Abstract
This study determined the changes in microRNA expression in mammalian Chinese hamster ovary (CHO) cells undergoing apoptosis induced by exposing the cells to nutrient-depleted media. The apoptosis onset was confirmed by reduced cell viability and caspase-3/7 activation. Microarray comparison of known mouse and rat microRNA’s in CHO cells exposed to fresh or depleted media revealed up-regulation of the mouse miR-297-669 cluster in CHO cells subjected to depleted media. Mmu-miR-466h was chosen for further analysis as the member of this cluster with the highest overexpression and its up-regulation in depleted media was confirmed with qRT-PCR.
Since microRNAs suppress mRNA translation, we hypothesized that up-regulated mmu-miR-466h inhibits anti-apoptotic genes and induces apoptosis. A combination of bioinformatics and experimental tools was used to predict and verify mmu-miR-466h anti-apoptotic targets. 8708 predicted targets were obtained from miRecords database and narrowed to 38 anti-apoptotic genes with DAVID NCBI annotation tool. Several genes were selected from this anti-apoptotic subset based on nucleotide pairing complimentarity between the mmu-miR-466h seed region and 3′ UTR of the target mRNAs. qRT-PCR analysis revealed reduced mRNA levels of bcl2l2, dad1, birc6, stat5a and smo genes in CHO cells exposed to depleted media. The inhibition of the mmu-miR-466h increased the expression levels of those genes and resulted in increased cell viability and decreased caspase-3/7 activation. The up-regulation of mmu-miR-466h in response to nutrients depletion causes the inhibition of several anti-apoptotic genes in unison. This suggests the pro-apoptotic role of mmu-miR-466h and its capability to modulate the apoptotic pathway in mammalian cells.
Introduction
Mammalian cells are widely used in biotechnology for the production of complex recombinant proteins due to their ability to convey the proper protein folding and the human-like post-translational modifications. Chinese hamster ovary (CHO) cells are the primary mammalian cell culture systems utilized for biopharmaceuticals production (Werner et al. 1998). One of the drawbacks in the CHO cell production system is low stress tolerance of the cells in bioreactors, which affects productivity. Different stress conditions in bioreactors such as nutrients and growth factors depletion, shear stresses, metabolites accumulation, and hypoxia may cause apoptosis by lowering the viable cells’ densities which has a negative effect on proteins yields and quality (Lim et al. 2010). The apoptosis cascade is initiated as a sequence of complex signaling networks starting inside or outside the cell. There are two major apoptosis-initiating pathways: intrinsic (mitochondria dependent and endoplasmic reticulum) and extrinsic (receptor mediated) pathways (Kroemer and Reed 2000; Rossi and Gaidano 2003; Thorburn 2004). These pathways initially activate the upstream cysteine aspartate proteases (caspases), Caspases-8,9,10,12, followed by the activation of downstream Caspases-3,6,7 which carry out the final steps of cell death (Barisic et al. 2003; Majors et al. 2007).
There are a number of possible stimuli that can activate the apoptosis pathways including UV irradiation, death ligands, chemical toxins and deprivation of nutrients (Mastrangelo and Betenbaugh 1998). It was previously reported that glucose and glutamine exhaustion as well as depletion of some essential amino acids induce apoptosis in mammalian cell cultures. Glucose is one of the main nutrients supporting growth and maintaining viability in mammalian cell cultures and its depletion can activate apoptosis cascade (Bialik et al. 1999; Mercille and Massie 1994; Moley and Mueckler 2000). There are several possible apoptotic scenarios resulting from glucose metabolism impairment. First, reduced glucose levels lead to ATP depletion which triggers the mitochondria-mediated apoptosis pathway. Secondly, decreased glycolysis resulting from glucose transport limitation causes oxidative stress and shift in a redox balance of the cell. This activates a signal transduction cascade which was shown to cause death of human cancer cell lines (Blackburn et al. 1999). In addition, glucose deprivation can result in increased expression of Hif-1alpha, stabilization of p53, and onset of p53-mediated apoptosis (Carmeliet et al. 1998)
The current approaches to optimize mammalian cell cultures are directed toward delay or reduction of apoptosis to minimize the extensive loss of viable cells and increase the production. There are two main strategies to inhibit or slow down the onset of apoptosis. One involves the manipulation of the outer cellular environment by media supplementation of growth factors, limiting nutrients, and hydrolysates (Majors et al. 2007; Zanghi et al. 1999). The second approach is intended on changing of the intracellular biochemical environment using genetic engineering to reallocate the tightly regulated balance of pro- and anti-apoptotic factors in favor of the anti-apoptotic proteins (Chiang and Sisk 2005; Lim et al. 2006; Wong et al. 2006). Increased viability and extended cultures in CHO cells were achieved by exogenous expression of Bcl-2 and Bcl-xL proteins known to reduce apoptosis by binding the bcl-2 pro-apoptotic factors and maintaining the integrity of mitochondrial membrane (Arden et al. 2007; Janumyan et al. 2003; Kim 2005). Also, the overexpression of viral domain-containing homologs of bcl-2 such as E1B19K and Aven was reported to inhibit the apoptosis in mammalian cell cultures (Figueroa et al. 2007; Mercille and Massie 1999).
MicroRNAs (miRs) were recently discovered as small, single stranded non-coding regulatory RNA molecules 18–25 nucleotides in length (Tang et al. 2009). They have been found to be highly conserved in genomes of animals, plants, fungi and viruses, and account for approximately 1% of the human genome (Lynam-Lennon et al. 2009). MiRs are usually transcribed as long primary microRNA transcripts (pri-miRs) several kb in length with a stem-loop structure containing a 5′-end cap and 3′-poly (A) tail (Cai et al. 2004). Pri-miRs undergo a two-step cleavage to produce the mature functional microRNA. The first cleavage step is facilitated in the nucleus by endonuclease RNase III called Drosha which generates a stem-loop miR precursor 70 nucleotides long called pre-microRNA (Lee et al. 2003). The latter is then transported to the cytoplasm via Exportin-5 and Ran-GTP mediated mechanism (Lynam-Lennon et al. 2009). The second miR maturation step is facilitated by another RNase III endonuclease, called Dicer in the cytoplasm which produces microRNA/microRNA* duplex of 18–25 nucleotides in length. The duplex is subsequently loaded to the RNA-induced silencing complex (RISC) which is activated upon unwinding and degradation of the complementary microRNA* strand (Schwarz et al. 2003). The single-stranded miR guides the RISC complex to its target mRNA. The miR to mRNA binding is restricted to the 3′-UTR region of target mRNA and 5′-end of targeting miR (5′ seed region between 2nd and 8th nucleotides of miR) (Lynam-Lennon et al. 2009). Upon its recognition, target mRNA either gets degraded via RNA interference process in case of sufficient miR-mRNA bases complimentarity, or sequestered and stored in cytoplasmic processing bodies away from the translational machinery in cases of insufficient complimentarity (Hutvagner and Zamore 2002; Lynam-Lennon et al. 2009). In both cases the miR-guided regulation affects the translated protein levels.
MiRs have been found to be involved in numerous cellular processes including cell development, differentiation, metabolism, proliferation and death. Changes in miRs expression profiles have been associated with breast, prostate, and lung cancers development and progression, and recently a number of miRs have been associated with the regulation of apoptosis (Jovanovic and Hengartner 2006; Lynam-Lennon et al. 2009). However, only few studies attempted to characterize the involvement of miRs in regulation of nutrients homeostasis in mammalian cell cultures (Lanceta et al. 2010; Liang et al. 2009; Maes et al. 2008). It was suggested that miRNAs should be implemented in bioprocess applications. MiRs can be over-expressed or inhibited with antagomirs using relatively simple constructs (Gammell 2007; Muller et al. 2008). One of the main advantages of over-expressing miRs instead of the exogenous regulatory protein(s) is the fact that they do not compete for the translational machinery in the host cells. Since each miR targets several genes it can globally regulate the entire pathways of cellular growth, death, stress tolerance and protein secretion. Therefore, if the miRNAs expressions profiles are linked to known phenotype, the manipulation of the respective miRs may introduce another dimension to genetic engineering of mammalian cell factories.
This study examines the changes in miRNA expression profiles in Chinese Hamster Ovary (CHO) cells under conditions induced by nutrients depletion. The CHO cell system was selected because it is a well-characterized mammalian system for apoptosis investigation and control. Previous studies on apoptosis characterization in mammalian cell lines used CHO cells as a model organism, and some pro and anti-apoptotic genes have been explored in CHO cells (Arden et al. 2007; Chigancas et al. 2002; Figueroa et al. 2004; Reynolds et al. 1994). The current study identifies and confirms the pro-apoptotic role of a novel microRNA, mmu-miR-466h, and reveals some of its molecular targets based on a combination of bioinformatics prediction and experimental changes in the cellular phenotype.
