Abstract
GTP cyclohydrolase I (GTPCH) catalyzes the first step in pteridine biosynthesis in Nocardia sp. strain NRRL 5646. This enzyme is important in the biosynthesis of tetrahydrobiopterin (BH4), a reducing cofactor required for nitric oxide synthase (NOS) and other enzyme systems in this organism. GTPCH was purified more than 5,000-fold to apparent homogeneity by a combination of ammonium sulfate fractionation, GTP-agarose, DEAE Sepharose, and Ultragel AcA 34 chromatography. The purified enzyme gave a single band for a protein estimated to be 32 kDa by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The molecular mass of the native enzyme was estimated to be 253 kDa by gel filtration, indicating that the active enzyme is a homo-octamer. The enzyme follows Michaelis-Menten kinetics, with a Km for GTP of 6.5 μM. Nocardia GTPCH possessed a unique N-terminal amino acid sequence. The pH and temperature optima for the enzyme were 7.8 and 56°C, respectively. The enzyme was heat stable and slightly activated by potassium ion but was inhibited by calcium, copper, zinc, and mercury, but not magnesium. BH4 inhibited enzyme activity by 25% at a concentration of 100 μM. 2,4-Diamino-6-hydroxypyrimidine (DAHP) appeared to competitively inhibit the enzyme, with a Ki of 0.23 mM. With Nocardia cultures, DAHP decreased medium levels of NO2− plus NO3−. Results suggest that in Nocardia cells, NOS synthesis of nitric oxide is indirectly decreased by reducing the biosynthesis of an essential reducing cofactor, BH4.
A tetrahydrobiopterin (BH4)-dependent bacterial nitric oxide synthase (NOS), designated NOSNoc, was first purified and characterized in Nocardia sp. strain NRRL 5646 in our laboratory (7). Continued pursuit of the possible roles of NOS resulted in the demonstration of guanylate cyclase (GC) activity in this bacterium (34). With viable Nocardia cells, additions of BH4 plus arginine enhanced GC activity and the amounts of cyclic GMP eightfold. This work established a novel role for NOSNoc in Nocardia similar to that known to exist in higher life forms.
The occurrence of a BH4 biosynthetic pathway in Nocardia was partially established by high-performance liquid chromatography (HPLC) product analysis. Mass spectrometry and nuclear magnetic resonance spectroscopy were used to confirm the identifications of neopterin and biopterin produced by crude cell extract conversions of GTP (34). The novelty and occurrence of a defined role for BH4 in Nocardia prompted us to examine the nature of GTP cyclohydrolase I (GTPCH; EC 3.5.4.16) involved in the biosynthesis of BH4 in this organism.
GTPCH catalyzes the conversion of GTP to dihydroneopterin triphosphate via a mechanistically complex ring expansion process (2, 27, 30, 32). This reaction is the first step in the biosynthetic pathway leading to the synthesis of the pteridine portion of tetrahydrofolate (FH4) in plants and some microorganisms (6) and of BH4 in mammals (29). FH4 serves as a coenzyme for a variety of one-carbon transfer reactions (26), while BH4 functions as an essential reducing cofactor for NOSs, glyceryl ether mono-oxygenases, and mammalian aromatic amino acid hydroxylases (23, 29, 36, 37). Bacterial GTPCHs from Escherichia coli (44), Bacillus subtilis (10), Lactobacillus plantarum (17), and Serratia indica (21) are all known in folate biosynthetic pathways. Streptomyces tubercidicus (46) contains a GTP-8-formylhydrolase involved in the formation of pyrrolopyrimidine nucleoside antibiotics, an enzyme that cleaves the diazole ring but does not cyclize the product to produce pteridines as the final product. Instead, pyrrolopyrimidine is formed (11). GTPCHs involved in BH4 biosynthesis have been purified to homogeneity from human, mouse, and rat tissues (15, 20, 33), as well as from Drosophila melanogaster (41). To date, no GTPCH enzymes have been characterized in actinomycetes.
