Introduction
There are many challenges associated with neonatal research. Neonates have limited blood volumes, are often too fragile for invasive procedures, and parental consent is difficult to obtain in studies involving experimental drugs or therapies. Hence, there is a strong need to develop new techniques for noninvasive neonatal monitoring in order to identify novel biomarkers that elucidate biological functions and predict disease states. Neonatal saliva overcomes many of the hurdles associated with research in neonates and offers the investigator a new and exciting sample source for exploring neonatal biology. As a product of whole blood filtration in the salivary glands, saliva contains water, electrolytes, proteins and genetic material. While the majority of cellular nucleic acids (DNA and RNA) contained in saliva originate from the buccal mucosa, cell-free nucleic acids originate from a wide variety of sources within the body. Thus, saliva has been deemed “the mirror of the body” with a biomarker profile that is similar to plasma [1–2].
Although dentists have long explored the diagnostic potential of saliva, the application of salivary analyses to neonates has been slow to emerge [3–4]. This delayed transition may be due to inherent technical challenges associated with limited neonatal salivary volumes, combined with a paucity of data delineating the clinical utility of neonatal salivary analyses. To address these issues, our research has focused on neonatal salivary gene expression, also known as the salivary transcriptome. This work highlights the enormous amount of informative, and potentially diagnostic, data available from neonatal saliva, and demonstrates how key developmental pathways may be targeted and explored. As part of our research, we have optimized acquisition protocols and extraction techniques to provide the researcher with the tools necessary to generate data on multiple diagnostic platforms from quantitatively limited salivary samples. This research is allowing us to explore physiological and pathological processes in premature neonates and utilize real-time clinical information derived from the salivary transcriptome to improve neonatal care and outcomes.
Methods
To examine biologically relevant information derived from the neonatal salivary transcriptome, serial whole saliva samples (50–200 µL) from premature infants (n=5; gestational age 28 to 32 weeks’) were collected during five distinct time points during their hospitalization: 1) shortly after birth prior to enteral nutrition; 2) at the start of enteral nutrition; 3) during the advancement of enteral nutrition; 4) at the start of oral feeding; and 5) at full or near full oral feeds. Total RNA was extracted, amplified and hybridized onto Affymetrix HG U133 2.0 Plus whole genomic microarrays for each baby for each time point (n=25). Arrays were normalized and underwent bioinformatic analyses to identify genes that were statistically significantly altering their expression over time. Genes identified underwent further analyses with the Ingenuity® Pathway Analyses (IPA) to better understand gene-gene relationships and important biological pathways associated with the normal physiological development of premature infants.
In order to optimize RNA extraction techniques, we compared RNA yield from two modified extraction protocols. Two saliva samples (10 – 100 µL) were serially collected from newborns (36 to 41 weeks’) (n=13). Total RNA was extracted from the cell-free salivary supernatant with either the 1) Qiagen RNAprotect® Saliva Mini Kit (Sample 1) or the 2) Qiagen QIAamp Viral RNA Mini Kit (Sample 2). Quantitative RT-PCR amplification for GAPDH was performed on extracted salivary samples. Statistical analyses were performed on mean threshold cycle (Ct) levels to compare RNA yield from each protocol.
Results
Comparative salivary analyses identified a subset of genes that statistically significantly increased (5,764) or decreased (3,522) their gene expression over time. Statistically significantly down-regulated expression was seen in embryonic development, connective tissue development and function, hematological system development and function, and organismal survival (10−14 < p <10−3). Conversely, genes associated with behavior, nervous system development, tissue development, organ development, and digestive system development were statistically significantly up-regulated (10−11 < p < 10−2) [5] (Figure 1). The gene lists highlighted a number of genes that appear to be associated with successful oral feeding in premature neonates. These genes included CCKAR, a known regulator of satiety, NPY2R, a neuronal regulator of food intake, GALR3, a modulator of feeding behavior, and a subset of genes associated with the development and proliferation of the trigeminal nerve (cranial nerve V). In addition, there were 49 genes associated with ‘digestive system development’ that were significantly upregulated over time in these premature infants (10−5 < p < 10−2). Finally, extraction with a modified protocol and the QIAamp Viral RNA Mini Kit yielded more RNA than a modified extraction protocol with the Qiagen RNAprotect® Saliva Mini Kit (p=0.008).