Materials and Methods
Cells Growth and Viability Studies
Chinese Hamster Ovary cells adapted to growth in suspension (CHO-S) were purchased from Invitrogen (Cat. No. 11619-012) and grown in commercial CD-CHO serum-free media (Cat. No. 10743-029) supplemented with 8mM of L-glutamine (Cat. No. 25030-164). Cells were grown in 37°C, 5% CO2 humidified incubator. For the growth kinetic studies, cells were seeded at the concentration 2×105 cells/ml in 40 ml of media in triplicates in 125ml vented shake flasks and shaken at 130 rpm. Cells were sampled daily. Cells were counted using Cedex (Innovatis AG) and viability was assessed using Trypan Blue exclusion method. Glucose and lactate concentrations were determined by YSI 2700 Select biochemistry analyzer (YSI Life Sciences).
Nutrient-depleted Media Generation and Apoptotic Assays
To generate the depleted media cells were seeded at 2×105 cells/ml in 40 ml of CD-CHO media and grown for about 9 days until they were exhausted of nutrients and significantly lost viability (less than 15% viability). Depleted media was then collected, sterile-filtered and stored for the further studies. The EnzChek® Caspase-3 Assay Kit #1 (Invitrogen Cat. No. E13183) was used to detect the onset of apoptosis by assaying for increases in Caspase-3 (and other DEVD-specific proteases such as Caspase-7) activity. Briefly: cells pellets were prepared in sterile eppendorf tubes. After cells lysis the samples were normalized to the same proteins levels by BCA protein assay (Thermo Scientific Cat. No 23227). The increase in caspase-3/7 activity was assessed based on its specificity for Asp-Glu-Val-Asp (DEVD). The proteolytic cleavage of Z-DEVD-AMC substrate yielded a bright fluorescent product (excitation/emission ~342/441nm) and fluorescence was continuously monitored in the SPECTRAmax GEMINI-XS spectrofluorometer. For the apoptotic studies, samples were taken at different time points, pelleted and frozen, and then assayed altogether for caspase-3/7 activity continuously. 3.3×106 cells/ml were incubated in spent media during the initial apoptosis onset assessement, and 4×105 cells/ml during mmu-miR-466h inhibition analysis.
RNA Isolation
Total RNA was isolated from the samples using TRIzol® Reagent (Invitrogen Cat. No. 15596-018) according to the manufacturer’s protocol and quantified based on its absorbance at 260nm in NanoDrop 1000 (Thermo Scientific).
miRNA Analysis Using Microarrays
The analysis was performed by the service provider (LC Sciences, Houston, TX, http://www.lcsciences.com) using μParaflo® microfluidics synthesis technology (Zhu et al. 2007). Briefly: The probe content included 714 combined mouse and rat miRNAs listed in Sanger miRBase Release 12.0. The probes for the respective miRs were spotted to the μParaflo® microfluidics biochips. The probes were chemically modified and had normalized Tm-s ensuring uniform hybridization on the chip to enhance the sensitivity and specificity of the probes. Fresh and depleted media derived RNA samples were labeled with Cy5 and Cy3 (and vice versa to account for possible labeling bias) and hybridized to the wells containing probes for the respective miR. The fluorescent intensities of the wells were determined with respect to the control’s values. Multiple control probes were included on chip for quality controls of chip production, sample labeling and assay conditions. Among the control probes, PUC2PM-20B and PUC2MM-20 were respectively perfect match and single-based match of 20-mer RNA positive control sequences spiked into RNA samples before labeling. The assay stringency was assessed from the intensity ratio of PUC2PM-20B and PUC2MM-20 which was larger than 30. The sampling was performed in quadruplicate and average fluorescent intensities of fresh and depleted media exposed RNA samples were compared for each microRNA. The t-test and ANOVA were carried out on the ratios of the average fluorescent intensities for all respective microRNAs to assess their differentiation significance. The analyzed data were presented in the Table form which contained the respective average fluorescent intensities in fresh and depleted media samples and p-values for all significantly differentiated miRs. The raw fluorescence data was shown as separate Cy3 and Cy5 chips images for each sample (including the inverse labeling). The maps of overlaid images for the respective fluorescent signals in fresh and depleted media samples were also generated (one is shown in Figure 3).
Figure 3.
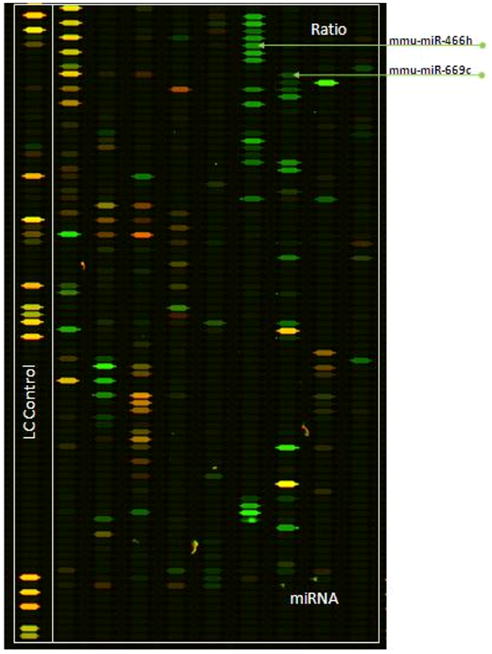
Map of the overlaid images of the respective microRNA fluorescent signals in nutrient-depleted and fresh media grown CHO-S cells. Each well represents a distinct microRNA. Depleted media derived RNA sample was labeled with Cy3 (green) and fresh media with Cy5 (red). The wells with probes for mmu-miR-466h and mmu-miR-669c are pointed at with arrows.
qRT-PCR Analysis of mRNA Levels
qRT-PCR analysis was performed in Prism 7900H Sequence Detector (Applied Biosystems) with 40 amplification cycles using TaqMan® chemistry according to the provided protocols for mRNAs detection. TaqMan® assays for bcl2l2, bcl2, birc6, cflar, dad1, naip7, smo, stat5a, and tegt genes were purchased from Applied Biosystems (AssayIDs:Mm00432054_m1, Mm_00477631_m1, Mm00464380_m1, Mm01255576_m1, Mm01319221_m1, Rn01757810_m1, Mm01162710_m1, Mm00839861_m1, Mm00509863_m1). The murine qRT-PCR TaqMan mRNA assays for the respective genes (if available) were selected with probes not discriminating from closely related species but restricted to at least 90 percent homology among mouse, human and rat in the probe-mRNA binding region. The data were normalized to 18S levels (Applied Biosystems Assay ID: Hs99999901_s1) in the respective sample, and 2−ΔΔCt method was performed for data interpretation. Measurements were performed in triplicates.
qRT-PCR Analysis of microRNA
The analysis was done using Prism 7900H Sequence Detector (Applied Biosystems) with 40 amplification cycles and TaqMan® chemistry according to the manufacturer’s protocols for miRNA detection. The mmu-miR-466h quantification was done using mmu-miR-466h assay with a stem loop primer specific for mmu-miR-466h. The assay was purchased from Applied Biosystems (Assay ID: AM002516). The 2−ΔΔCt analysis was done to estimate mmu-miR-466h differentiation. Mmu-miR-let-7c was used as internal endogenous control based on its least differentiation levels based on microarray statistical analysis (Assay ID: AM000379). SnoRNA202 (Assay ID: AM001232) was used as the second control. The mmu-miR-466h preamplification was performed in all samples to increase mmu-miR-466h levels in total RNA samples (it was barely detectable in fresh media samples due to low signal) for better qRT-PCR detection. The preamplification (10 cycles) was done using Applied Systems TaqMan® PreAmp Kit (Part No. 4384267) in PCR Thermal cycler (Applied Biosystems) after miRNA reverse transcription and before qRT-PCR reads. TaqMan® PreAmp Master Mix preamplifies small amounts of cDNA without introducing amplification bias to the sample and provides a very high correlation coefficients between amplified and unamplified cDNA (Li et al. 2008). qRT-PCR measurements were performed in quadruplicates.
Anti-miR™ Transfections and mmu-miR-466h Inhibition Studies
Transfections were performed using HiPerFect transfection reagent from Qiagen (Cat. No. 301704). To optimize transfection efficiency CHO-S cells were first transfected with FAM™ dye-labeled Anti-miR™ Negative Control #1 (Applied Biosystems ID: AM 17012) according to Qiagen miR transfection protocol for suspension cells. The optimal final concentration of the inhibitor was 162.5nM combined with the recommended HiPerFect reagent amount. Mmu-miR-466h was inhibited using the chemically modified single-stranded Anti-miR™ inhibitor specific for mmu-miR-466h (Applied Biosystems ID: AM12941). The inhibition was done on Corning Costar® 6-well plates (Cat. No. 3506). Briefly: ~106 cells/well were seeded in 400μl of CD-CHO medium. The miR-466h inhibitor or Anti-miR™ Negative Control #1 (a random sequence of single stranded chemically modified miR inhibitor–like molecule validated to not produce any effect on known microRNAs function-Applied Biosystems ID: AM17001) were incubated with HiPerFect reagent in 400μl of CD-CHO medium for 10min at room temperature. The transfection complexes were then added to the cells drop wise and the plates were shaken at 130rpm in 37°C, 5% CO2 humidified incubator. After 6 hours of incubation 1.6ml of CD-CHO media was added to each well. The cells were pelleted on the next day and resuspended in spent media. After designated point of depleted media exposure, the cells were pelleted for total RNA isolation or caspase-3/7 activity studies. For viability and Caspase activity comparison between spent media grown CHO-S cells with and without miR-466h inhibition, cells were resuspended in depleted media at least 22h after transfections, and assayed for caspases-3/7 activity and viability together with fresh media grown cells at different time points.