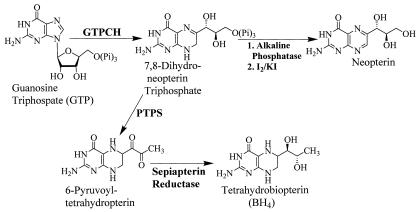
In higher organisms, de novo BH4 biosynthesis has been extensively characterized, and the activity of GTPCH is regulated by a variety of mechanisms (16, 19, 37, 38, 42, 45). Dihydroneopterin triphosphate, the first product of GTPCH cleavage of GTP is ultimately converted to BH4 by the sequential action of 6-pyruvoyltetrahydropterin synthase (EC 4.6.1.10) and sepiapterin reductase (EC 1.1.1.153) (37) (Fig. 1). The process of biosynthesis of BH4 in bacteria is much less known, and the functions of BH4 in prokaryotes are not widely understood. Putative genes encoding BH4 biosynthetic proteins have been detected in the genomes of B. subtilis (22) and a Synechocystis sp. (18). A sepiapterin reductase producing l-threo-dihydrobiopterin was purified from Chlorobium tepidum (8). However, few studies on the characterization of GTPCHs involved in prokaryotic BH4 biosynthesis have been reported (25).
FIG. 1.
Pathways for BH4 biosynthesis and activity assay for GTPCH. PTPS, 6R-pyruvoyltetrahydropterin.
This paper describes the purification and biochemical characterization of GTPCH from Nocardia sp. strain NRRL 5646, compares its properties with those of known GTPCHs, and probes its possible function in Nocardia NO synthesis.
MATERIALS AND METHODS
Materials.
Molecular weight standards for gel electrophoresis and polyvinylidene difluoride membranes were purchased from Bio-Rad (Hercules, Calif.); the protein microassay kit was purchased from Pierce (Rockford, Ill.). Derivatives of biopterin, pterin, folate, and nucleotides, dithioerythritol (DTE), phenylmethylsulfonyl fluoride, molecular markers for gel permeation chromatography, alkaline phosphatase (from bovine intestinal mucosa), GTP-agarose, DEAE Sepharose (DFF-100), Ultragel AcA 34, 2,4-diamino-6-hydroxypyrimidine (DAHP), N-methyl-l-arginine (LNMA), and other chemicals were purchased from Sigma Chemical Co. (St. Louis, Mo.).
Buffers used in the preparation of cell extracts and in enzyme purification were buffer A (0.05 M Tris, 0.05 M KCl, 2.5 mM EDTA, and 10% glycerin [vol/vol] [pH 7.8]), buffer B (25 mM Tris, 50 mM KCl, 1.25 mM EDTA, and 5% glycerin [pH 7.8]), and buffer C (0.05 M Tris, 0.3 M KCl, 10% glycerin, 2.5 mM EDTA, and 2.5 mM DTE [pH 7.8]).
Cultivation and preparation of cell extracts.
Nocardia sp. strain NRRL 5646 was maintained in the University of Iowa College of Pharmacy culture collection and was grown and maintained on slants of Sabouraud dextrose agar or sporulating agar (American Type Culture Collection no. 5 medium). Shaken flask cultures were grown by a standard two-stage incubation protocol (3) in 200 ml of sterile medium held in stainless steel-capped 1-liter DeLong culture flasks. The medium contained (wt/vol) 2% glucose, 0.5% yeast extract, 0.5% soybean meal, 0.5% NaCl, 0.5% K2HPO4, and 0.25% phenylalanine in distilled water and was adjusted to pH 7.0 with 6 N HCl before being autoclaved at 121°C for 20 min. The same medium with glucose substituted for phenylalanine served as a phenylalanine-free control. Cultures were begun by suspending growth from slants to a first-stage culture that was incubated with shaking at 250 rpm at 28°C on a New Brunswick Scientific Innova 5000 Gyrotory tier shaker. A 10% inoculum (first-stage culture, 72 h) was used to begin the second-stage culture, which was incubated as described before, harvested 48 h later, and filtered through four folds of cheesecloth to remove remaining soybean meal solids. The filtrate was centrifuged at 8,000 × g for 20 min at 4°C, and the resulting pellet was washed twice with 0.9% (wt/vol) NaCl and pelleted once again. Cell pellets were stored at −20°C until needed. Typical wet-weight cell yields by this cultivation process were approximately 20 g/liter.
