. Cameron, J. C. Thornton, and W. W. Kay. 2001. An efficacious recombinant subunit vaccine against the salmonid rickettsial pathogen Piscirickettsia salmonis. Vaccine 19:2337-2344. [DOI] [PubMed] [Google Scholar]
- 24.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 25.Ling, S. H. M., X. H. Wang, T. M. Lim, and K. Y. Leung. 2001. Green fluorescent protein-tagged Edwardsiella tarda reveals portal of entry in fish. FEMS Microbiol. Lett. 194:239-243. [DOI] [PubMed] [Google Scholar]
- 26.Ling, S. H. M., X. H. Wang, L. Xie, T. M. Lim, and K. Y. Leung. 2000. Use of green fluorescent protein (GFP) to study the invasion pathways of Edwardsiella tarda in in vivo and in vitro fish models. Microbiology 146:7-19. [DOI] [PubMed] [Google Scholar]
- 27.Lorenzen, N., E. Lorenzen, K. Einer-Jensen, and S. E. LaPatra. 2002. DNA vaccines as a tool for analysing the protective immune response against rhabdoviruses in rainbow trout. Fish Shellfish Immunol. 12:439-453. [DOI] [PubMed] [Google Scholar]
- 28.Lutwyche, P., M. M. Exner, R. E. W. Hancock, and T. J. Trust. 1995. A conserved Aeromonas salmonicida porin provides protective immunity to rainbow trout. Infect. Immun. 63:3137-3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mahan, M. J., J. M. Slauch, and J. J. Mekalanos. 1993. Selection of bacterial virulence genes that are specifically induced in host tissue. Science 259:686-688. [DOI] [PubMed] [Google Scholar]
- 30.Mahan, M. J., J. W. Tobias, J. M. Slauch, P. C. Hanna, R. J. Collier, and J. J. Mekalanos. 1995. Antibiotic-based selection for bacterial genes that are specifically induced during infection of a host. Proc. Natl. Acad. Sci. USA 92:669-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mekalanos, J. J. 1992. Environmental signals controlling expression of virulence determinants in bacteria. J. Bacteriol. 174:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mikulskis, A. V., I. Delor, V. H. Thi, and G. R. Cornelis. 1994. Regulation of the Yersinia enterocolitica enterotoxin yst gene. Influence of growth phase, temperature, osmolaryty, pH, and bacterial host factors. Mol. Microbiol. 14:905-915. [DOI] [PubMed] [Google Scholar]
- 33.Miller, J. H. 1972. Experiments in molecular genetics, p. 354. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 34.Miyoshi, S.-I., and S. Shinoda. 2000. Microbial metalloproteases and pathogenesis. Microbiol. Infect. 2:91-98. [DOI] [PubMed] [Google Scholar]
- 35.Pepe, J. C., J. L. Badger, and V. L. Miller. 1994. Growth phase and low pH affect the thermal regulation of the Yersinia enterocolitica inv gene. Mol. Microbiol. 11:123-135. [DOI] [PubMed] [Google Scholar]
- 36.Revell, P. A., and V. L. Miller. 2001. Yersinia virulence: more than a plasmid. FEMS Microbiol. Lett. 201:159-164. [DOI] [PubMed] [Google Scholar]
- 37.Rodgers, C. J. 1992. Development of a selective-differential medium for the isolation of Yersinia ruckeri and its application in epidemiological studies. J. Fish Dis. 15:243-254. [Google Scholar]
- 38.Romalde, J. L., J. L. Barja, B. Magariños, and A. E. Toranzo. 1994. Starvation-survival processes of the bacterial fish pathogen Yersinia ruckeri. Syst. Appl. Microbiol. 17:161-168. [Google Scholar]
- 39.Romalde, J. L., R. F. Conchas, and A. E. Toranzo. 1991. Evidence that Yersinia ruckeri possesses a high affinity iron uptake system. FEMS Microbiol. Lett. 80:121-126. [DOI] [PubMed] [Google Scholar]
- 40.Romalde, J. L., M. L. Lemos, R. F. Conchas, I. Bandín, and A. E. Toranzo. 1990. Adhesive properties and other virulence factors in Yersinia ruckeri, p. 123-139. In T. C. Cheng and F. O. Perkins (ed.), Pathology in marine science. Academic Press, Inc., New York, N.Y.
- 41.Romalde, J. L., Y. Santos, and A. E. Toranzo. 1992. Presence of skin permeability factors in the extracellular products of Yersinia ruckeri. Curr. Microbiol. 24:263-267. [Google Scholar]
- 42.Romalde, J. L., and A. E. Toranzo. 1993. Pathological activities of Yersinia ruckeri, the enteric redmouth (ERM) bacterium. FEMS Microbiol. Lett. 112:291-300. [DOI] [PubMed] [Google Scholar]
- 43.Ross, A. J., R. R. Rucker, and W. H. Ewing. 1966. Description of a bacterium associated with redmouth disease of rainbow trout (Salmo gairdneri). Can. J. Microbiol. 12:763-770. [DOI] [PubMed] [Google Scholar]
- 44.Schemiel, D. H., G. M. Young, and V. L. Miller. 2000. The Yersinia enterocolitica phospholipase gene yplA is part of the flagellar regulon. J. Bacteriol. 182:2314-2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Secades, P., and J. A. Guijarro. 1999. Purification and characterization of an extracellular protease from the fish pathogen Yersinia ruckeri and effect of culture conditions on production. Appl. Environ. Microbiol. 65:3969-3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Secades, P., B. Alvarez, and J. A. Guijarro. 2001. Purification and characterization of a psychrophilic calcium induced, growth-phase-dependent metalloprotease from the fish pathogen Flavobacterium psychrophilum. Appl. Environ. Microbiol. 67:2436-2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stevenson, R. M. W. 1997. Immunization with bacterial antigens: yersiniosis. Dev. Biol. Stand. 90:117-124. [PubMed] [Google Scholar]
- 48.Straley, S. C., and R. D. Perry. 1995. Environmental modulation of gene expression and pathogenesis in Yersinia. Trends Microbiol. 3:310-317. [DOI] [PubMed] [Google Scholar]
- 49.Van Roessel, P., and A. H. Brand. 2002. Imaging into the future: visualizing gene expression and protein interactions with fluorescent proteins. Nat. Cell Biol. 4:E15-E20. [DOI] [PubMed] [Google Scholar]
- 50.Woods, R. G., M. Burger, C. A. Beven, and I. R. Beacham. 2001. The aprX-lipA operon of Pseudomonas fluorescens B52: a molecular analysis of metalloprotease and lipase production. Microbiology 147:345-354. [DOI] [PubMed] [Google Scholar]