Results
Growth Rates and Identification of Apoptosis Onset During Incubation in Nutrient Depleted Media
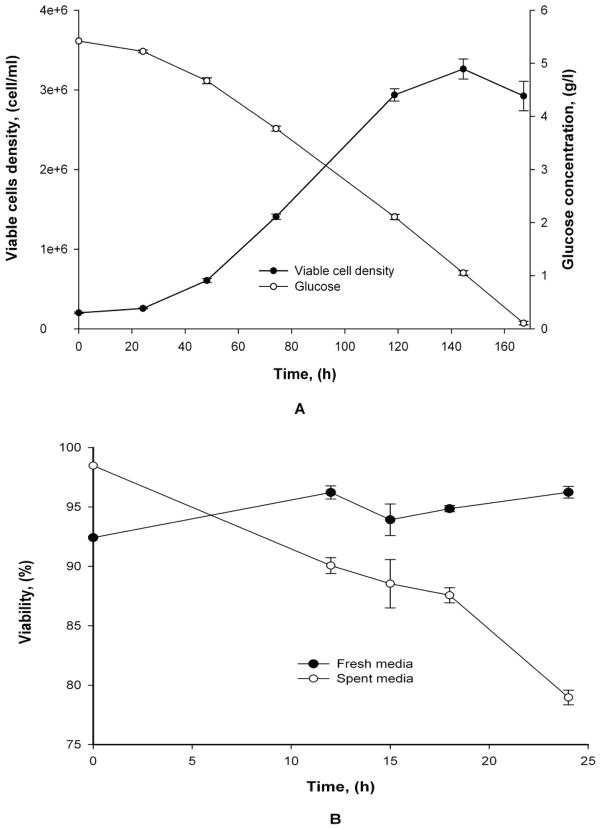
Growth and viability of CHO cells in fresh and nutrient-depleted media are shown in Figure 1. Cells were grown in fresh media to a concentration of 3.3×106 cells/ml, depleting most of the glucose (Figure 1A). At that concentration cells were resuspended in nutrient-depleted media obtained from 9 day old cultures. In the depleted media, cells exhibited a steady state decline in viability between 15 and 24 hours (Figure 1B) with almost no viable cells detected after 39 hours (data not shown). In contrast, when the cells were resuspended in fresh media at the same density, they maintained viability above 93 percent (Figure 1B).
Figure 1.
Growth and glucose depletion of CHO cells. (A)Fresh media growth, (B) Viability comparison
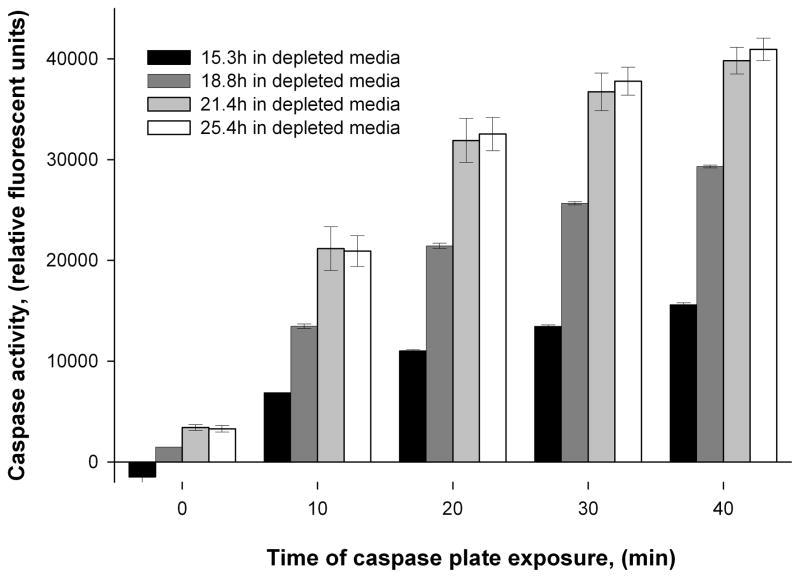
In order to evaluate if the apoptosis cascade was activated during the exposure of the cells to the depleted media the caspase-3/7 activity was assessed as shown in Figure 2. The time points for caspase-3/7 activity assessment were selected based on the viability decline (Figure 1B) when the cells were exposed to depleted media. The caspase-3/7 activity was found to correlate with the depleted media exposure time with the highest activity obtained for cell samples taken at 21.4 and 25.4 hours. The enhanced caspase-3/7 activities are a good indicator of the onset of apoptotic events since it follows the initial apoptotic stimuli and preliminary stages of pathway activation (Arden and Betenbaugh 2004; Majors et al. 2007). For the current study, it was important to prevent the complete execution phase of apoptosis orchestrated by Caspases-3/7 and 6 in order to avoid degradation of nucleic acids and proteins used for subsequent analysis. Therefore, a 24 hours exposure (when the viability was in the range of 80%) was selected as the point for microRNA expression analyses in fresh and nutrient-depleted media.
Figure 2.
Caspase-3/7 activity in CHO cells exposed to nutrient-depleted media. Assay was read continuously for 40 min in 10 min intervals.
Comparison of microRNA Profiles
Microarrays comparison was performed to examine the levels of microRNA (miR) expression in CHO cells exposed to fresh and nutrient- depleted media for 24 hours. Since there are no data available on microRNA for CHO (Chinese hamster ovary) cells, the screening was done for all known mouse and rat miRs (total of 714 microRNAs) given the conservation among closely related species (Babak et al. 2004; Gammell et al. 2007; Li and Ruan 2009). It is possible that because of non-specificity of the miR screening, some expressed miRs were not detected Shown in Figure 3 are superimposed images for the respective average miR fluorescent signals in RNA samples taken from cells grown in depleted or fresh media. Out of 300 combined mouse and rat miRs detected in CHO cells 70 miRs were found with more than 95 percent confidence (p<0.05) to be differentially expressed (were up or down-regulated at least 2 times) in the cells exposed to depleted media compared to cells exposed to fresh media. Sixty five out of the 70 differentially expressed miRs were found to be up-regulated in CHO cells exposed to nutrient-depleted media.
Due to the high number of differentially expressed miRs obtained from microarray data and the information available about miRs clustering, especially regulating multi-faceted cellular processes such as apoptosis (Mendell 2005; Vecchione and Croce 2009; Yue and Tigyi 2010), we evaluated if the up-regulated miRs were a part of an already identified cluster. The analysis revealed that the members of the mouse 297-669 miR cluster were up-regulated.
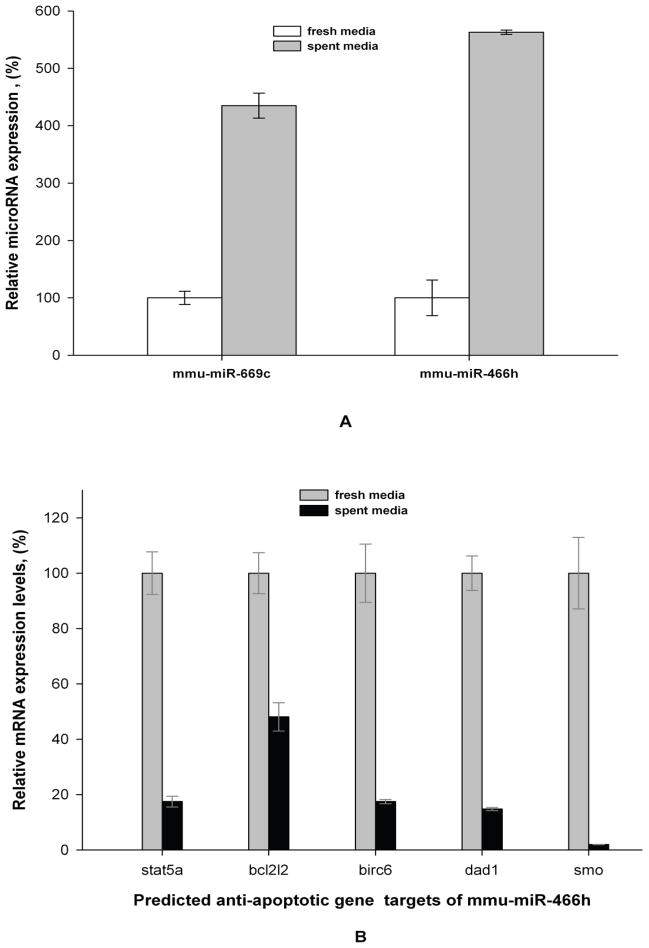
This cluster is composed of 28 miRs (according to Sanger miRBase Release 12.0), located in intron 10 of the mouse Sfmbt2 gene on Chromosome 2. The 18 detected members of mouse 297-669 miR cluster are listed in Table 1. All of them had low detection signals in CHO cells exposed to fresh media but were up-regulated in the depleted media with p-values at or below 0.05. Mmu-miR-466h was selected for subsequent analysis since its relative up-regulation was the highest among the members of the 297-669 miR cluster (Table 1). TaqMan qRT-PCR was performed on this miR to test for false positives which could be introduced by nonspecific microarray hybridization (Figure 4A). Mmu-miR-466h was found to be up-regulated 5.6 times in CHO cells grown in depleted media compared to its level in fresh media.