For preparation of cell extracts, 84 g (wet weight) of cell pellet was suspended in 420 ml of cold buffer A containing 0.2 mM phenylmethylsulfonyl fluoride and 5 mM DTE. This cell suspension was disrupted by passing it twice through a French pressure cell at 12,000 lb/in2. The cell homogenate was centrifuged at 100,000 × g for 60 min at 4°C. The resulting supernatant (390 ml) was the cell extract. All subsequent enzyme purification steps were conducted at 4°C.
Enzyme assay.
The enzyme assay was based on the method of Viveros et al. (40) and modified significantly to reduce the time required to process samples. Unless specified otherwise, the standard reaction mixture contained enzyme, 500 μM GTP, and buffer A in a final volume of 100 μl. The reaction was carried out at 37°C for 60 min in darkness. To quantitatively analyze the formation of dihydroneopterin triphosphate from GTP, the enzyme reaction was terminated by adding 0.59 ml of buffer A and 0.1 ml of acidic iodine solution (1% I2 and 2% KI in 1 N HCl). After the mixture had been kept at room temperature for 15 min, the resulting insoluble material was removed by centrifugation at 20,000 × g for 4 min. Excess iodine remaining in mixtures was reduced by addition of 100 μl of 2.0% ascorbic acid in buffer A. After addition of 110 μl of 1 N NaOH, samples were incubated with 10 U of alkaline phosphatase at 37°C for 45 min. For HPLC, 20-μl samples were injected onto an Econosil C18 column (10 μm, 4.6 × 250 mm; Alltech, Inc.), where neopterin was eluted isocratically with a solvent of 25 mM sodium phosphate (pH 7.0) at a flow rate of 1 ml/min and then detected spectrofluorometrically with an excitation wavelength of 350 nm and an emission wavelength of 450 nm. The retention volume for neopterin in this system was 6.7 ml. One unit of the enzyme was defined as the amount of enzyme that catalyzed the formation of 1 nmol of dihydroneopterin triphosphate per h at 37°C.
Protein determination.
Protein concentrations were measured by the Bradford protein microassay (5) with bovine serum albumin used as the standard.
Enzyme purification and characterization.(i) Ammonium sulfate fractionation.
Cell extracts were brought to 35% saturation in ammonium sulfate with stirring for 30 min and then allowed to remain still for 1 h before being centrifuged at 20,000 × g for 20 min to remove precipitated material. Additional ammonium sulfate was added to 60% saturation, and the resulting precipitate was recovered by centrifugation at 20,000 × g for 25 min. This precipitate, containing the enzyme, was dissolved in 15 ml of buffer B and dialyzed with a Spectra/Por Molecularporous membrane (molecular weight cutoff, 12,000 to 14,000 ; Spectrum) against 1,600 ml of the same buffer for 16 h with two changes of buffer.
(ii) GTP-agarose affinity chromatography.
The dialyzed ammonium sulfate fraction was centrifuged at 30,000 × g for 10 min to remove precipitated substances and was then diluted with buffer B to a final volume of 104 ml, which was applied to a column of GTP-agarose (0.8 × 10 cm; bed volume [Vt], 5 ml) preequilibrated with buffer B. The column was developed sequentially with 25 mM Tris (pH 7.8) containing 0.25 M KCl, 1.25 mM EDTA, 5% glycerin, and 2.5 mM DTE (30 ml, at a rate of 0.4 ml/min); buffer B containing 2.5 mM DTE (20 ml, at a flow rate of 0.5 ml/min); and buffer B containing 3.8 mM GTP and 2.5 mM DTE (40 ml, at a flow rate of 0.2 ml/min). The active fractions obtained in the last column eluate were combined and concentrated to 5 ml in an Amicon concentrator (PM-30 membrane).