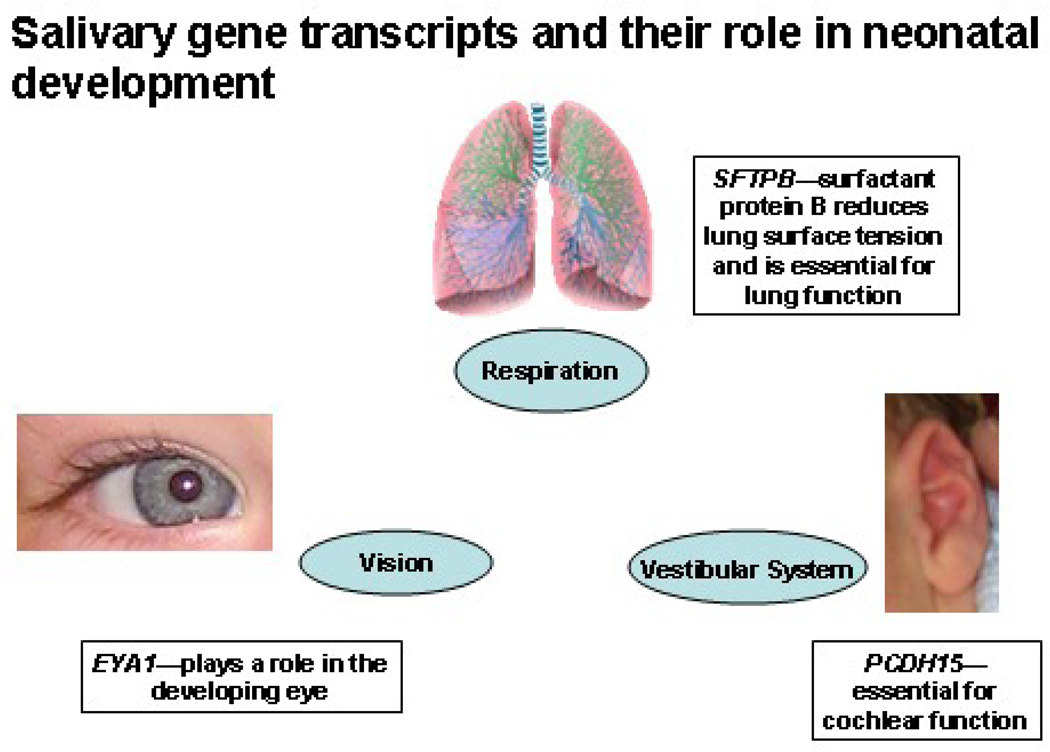
Figure 1.
The neonatal salivary transcriptome provides a surprising window into multiorgan development. Comparative salivary gene expression analyses unexpectedly detected up-regulation of gene transcripts involved in the developing lung, eye, and ear.
Discussion
Saliva represents an ideal biofluid for neonatal monitoring. It is readily available, may be noninvasively and repeatedly obtained, and contains a wealth of information that may be examined in the context of normal physiological development and unique disease sequelae. Our group was the first to analyze neonatal salivary gene expression in relation to gestational age and feeding status [5]. As premature infants learned to orally feed, we identified significant gene expression alterations in key modulators of feeding behavior, oral musculature and innervation. This work lays the foundation for a salivary diagnostic panel to predict successful oral feeding in premature neonates and reduce the multiple comorbidities associated with early oral feeding trials such as choking, aspiration, and hypoxia. Our analyses also provided us with a significant window into the premature infant’s developing gastrointestinal system. Genes identified in this healthy cohort can now be compared to the salivary transcriptomes of infants who develop pathological gastrointestinal conditions, such as feeding intolerance or necrotizing enterocolitis, a severe and often fatal disease resulting in inflammation and necrosis of the bowel. This ability to monitor and compare gene expression changes in relation to neonatal disease sequelae holds significant potential for improving clinical care.
Despite the ease to which it may be obtained and the wealth of information it contains, saliva remains an underutilized biofluid in neonatology. Our research has shown that technical limitations associated with quantitatively limited salivary samples may be overcome, and that the salivary transcriptome represents a powerful diagnostic platform to better understand neonatal biology. It is time that neonatologists incorporate saliva into their research in order to more effectively and noninvasively monitor normal physiology, development, and aberrant pathological pathways in the neonatal population.
Acknowledgements
This work was supported the National Institute of Health NICHD K08HD059819-02 and the Charles H. Hood Foundation. I would like to thank the technical support of Jessica Dietz and the mentorship of Drs. Diana Bianchi and Kirby Johnson.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wong DT. Salivary Diagnostics: Amazing as it might seem, doctors can detect and monitor diseases using molecules found in a sample of spit. Am Sci. 2008;96:37–43. doi: 10.1511/2008.69.3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Segal A, Wong DT. Salivary diagnostics: enhancing disease detection and making medicine better. Eur J Dent Educ. 2008;12:22–29. doi: 10.1111/j.1600-0579.2007.00477.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Su S, Arellano M, Boontheung P, Wang J, Zhou H, Jiang J, Elashoff D, Wei R, Loo JA, Wong DT. Salivary proteomics for oral cancer biomarker discover. Clin Cancer Res. 2008;12:6246–6252. doi: 10.1158/1078-0432.CCR-07-5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li Y, St John MA, Zhou X, Kim Y, Sinha U, Jordan RC, Eisele D, Abemayor E, Elashoff D, Park NH, Wong DT. Salivary transcriptome diagnostics for oral cancer detection. Clin Cancer Res. 2004;10:8442–8450. doi: 10.1158/1078-0432.CCR-04-1167. [DOI] [PubMed] [Google Scholar]
- 5.Maron JL, Johnson KL, Rocke DM, Cohen MG, Liley AJ, Bianchi DW. Neonatal salivary analysis reveals global developmental gene expression changes in the premature neonate. Clin Chem. 2010;56:409–416. doi: 10.1373/clinchem.2009.136234. [DOI] [PMC free article] [PubMed] [Google Scholar]