Table 1.
Microarray results for mouse 297-669 miR cluster in fresh and nutrient-depleted media exposed CHO-S cells. Relative fluorescent signals for all detected (homologous) members of this cluster were higher in depleted media exposed CHO-S cells than in fresh media exposed CHO-S cells after 24h. All detected sequences of mature miRs variants such as 5′-end pre-miR derived (marked 5p) and 3′-end pre-miR derived (marked 3p), and minor miRs variants (marked *) are shown.
| Mouse miR ID (mmu-) | Variant | Mature miR sequence | Fresh media relative fluorescent signal | Depleted media relative fluorescent signal | Up-regulation in depleted media (times) | p-value |
|---|---|---|---|---|---|---|
| miR-297a | - | AUGUAUGUGUGCAUGUGCAUGU | 14±4 | 1412±220 | 101±13 | 0.039 |
| miR-466a | 3p | UAUACAUACACGCACACAUAAGA | 8±1 | 203±30 | 25±1 | 0.050 |
| miR-466b | 3-3p | AAUACAUACACGCACACAUAAGA | 5±1 | 209±39 | 42±1 | 0.035 |
| miR-466d | 3p | UGUGUGUGCGUACAUGUACAUG | 10±1 | 648±122 | 65±6 | 0.039 |
| 5p | UAUACAUACACGCACACAUAG | 8±2 | 552±92 | 69±6 | 0.050 | |
| miR-466f | 3p | CAUACACACACACAUACACAC | 19±1 | 5056±105 | 266±8 | 0.009 |
| 5p | UACGUGUGUGUGCAUGUGCAUG | 102±13 | 22200±5982 | 218±31 | 0.036 | |
| miR-466g | - | AUACAGACACAUGCACACACA | 51±9 | 14935±4286 | 293±32 | 0.038 |
| miR-466h | - | UGUGUGCAUGUGCUUGUGUGUA | 29±4 | 13105±2366 | 452±19 | 0.023 |
| miR-467a | minor | AUAUACAUACACACACCUACAC | 44±1 | 9567±2001 | 217±40 | 0.025 |
| miR-467b | minor | AUAUACAUACACACACCAACAC | 72±8 | 14451±2817 | 201±17 | 0.027 |
| miR-467e | minor | AUAUACAUACACACACCUAUAU | 14±2 | 1056±38 | 75±8 | 0.022 |
| miR-669a | - | AGUUGUGUGUGCAUGUUCAUGU | 7±0 | 57±3 | 8±0 | 0.016 |
| miR-669b | - | AGUUUUGUGUGCAUGUGCAUGU | 8±0 | 46±1 | 6±0 | 0.003 |
| miR-669c | - | AUAGUUGUGUGUGGAUGUGUGU | 23±4 | 2870±299 | 125±9 | 0.023 |
| miR-669d | - | ACUUGUGUGUGCAUGUAUAUGU | 7±0 | 1511±463 | 216±66 | 0.038 |
| miR-669e | - | UGUCUUGUGUGUGCAUGUUCAU | 12±0 | 4090±971 | 341±81 | 0.027 |
| miR-669f | - | CAUAUACAUACACACACACGUAU | 44±1 | 13444±4267 | 306±90 | 0.037 |
| miR-669h | 3p | UAUGCAUAUACACACAUGCACA | 8±0 | 326±6 | 41±1 | 0.007 |
| miR-669i | - | UGCAUAUACACACAUGCAUAC | 18±1 | 39±1 | 2±0 | 0.048 |
Figure 4.
qRT-PCR comparison of mmu-miR-466h, mmu-miR-669c, and mRNA levels for mmu-miR-466h predicted targets in fresh and depleted media after 24h. (A) Mmu-miR-466h and mmu-miR-669c levels. The TaqMan microRNA assays were used for both miRs with sno202 and let-7c as controls for 2−ΔΔCt analysis. (B) mRNA levels for mmu-miR-466h predicted gene targets. Respective TaqMan mRNA assays were used to assess mRNA levels with 18S as control for 2−ΔΔCt analysis.
The up-regulation of one other member of the 297-669 cluster, mmu-miR-669c, was also verified by TaqMan qRT-PCR. This miR was found to be up-regulated 4.4 times in depleted media (Figure 4A). Mmu-miR-466h and mmu-miR-669c are designated with arrows on the microarray fluorescence map in Figure 3.
Identification of mmu-miR-466h Targets
It is likely that the over-expression of mmu-miR-466h following exposure to nutrient-depleted media promotes apoptosis by suppressing of its anti-apoptotic gene targets. Since there are no verified targets for mmu-miR-466h, the miRecords database which incorporates 11 microRNA targets prediction engines, was used to scan for all mmu-miR-466h possible targets (Xiao et al. 2009). All 8708 predicted gene targets for mmu-miR-466h were then entered into the DAVID NCBI (National Center for Biotechnology Information) annotation tool which classified all genes according to their biological roles. Thirty eight distinct anti-apoptotic genes were identified as potential mmu-miR-466h targets.
To narrow the potential target list, the 38 identified genes were further constrained by verifying their consistency across three prediction engines, miRanda, PITA, and RNAhybrid (Enright et al. 2003; Kertesz et al. 2007; Rehmsmeier et al. 2004), and then by analyzing the binding complementarity between mRNAs’ 3′-UTRs and 2–8 nucleotides of mmu-miR-466h (Kuhn et al. 2008; Yue et al. 2009). From this evaluation, nine potential anti-apoptotic genes were identified. The list of the nine putative gene targets, their known anti-apoptotic roles, and possible mmu-miR-466h binding sites are presented in Table 2.
Table 2.
Refined set of mmu-miR-466h possible targets. Table shows possible binding site(s) of mmu-miR-466h seed region(s) and mRNA 3′-UTRs of the respective target gene. Brief description of the known anti-apoptotic roles for those genes is given
| Mouse gene symbol | mmu-miR-466h binding site(s) in mRNA 3′-UTR (miR-466h binding nucleotides #) | Anti-apoptotic role of targeted gene |
|---|---|---|
| bcl2 | GCACAC (2–7) | Bcl2 is outer mitochondrial membrane protein which suppresses apoptosis either by preventing release of cytochrome C from mitochondria or by binding apoptosis activating factor (Apaf-1) |
| bcl2l2 | GCACAC(2–7) TGCACA(3–8) |
Bcl2l2 (bcl-w) inhibits formation of permeability transition pore and subsequent release of cytochrome C by binding to bax |
| birc6 | GCACA (3–7) | Birc6 (BRUCE, Apollon) can inhibit apoptosome by binding to Caspase-9 and occupying its active-site pocket. It can function as E2 ubiquitin conjugase for Caspase-9 and Smac/Diablo. Also, in response to loss of BIRC6 function p53 activates PIDD/Caspase-2 and bax/bak resulting in mitochondrial apoptosis |
| cflar | TGCACAC(2–8), 3 of TGCACA(3–8) GCACAC (2–7) |
Caspase-8 and FADD-like apoptosis regulator. Acts as an inhibitor of TNFRSF6 mediated apoptosis. It inhibits procaspase-8 cleavage and Caspase-8 activation. |
| dad1 | 2 of TGCACA (3–8) | Defender against cell death1. It is a component of N-oligosaccharyl transferase catalyzing transfer of oligosaccharide from lipid-linked donor to nascent polypeptide chain. Loss of dad1 was reported to trigger apoptosis |
| naip7 | 2 of GCACAC(2–7) | Naip7 (birc1g) Baculoviral iap repeat-containing 1g protein. BIR domains are known to inhibit apoptosis by direct inhibition of the caspase family of proteases |
| smo | TGCACAC (2–8) GCACAC (2–7) |
Part of Hedgehog signaling pathway. Activated smo uninhibits gli-1 transcriptional factor which stimulates up-regulation of bcl2 |
| stat5a | GCACAC (2–7) | Part of Jak-Stat signaling pathway. Stat5a dimers are transcriptional factors for bcl-xL and bcl2 genes |
| tegt | TGCACAC (2–8) | Tegt is a multipass membrane protein. Interacts with bcl2 and bcl-xL and inhibits bax |
To evaluate if the predicted mmu-miR-466h target genes were altered in nutrient-depleted media conditions, the mRNA levels for those genes were compared to the levels in fresh media using murine qRT-PCR analysis (Kantardjieff et al. 2009; Nissom 2007; Trummer et al. 2008). The relative expression of mRNA for bcl2l2, birc6, dad1, stat5a, and smo genes was lowered by levels ranging from 2.1 to 67 times in depleted medium as shown in Figure 4B. However, the mRNA of bcl2, cflar, naip7, and tegt genes were not detected in either fresh or depleted media, which could be the result of either no expression or no detection since mouse or rat probes were used for qRT-PCR analysis.