(iii) DEAE Sepharose ion-exchange chromatography.
The active enzyme concentrate from the GTP-agarose column was loaded onto a DEAE Sepharose (DFF-100) column (1.5 × 20 cm; Vt, 12 ml) that had been equilibrated with buffer A. After being washed with 30 ml of buffer A, the column was developed with a linear gradient of buffer A containing 0.0 to 0.3 M KCl over a total volume of 100 ml at a flow rate of 0.4 ml/min. Active fractions (0.16 to 0.19 M KCl) were combined and concentrated with a PM-30 membrane to a final volume of 2 ml.
(iv) Ultragel AcA 34 gel permeation chromatography.
The enzyme preparation from DEAE Sepharose was further concentrated with Microcon YM-10 filter to a final volume of 0.5 ml before being loaded onto a column of Ultragel AcA 34 (1 × 50 cm; Vt, 45 ml) equilibrated with buffer C. The column was eluted with the same buffer at a flow rate of 6 ml/h, and fractions 18 to 21 (4 ml) containing active enzyme were combined for subsequent analysis.
(v) SDS-PAGE.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed with a Bio-Rad mini-protein II dual slab cell with a discontinuous buffer system (24) and a 12% separation gel. Gels were stained with 0.1% Coomassie brilliant blue R-250. The molecular mass markers included phosphorylase b (97.4 kDa), serum albumin (66.2 kDa), ovalbumin (45 kDa), carbonic anhydrase (31 kDa), trypsin inhibitor (21.5 kDa), and lysozyme (14.4 kDa).
(vi) Native molecular mass.
The molecular weight of the native, nondenatured enzyme was determined by gel permeation over a column of Ultragel AcA 34 (1 × 50 cm; Vt, 45 ml) equilibrated with buffer C at a flow rate of 6 ml/h. Molecular mass standards included blue dextran (2,000 kDa), apoferritin (443 kDa), β-amylase (200 kDa), alcohol dehydrogenase (150 kDa), bovine serum albumin (66 kDa), and carbonic anhydrase (29 kDa).
(vii) N-terminal amino acid sequence.
For analysis of the N-terminal amino acid sequence, 10 μg of purified enzyme was subjected to SDS-PAGE. After electrophoresis, electrotransfer of proteins to a polyvinylidene difluoride membrane was performed according to the method of Towbin et al. (39). N-terminal amino acid microsequencing was determined by Edman degradation using a 475A automated sequencer (Applied Biosystems, Inc.) in the protein structure facility at the University of Iowa. The N-terminal amino acid sequence was determined twice by using samples obtained from two different enzyme purifications.
(viii) Temperature optima, pH, and thermal stability.
For temperature optima experiments, the samples were incubated in buffer A at various temperatures (4 to 60°C) for 1 h before being stopped and analyzed for enzyme activity. For determinations of optimum pH, three different buffers were used over a range of pH 6 to 10.4. These included 50 mM sodium phosphate, 50 mM Tris-HCl, and 50 mM glycine-NaOH. The effect of temperature on the stability of purified GTPCH was conducted by preincubating the enzyme at 37, 50, 60, 70, and 80°C. Samples were taken at predetermined intervals and assayed for residual enzyme activity. For all the above experiments, each determination was conducted in duplicate.
(ix) Effects of nucleotides, metal ions, pterins, and DAHP on GTPCH activity.
Various nucleotides and metal ions were added to standard reaction mixtures to give a final concentration of 1 to 2 mM and 1 to 5 mM for nucleotides and metal ions, respectively. Resulting enzyme activities were compared to those of the standard enzyme reaction as a control. As with others in the literature, limited quantities of pure GTPCH (40 μg from a typical purification) (Table 1) precluded taking more than single measurements for each metal and nucleotide.