Inhibition of mmu-miR-466h
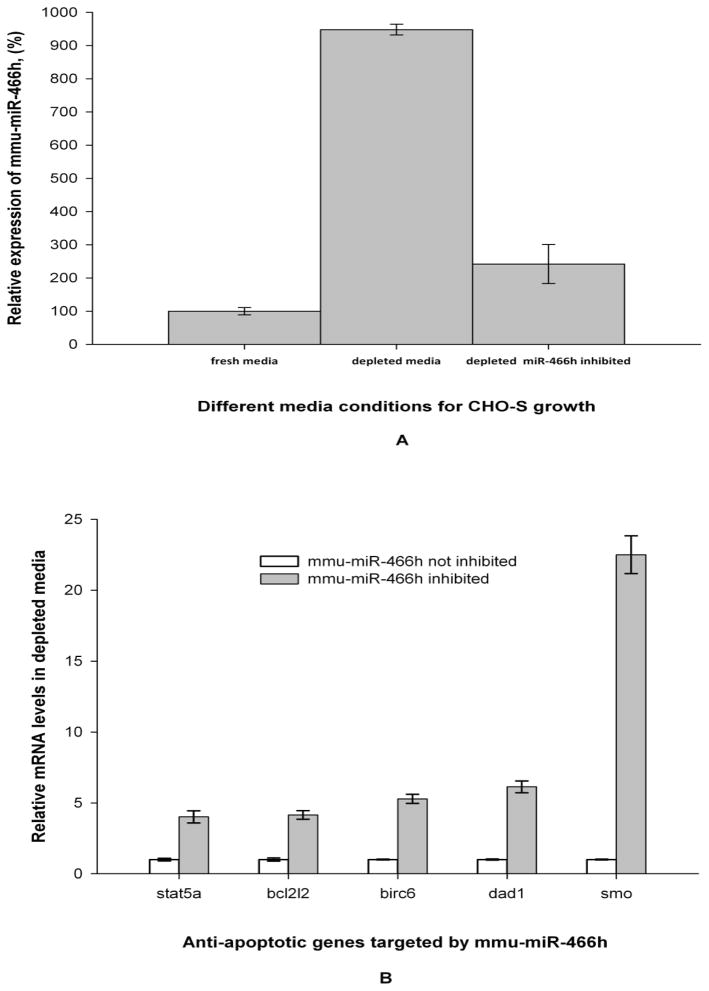
To evaluate the effects of mmu-miR-466h on its target genes, an anti-miR-466h (chemically modified single stranded oligonucleotide specific for mmu-miR-466h) was added to the culture media. The levels of mmu-miR-466h in fresh media and in the depleted media, with or without anti-miR-466h, are seen in Figure 5A. Compared with fresh media, mmu-miR-466h levels were 10 times higher in depleted media in the absence of the inhibitor, and were reduced by a factor of 4.5 in depleted media containing anti-miR-466h. The addition of the irrelevant anti-miR as a negative control to the depleted media did not alter the level of up-regulation of mmu-miR-466h (data not shown).
Figure 5.
Relative levels of mmu-miR-466h and its target genes in fresh and nutrient-depleted media with and without mmu-miR-466h inhibition. (A) Mmu-miR-466h levels. TaqMan microRNA qRT-PCR analysis was used to assess mmu-miR-466h levels at 23.5h in different media conditions with let-7c as control for 2−ΔΔCt analysis. (B) Relative mRNA levels of bcl2l2, birc6, dad1, stat5a, and smo genes in depleted media with and without mmu-miR-466h inhibition based on TaqMan qRT-PCR data. 18S levels were used as control for 2−ΔΔCt analysis.
To determine if the mmu-miR-466h inhibition in depleted media affected the expression of bcl2l2, birc6, dad1, stat5a, and smo genes, their mRNA levels were compared in depleted media in the presence or the absence of anti-miR-466h. The mRNA level for each gene increased between 4 and 23 fold in the presence of anti-miR-466h with the greatest relative increases observed in smo and dad1 genes (Figure 5B). Addition of the irrelevant anti-miR as a negative control at the same concentration as anti-miR-466h to the depleted media generated the respective mRNA levels that were comparable to their levels without inhibitor (data not shown).
Apoptotic Assessment of CHO-S Cells Grown in Depleted Media with and without anti-miR-466h
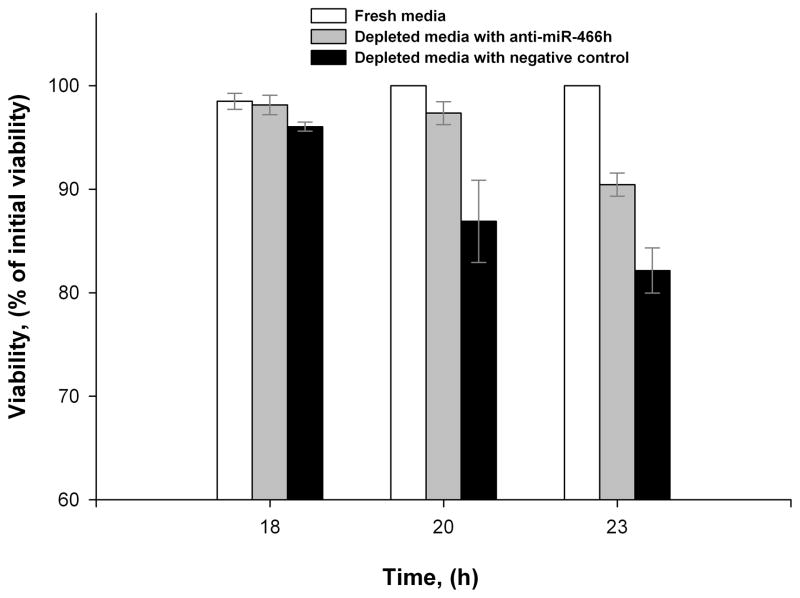
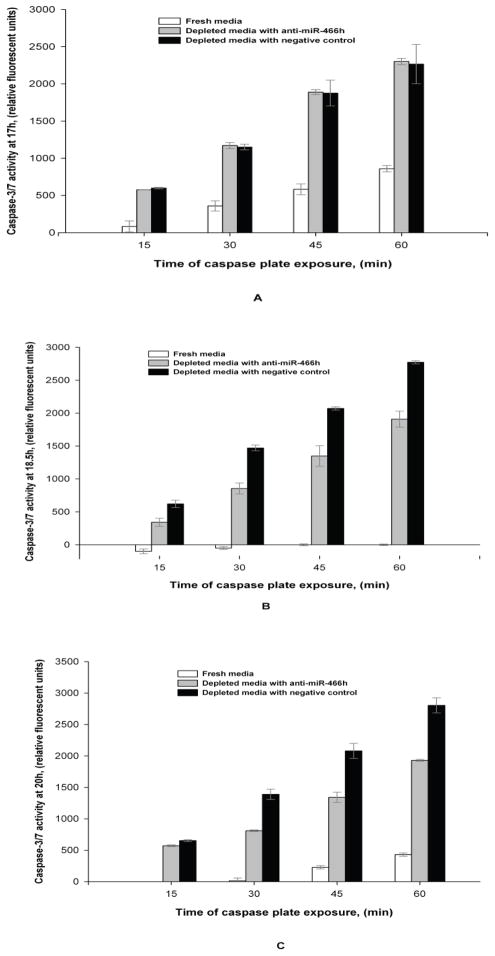
To examine a possible pro-apoptotic role of mmu-miR-466h, cell viability and caspase-3/7 activity were monitored in CHO cells exposed to fresh and depleted media with and without anti-miR-466h. As shown in Figure 6, cell viability in the depleted media started to decline after about 18 hours and fell to 81% by 23 hours. However, when the cells were treated with anti -miR-466h, the cell viability was higher at both the 20 and 23 hours time points. As for Caspase activity, samples were analyzed after 17, 18.5 and 20 hours of incubation in depleted media since Caspase activation often precedes the decline in cell viability (Nivitchanyong et al. 2007). Caspase -3/7 activities increased in the spent medium at all evaluated time points. There was no significant difference in Caspase-3/7 levels in CHO cells after 17 hours exposure to depleted media with or without anti-miR-466h (Figure 7A), but when evaluated after 18.5 and 20 hours exposure, the Caspase-3/7 activity was significantly reduced when anti-miR-466h was present (Figure 7B and 7C), which is consistent with the viability profiles shown in Figure 6.
Figure 6.
Viability comparison of CHO-S cells grown in fresh media and exposed to nutrient-depleted media with and without mmu-miR-466h inhibition.
Figure 7.
Comparison of Caspase-3/7 activity at different time points during fresh media incubation and nutrient-depleted media incubation with and without mmu-miR-466h inhibition. Depleted media incubated CHO cells were transfected with anti-miR-466h or anti-miR negative control 22h before depleted media exposure. Caspase plate was read continuously for 60 min in 15 min intervals. (A) 17h incubation, (B) 18.5h incubation, (C) 20h incubation.
Discussion
MicroRNAs were found to be involved in various cellular processes including apoptosis and its deregulation in cancer (Cheng et al. 2005; Hayashita et al. 2005; Kumar et al. 2007; Lynam-Lennon et al. 2009; Roldo et al. 2006). MicroRNAs involvement has been associated with regulation of intrinsic and extrinsic apoptotic pathways (Garofalo et al. 2008; Mott et al. 2007). Taking into consideration the complex nature and combined action of the apoptotic signals, it is not surprising that apoptosis would include several master regulators.
The purpose of this study was to identify microRNAs involved in apoptosis caused by nutrients depletion in mammalian cell culture and consequently attempt to affect this programmed cell death to create cells that are less sensitive to media changes and nutrient depletion. Group of microRNAs was found to be up-regulated when the cells were exposed to nutrient-depleted media; the group is located within intron 10 of the mouse Sfmbt2 gene and is clustered as 297-669 miRs. Since clustered miRs are known to be transcribed together as polycistronic transcrips to regulate mRNA of genes with similar functions, the 297-669 miR cluster may be transcribed as an intact pri-miR unit to stimulate apoptosis caused by nutrients depletion (Faraoni et al. 2009; Hayashita et al. 2005; Liang et al. 2009). Because of its highest level of up-regulation, mmu-miR-466h was selected for further analysis. The pro-apoptotic role of this miR was examined by investigating its effects on several anti-apoptotic genes. Currently, there is no clear agreement on the criteria for miRs targets validation (Kuhn et al. 2008). Given the considerable number of mmu-miR-466h possible targets and the intent to investigate its effects on the apoptotic phenotype we used two main criteria for mmu-miR-466h targets validation: qRT-PCR verification of mmu-miR-466h and its target genes co-expression, and the effects of mmu-miR-466h on its target biological function (Kuhn et al. 2008). Using a combination of bioinformatics and experimental approaches it was found that the up-regulation of this microRNA was associated with the down-regulation of the anti-apoptotic genes bcl2l2, dad1, birc6, stat5a and smo. These genes have been previously linked to nutrient deprivation in mammalian cells. Dad1 is a subunit of oligosaccharide transferase complex in endoplasmic reticulum which aids N-linked glycosylation of the proteins (Makishima et al. 2000; Sanjay et al. 1998). Glucose deprivation leads to the inhibition of N-linked glycosylation with lowered levels of dad1 (Makishima et al. 2000; Yoshimi et al. 2000), accumulation of improperly folded proteins in the endoplasmic reticulum (ER), and the ER apoptosis activation via unfolded protein response (Fribley et al. 2009). Bcl2l2 inhibits apoptosis by binding Bax which translocates to mitochondria and induces cytochrome C release in response to both oxidative stress and ATP levels reduction caused by glucose deprivation (Majors et al. 2007; Moley and Mueckler 2000). Bcl2l2 was found to be negatively regulated by mmu-miR-497 during ischemic conditions which could result in neuronal cell death (Yin et al.2010 ). Birc6 is known to protect the cells from glucose deprivation-induced hypoxic stress and the resulting p53 associated apoptosis (Aharinejad et al. 2008). Smo and stat5a promote the transcription of bcl2 and bcl-xL genes (Chen et al. 2008; Fujinaka et al. 2007; Xu et al. 2009a; Xu et al. 2009b), which protect the cells from apoptosis resulting from metabolic oxidative stress and reduction of ATP levels due to nutrient deprivation (Lee et al. 1997; Majors et al. 2007).
Inactivation of mmu-miR-466h by transient transfection with anti-miR-466h during exposure to nutrient-depleted media increased the levels of bcl2l2, dad1, birc6, stat5a, and smo genes four to twenty three fold. It is, therefore, likely that mmu-miR-466h plays a role in apoptosis induction caused by nutrient depletion through the simultaneous inhibition of several genes. However, it is possible that the levels of bcl2l2, dad1, birc6, stat5a, and smo were indirectly affected by mmu-miR-466h activity. MiRs are known to regulate cellular pathways by controlling the levels of multiple genes (Akao et al. 2006; Crawford et al. 2009; Yin et al. 2010 ). Mmu-miR-466h affects the apoptotic pathway by targeting at least five anti-apoptotic genes in unison, but the inhibition of mmu-miR-466h did not entirely save CHO cells from apoptosis onset when the cells were exposed to nutrient-depleted media. Compared with the negative control, the cells containing the anti-miR-466h showed a decrease in caspase-3/7 levels and an increase in cell viability. Although the relative up-regulation of mmu-miR-466h was the highest among all members of mouse 297-669 miR cluster and its pro-apoptotic activity became evident, other members of this cluster were also up-regulated and may be essential to complement the pro-apoptotic activity of mmu-miR-466h. One member of this cluster, mmu-miR-669c, has been previously associated with glutathione metabolism and oxidative stress in the aging mouse (Lanceta et al.2010; Maes et al. 2008).
This report is the first that links mmu-miR-466h to apoptosis, further understanding of its pro-apoptotic role and the role of the other members of 297-669 miR cluster can clarify the regulatory role of this cluster in mammalian cell death and other cellular processes. Despite the incomplete knowledge about the exact mechanism of mmu-miR-466h activity, the benefits of mmu-miR-466h induction or suppression in mammalian cell cultures are encouraging. It would be worthwhile to investigate the effects of this microRNA (and perhaps the whole 297-669 cluster) on the phenotypes related to deregulations in apoptotic program and metabolism for diseases such as cancer and diabetes. Considering the low expression levels of 297-669 miR cluster in mammalian cells during normal growth and that it has not been reported to be differentially expressed in cancer cells compared to healthy cells, it is possible that this cluster is associated with cell death processes in cancer cells. Stable suppression of mmu-miR-466h activity (and other members of 297-669 cluster) may generate apoptosis-resistant cell lines since single miR has a potential to regulate multiple gene targets. Additional studies searching for other genes affected by mmu-miR-466h and 297-669 miR cluster will be important for creating “stress tolerant” cell lines that are better suitable for biologicals production.
Acknowledgments
Funding was provided by the intramural program of the National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health.
References
- Aharinejad S, Andrukhova O, Lucas T, Zuckermann A, Wieselthaler G, Wolner E, Grimm M. Programmed cell death in idiopathic dilated cardiomyopathy is mediated by suppression of the apoptosis inhibitor Apollon. Ann Thorac Surg. 2008;86(1):109–14. doi: 10.1016/j.athoracsur.2008.03.057. discussion 114. [DOI] [PubMed] [Google Scholar]
- Akao Y, Nakagawa Y, Naoe T. let-7 microRNA functions as a potential growth suppressor in human colon cancer cells. Biol Pharm Bull. 2006;29(5):903–6. doi: 10.1248/bpb.29.903. [DOI] [PubMed] [Google Scholar]
- Arden N, Betenbaugh MJ. Life and death in mammalian cell culture: strategies for apoptosis inhibition. Trends Biotechnol. 2004;22(4):174–80. doi: 10.1016/j.tibtech.2004.02.004. [DOI] [PubMed] [Google Scholar]
- Arden N, Majors BS, Ahn SH, Oyler G, Betenbaugh MJ. Inhibiting the apoptosis pathway using MDM2 in mammalian cell cultures. Biotechnol Bioeng. 2007;97(3):601–14. doi: 10.1002/bit.21254. [DOI] [PubMed] [Google Scholar]
- Babak T, Zhang W, Morris Q, Blencowe BJ, Hughes TR. Probing microRNAs with microarrays: tissue specificity and functional inference. RNA. 2004;10(11):1813–9. doi: 10.1261/rna.7119904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barisic K, Petrik J, Rumora L. Biochemistry of apoptotic cell death. Acta Pharm. 2003;53(3):151–64. [PubMed] [Google Scholar]
- Bialik S, Cryns VL, Drincic A, Miyata S, Wollowick AL, Srinivasan A, Kitsis RN. The mitochondrial apoptotic pathway is activated by serum and glucose deprivation in cardiac myocytes. Circ Res. 1999;85(5):403–14. doi: 10.1161/01.res.85.5.403. [DOI] [PubMed] [Google Scholar]
- Blackburn RV, Spitz DR, Liu X, Galoforo SS, Sim JE, Ridnour LA, Chen JC, Davis BH, Corry PM, Lee YJ. Metabolic oxidative stress activates signal transduction and gene expression during glucose deprivation in human tumor cells. Free Radic Biol Med. 1999;26(3–4):419–30. doi: 10.1016/s0891-5849(98)00217-2. [DOI] [PubMed] [Google Scholar]
- Cai X, Hagedorn CH, Cullen BR. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA. 2004;10(12):1957–66. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmeliet P, Dor Y, Herbert JM, Fukumura D, Brusselmans K, Dewerchin M, Neeman M, Bono F, Abramovitch R, Maxwell P, et al. Role of HIF-1alpha in hypoxia-mediated apoptosis, cell proliferation and tumour angiogenesis. Nature. 1998;394(6692):485–90. doi: 10.1038/28867. [DOI] [PubMed] [Google Scholar]
- Chen XL, Cao LQ, She MR, Wang Q, Huang XH, Fu XH. Gli-1 siRNA induced apoptosis in Huh7 cells. World J Gastroenterol. 2008;14(4):582–9. doi: 10.3748/wjg.14.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng AM, Byrom MW, Shelton J, Ford LP. Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Res. 2005;33(4):1290–7. doi: 10.1093/nar/gki200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang GG, Sisk WP. Bcl-x(L) mediates increased production of humanized monoclonal antibodies in Chinese hamster ovary cells. Biotechnol Bioeng. 2005;91(7):779–92. doi: 10.1002/bit.20551. [DOI] [PubMed] [Google Scholar]
- Chigancas V, Batista LF, Brumatti G, Amarante-Mendes GP, Yasui A, Menck CF. Photorepair of RNA polymerase arrest and apoptosis after ultraviolet irradiation in normal and XPB deficient rodent cells. Cell Death Differ. 2002;9(10):1099–107. doi: 10.1038/sj.cdd.4401072. [DOI] [PubMed] [Google Scholar]
- Crawford M, Batte K, Yu L, Wu X, Nuovo GJ, Marsh CB, Otterson GA, Nana-Sinkam SP. MicroRNA 133B targets pro-survival molecules MCL-1 and BCL2L2 in lung cancer. Biochem Biophys Res Commun. 2009;388(3):483–9. doi: 10.1016/j.bbrc.2009.07.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS. MicroRNA targets in Drosophila. Genome Biol. 2003;5(1):R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faraoni I, Antonetti FR, Cardone J, Bonmassar E. miR-155 gene: a typical multifunctional microRNA. Biochim Biophys Acta. 2009;1792(6):497–505. doi: 10.1016/j.bbadis.2009.02.013. [DOI] [PubMed] [Google Scholar]
- Figueroa B, Jr, Ailor E, Osborne D, Hardwick JM, Reff M, Betenbaugh MJ. Enhanced cell culture performance using inducible anti-apoptotic genes E1B-19K and Aven in the production of a monoclonal antibody with Chinese hamster ovary cells. Biotechnol Bioeng. 2007;97(4):877–92. doi: 10.1002/bit.21222. [DOI] [PubMed] [Google Scholar]
- Figueroa B, Jr, Chen S, Oyler GA, Hardwick JM, Betenbaugh MJ. Aven and Bcl-xL enhance protection against apoptosis for mammalian cells exposed to various culture conditions. Biotechnol Bioeng. 2004;85(6):589–600. doi: 10.1002/bit.10913. [DOI] [PubMed] [Google Scholar]
- Fribley A, Zhang K, Kaufman RJ. Regulation of apoptosis by the unfolded protein response. Methods Mol Biol. 2009;559:191–204. doi: 10.1007/978-1-60327-017-5_14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujinaka Y, Takane K, Yamashita H, Vasavada RC. Lactogens promote beta cell survival through JAK2/STAT5 activation and Bcl-XL upregulation. J Biol Chem. 2007;282(42):30707–17. doi: 10.1074/jbc.M702607200. [DOI] [PubMed] [Google Scholar]
- Gammell P. MicroRNAs: recently discovered key regulators of proliferation and apoptosis in animal cells: Identification of miRNAs regulating growth and survival. Cytotechnology. 2007;53(1–3):55–63. doi: 10.1007/s10616-007-9049-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gammell P, Barron N, Kumar N, Clynes M. Initial identification of low temperature and culture stage induction of miRNA expression in suspension CHO-K1 cells. J Biotechnol. 2007;130(3):213–8. doi: 10.1016/j.jbiotec.2007.04.020. [DOI] [PubMed] [Google Scholar]
- Garofalo M, Quintavalle C, Di Leva G, Zanca C, Romano G, Taccioli C, Liu CG, Croce CM, Condorelli G. MicroRNA signatures of TRAIL resistance in human non-small cell lung cancer. Oncogene. 2008;27(27):3845–55. doi: 10.1038/onc.2008.6. [DOI] [PubMed] [Google Scholar]
- Hayashita Y, Osada H, Tatematsu Y, Yamada H, Yanagisawa K, Tomida S, Yatabe Y, Kawahara K, Sekido Y, Takahashi T. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 2005;65(21):9628–32. doi: 10.1158/0008-5472.CAN-05-2352. [DOI] [PubMed] [Google Scholar]
- Hutvagner G, Zamore PD. A microRNA in a multiple-turnover RNAi enzyme complex. Science. 2002;297(5589):2056–60. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
- Janumyan YM, Sansam CG, Chattopadhyay A, Cheng N, Soucie EL, Penn LZ, Andrews D, Knudson CM, Yang E. Bcl-xL/Bcl-2 coordinately regulates apoptosis, cell cycle arrest and cell cycle entry. EMBO J. 2003;22(20):5459–70. doi: 10.1093/emboj/cdg533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovanovic M, Hengartner MO. miRNAs and apoptosis: RNAs to die for. Oncogene. 2006;25(46):6176–87. doi: 10.1038/sj.onc.1209912. [DOI] [PubMed] [Google Scholar]
- Kantardjieff A, Nissom PM, Chuah SH, Yusufi F, Jacob NM, Mulukutla BC, Yap M, Hu WS. Developing genomic platforms for Chinese hamster ovary cells. Biotechnol Adv. 2009;27(6):1028–35. doi: 10.1016/j.biotechadv.2009.05.023. [DOI] [PubMed] [Google Scholar]
- Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microRNA target recognition. Nat Genet. 2007;39(10):1278–84. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- Kim R. Unknotting the roles of Bcl-2 and Bcl-xL in cell death. Biochem Biophys Res Commun. 2005;333(2):336–43. doi: 10.1016/j.bbrc.2005.04.161. [DOI] [PubMed] [Google Scholar]
- Kroemer G, Reed JC. Mitochondrial control of cell death. Nat Med. 2000;6(5):513–9. doi: 10.1038/74994. [DOI] [PubMed] [Google Scholar]
- Kuhn DE, Martin MM, Feldman DS, Terry AV, Jr, Nuovo GJ, Elton TS. Experimental validation of miRNA targets. Methods. 2008;44(1):47–54. doi: 10.1016/j.ymeth.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet. 2007;39(5):673–7. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- Lanceta J, Prough RA, Liang R, Wang E. MicroRNA group disorganization in aging. Exp Gerontol. 2010;45(4):269–78. doi: 10.1016/j.exger.2009.12.009. [DOI] [PubMed] [Google Scholar]
- Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425(6956):415–9. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- Lee YJ, Galoforo SS, Berns CM, Tong WP, Kim HR, Corry PM. Glucose deprivation-induced cytotoxicity in drug resistant human breast carcinoma MCF-7/ADR cells: role of c-myc and bcl-2 in apoptotic cell death. J Cell Sci. 1997;110 ( Pt 5):681–6. doi: 10.1242/jcs.110.5.681. [DOI] [PubMed] [Google Scholar]
- Li J, Smyth P, Cahill S, Denning K, Flavin R, Aherne S, Pirotta M, Guenther SM, O’Leary JJ, Sheils O. Improved RNA quality and TaqMan Pre-amplification method (PreAmp) to enhance expression analysis from formalin fixed paraffin embedded (FFPE) materials. BMC Biotechnol. 2008;8:10. doi: 10.1186/1472-6750-8-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Ruan K. MicroRNA detection by microarray. Anal Bioanal Chem. 2009;394(4):1117–24. doi: 10.1007/s00216-008-2570-2. [DOI] [PubMed] [Google Scholar]
- Liang R, Bates DJ, Wang E. Epigenetic Control of MicroRNA Expression and Aging. Curr Genomics. 2009;10(3):184–93. doi: 10.2174/138920209788185225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim SF, Chuan KH, Liu S, Loh SO, Chung BY, Ong CC, Song Z. RNAi suppression of Bax and Bak enhances viability in fed-batch cultures of CHO cells. Metab Eng. 2006;8(6):509–22. doi: 10.1016/j.ymben.2006.05.005. [DOI] [PubMed] [Google Scholar]
- Lim Y, Wong NS, Lee YY, Ku SC, Wong DC, Yap MG. Engineering mammalian cells in bioprocessing - current achievements and future perspectives. Biotechnol Appl Biochem. 2010;55(4):175–89. doi: 10.1042/BA20090363. [DOI] [PubMed] [Google Scholar]
- Lynam-Lennon N, Maher SG, Reynolds JV. The roles of microRNA in cancer and apoptosis. Biol Rev Camb Philos Soc. 2009;84(1):55–71. doi: 10.1111/j.1469-185X.2008.00061.x. [DOI] [PubMed] [Google Scholar]
- Maes OC, An J, Sarojini H, Wang E. Murine microRNAs implicated in liver functions and aging process. Mech Ageing Dev. 2008;129(9):534–41. doi: 10.1016/j.mad.2008.05.004. [DOI] [PubMed] [Google Scholar]
- Majors BS, Betenbaugh MJ, Chiang GG. Links between metabolism and apoptosis in mammalian cells: applications for anti-apoptosis engineering. Metab Eng. 2007;9(4):317–26. doi: 10.1016/j.ymben.2007.05.003. [DOI] [PubMed] [Google Scholar]
- Makishima T, Yoshimi M, Komiyama S, Hara N, Nishimoto T. A subunit of the mammalian oligosaccharyltransferase, DAD1, interacts with Mcl-1, one of the bcl-2 protein family. J Biochem. 2000;128(3):399–405. doi: 10.1093/oxfordjournals.jbchem.a022767. [DOI] [PubMed] [Google Scholar]
- Mastrangelo AJ, Betenbaugh MJ. Overcoming apoptosis: new methods for improving protein-expression systems. Trends Biotechnol. 1998;16(2):88–95. doi: 10.1016/s0167-7799(97)01159-1. [DOI] [PubMed] [Google Scholar]
- Mendell JT. MicroRNAs: critical regulators of development, cellular physiology and malignancy. Cell Cycle. 2005;4(9):1179–84. doi: 10.4161/cc.4.9.2032. [DOI] [PubMed] [Google Scholar]
- Mercille S, Massie B. Induction of apoptosis in nutrient-deprived cultures of hybridoma and myeloma cells. Biotechnol Bioeng. 1994;44(9):1140–54. doi: 10.1002/bit.260440916. [DOI] [PubMed] [Google Scholar]
- Mercille S, Massie B. Apoptosis-resistant E1B-19K-expressing NS/0 myeloma cells exhibit increased viability and chimeric antibody productivity under perfusion culture conditions. Biotechnol Bioeng. 1999;63(5):529–43. doi: 10.1002/(sici)1097-0290(19990605)63:5<529::aid-bit3>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- Moley KH, Mueckler MM. Glucose transport and apoptosis. Apoptosis. 2000;5(2):99–105. doi: 10.1023/a:1009697908332. [DOI] [PubMed] [Google Scholar]
- Mott JL, Kobayashi S, Bronk SF, Gores GJ. mir-29 regulates Mcl-1 protein expression and apoptosis. Oncogene. 2007;26(42):6133–40. doi: 10.1038/sj.onc.1210436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller D, Katinger H, Grillari J. MicroRNAs as targets for engineering of CHO cell factories. Trends Biotechnol. 2008;26(7):359–65. doi: 10.1016/j.tibtech.2008.03.010. [DOI] [PubMed] [Google Scholar]
- Nissom PM. Specific detection of residual CHO host cell DNA by real-time PCR. Biologicals. 2007;35(3):211–5. doi: 10.1016/j.biologicals.2006.09.001. [DOI] [PubMed] [Google Scholar]
- Nivitchanyong T, Martinez A, Ishaque A, Murphy JE, Konstantinov K, Betenbaugh MJ, Thrift J. Anti-apoptotic genes Aven and E1B-19K enhance performance of BHK cells engineered to express recombinant factor VIII in batch and low perfusion cell culture. Biotechnol Bioeng. 2007;98(4):825–41. doi: 10.1002/bit.21479. [DOI] [PubMed] [Google Scholar]
- Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10(10):1507–17. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds JE, Yang T, Qian L, Jenkinson JD, Zhou P, Eastman A, Craig RW. Mcl-1, a member of the Bcl-2 family, delays apoptosis induced by c-Myc overexpression in Chinese hamster ovary cells. Cancer Res. 1994;54(24):6348–52. [PubMed] [Google Scholar]
- Roldo C, Missiaglia E, Hagan JP, Falconi M, Capelli P, Bersani S, Calin GA, Volinia S, Liu CG, Scarpa A, et al. MicroRNA expression abnormalities in pancreatic endocrine and acinar tumors are associated with distinctive pathologic features and clinical behavior. J Clin Oncol. 2006;24(29):4677–84. doi: 10.1200/JCO.2005.05.5194. [DOI] [PubMed] [Google Scholar]
- Rossi D, Gaidano G. Messengers of cell death: apoptotic signaling in health and disease. Haematologica. 2003;88(2):212–8. [PubMed] [Google Scholar]
- Sanjay A, Fu J, Kreibich G. DAD1 is required for the function and the structural integrity of the oligosaccharyltransferase complex. J Biol Chem. 1998;273(40):26094–9. doi: 10.1074/jbc.273.40.26094. [DOI] [PubMed] [Google Scholar]
- Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115(2):199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- Tang Y, Zheng J, Sun Y, Wu Z, Liu Z, Huang G. MicroRNA-1 regulates cardiomyocyte apoptosis by targeting Bcl-2. Int Heart J. 2009;50(3):377–87. doi: 10.1536/ihj.50.377. [DOI] [PubMed] [Google Scholar]
- Thorburn A. Death receptor-induced cell killing. Cell Signal. 2004;16(2):139–44. doi: 10.1016/j.cellsig.2003.08.007. [DOI] [PubMed] [Google Scholar]
- Trummer E, Ernst W, Hesse F, Schriebl K, Lattenmayer C, Kunert R, Vorauer-Uhl K, Katinger H, Muller D. Transcriptional profiling of phenotypically different Epo-Fc expressing CHO clones by cross-species microarray analysis. Biotechnol J. 2008;3(7):924–37. doi: 10.1002/biot.200800038. [DOI] [PubMed] [Google Scholar]
- Vecchione A, Croce CM. Apoptomirs: small molecules have gained the license to kill. Endocr Relat Cancer. 2009;17(1):F37–50. doi: 10.1677/ERC-09-0163. [DOI] [PubMed] [Google Scholar]
- Werner RG, Noe W, Kopp K, Schluter M. Appropriate mammalian expression systems for biopharmaceuticals. Arzneimittelforschung. 1998;48(8):870–80. [PubMed] [Google Scholar]
- Wong DC, Wong KT, Nissom PM, Heng CK, Yap MG. Targeting early apoptotic genes in batch and fed-batch CHO cell cultures. Biotechnol Bioeng. 2006;95(3):350–61. doi: 10.1002/bit.20871. [DOI] [PubMed] [Google Scholar]
- Xiao F, Zuo Z, Cai G, Kang S, Gao X, Li T. miRecords: an integrated resource for microRNA-target interactions. Nucleic Acids Res. 2009;37(Database issue):D105–10. doi: 10.1093/nar/gkn851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Chen S, Chen H, Xiao Q, Hsu CY, Michael D, Bao J. STAT5 mediates antiapoptotic effects of methylprednisolone on oligodendrocytes. J Neurosci. 2009a;29(7):2022–6. doi: 10.1523/JNEUROSCI.2621-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu XF, Guo CY, Liu J, Yang WJ, Xia YJ, Xu L, Yu YC, Wang XP. Gli1 maintains cell survival by up-regulating IGFBP6 and Bcl-2 through promoter regions in parallel manner in pancreatic cancer cells. J Carcinog. 2009b;8:13. doi: 10.4103/1477-3163.55429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin KJ, Deng Z, Huang H, Hamblin M, Xie C, Zhang J, Chen YE. miR-497 regulates neuronal death in mouse brain after transient focal cerebral ischemia. Neurobiol Dis. 2010;38(1):17–26. doi: 10.1016/j.nbd.2009.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimi M, Sekiguchi T, Hara N, Nishimoto T. Inhibition of N-linked glycosylation causes apoptosis in hamster BHK21 cells. Biochem Biophys Res Commun. 2000;276(3):965–9. doi: 10.1006/bbrc.2000.3565. [DOI] [PubMed] [Google Scholar]
- Yue D, Liu H, Huang Y. Survey of Computational Algorithms for MicroRNA Target Prediction. Curr Genomics. 2009;10(7):478–92. doi: 10.2174/138920209789208219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue J, Tigyi G. Conservation of miR-15a/16-1 and miR-15b/16-2 clusters. Mamm Genome. 2010;21(1–2):88–94. doi: 10.1007/s00335-009-9240-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanghi JA, Fussenegger M, Bailey JE. Serum protects protein-free competent Chinese hamster ovary cells against apoptosis induced by nutrient deprivation in batch culture. Biotechnol Bioeng. 1999;64(1):108–19. doi: 10.1002/(sici)1097-0290(19990705)64:1<108::aid-bit12>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- Zhu Q, Hong A, Sheng N, Zhang X, Matejko A, Jun KY, Srivannavit O, Gulari E, Gao X, Zhou X. microParaflo biochip for nucleic acid and protein analysis. Methods Mol Biol. 2007;382:287–312. doi: 10.1007/978-1-59745-304-2_19. [DOI] [PubMed] [Google Scholar]