TABLE 1.
Purification of Nocardia sp. GTPCH
| Step | Total protein (mg) | Total activity (U)a | Sp act (U/mg) | Yield (%) | Purification (n-fold) |
|---|---|---|---|---|---|
| Crude extract | 2,184 | 154.5 | 0.07 | 100 | 1 |
| (NH4)2SO4 | 647 | 101.4 | 0.16 | 65.6 | 2.2 |
| GTP-agarose | 1.88 | 85.8 | 45.6 | 55.5 | 64.5 |
| DEAE Sepharose | 0.12 | 22.8 | 190 | 14.7 | 2,693 |
| Ultragel AcA 34 | 0.04 | 14.9 | 373 | 9.6 | 5,246 |
One unit is defined as the amount of the enzyme that catalyzed the formation of 1 nmol of dihydroneopterin triphosphate per h at 37°C.
To determine the effects of pterin derivatives on GTPCH activity, concentrations of 100 μM BH4, BH2 and biopterin, FH4, FH2 and folate, pterin, and pterin-6-carboxylic acid were preincubated with standard incubation mixtures for 5 min before adding GTP to start the enzyme reaction. Inhibition of GTPCH activity by DAHP was examined at four different concentrations of DAHP (0, 1, 3, and 10 mM) versus various concentrations of GTP (10 μM to 110 μM) in standard reaction mixtures.
All assays for kinetic constant determination and DAHP inhibition were conducted in duplicate. Each data point was the mean of two assays. Kinetic constants were obtained by fitting experimental data with Cleland's kinetics program (9). The inhibition mechanism was established by comparing the fitting of experimental data to different models and confirmed by graphic analysis.
(x) Effects of DAHP and LNMA on NO2− plus NO3− synthesis by Nocardia cultures.
For whole-cell inhibition studies, cultures were grown in 25 ml of sterile medium in 125-ml stainless steel-capped DeLong flasks. The medium and incubation protocols were as described before. Concentrations of 1 to 10 mM DAHP were added into 24-h old stage II cultures that were incubated at 28°C for an additional 24 h. Thereafter, the supernatants were obtained by centrifugation at 20,000 × g and 4°C for 20 min and used to determine NO2− plus NO3− concentrations reflective of nitric oxide levels. The cell pellets were used for cell mass determinations. Typical cell mass was 20 g/liter, and DAHP at concentrations of up to 10 mM and 0.5 mM LNMA had no effect on cell growth relative to controls.
Nocardia supernatant samples were assayed for the stable end products of NO oxidation, total nitrate plus nitrite (NO2− plus NO3−), by using an automated procedure based on the Griess reaction (13, 14). An essential step in the measurements of NO2− plus NO3− required that culture supernatants be ultrafiltered through Microcon YM-3 filters (Millipore Corp., Bedford, Mass.) to reduce otherwise high blank readings. The resulting filtrates were analyzed for NO2− plus NO3− concentrations with a nitrate-nitrite colorimetric assay kit in 96-well plates. For each assay, 40 μl of ultrafiltrate was diluted to 80 μl by using the assay buffer solution, followed by adding 10 μl of enzyme cofactor mixture and 10 μl of nitrate reductase mixture. The 96-well plate was covered and incubated at room temperature for 3 h. After incubation, 50 μl of Griess reagent R1 (1% sulfanilamide) was added, followed immediately by the addition of 50 μl of Griess reagent R2 [0.1% N-(1-naphthyl)ethylenediamine]. After color development for 10 min, sample absorbances were determined at 540 nm using a microplate reader (Molecular Devices, Menlo Park, Calif.). The results were expressed as means ± standard deviations of three observations. Student's unpaired t tests were used to assess the statistical significance of differences at a P value of less than 0.05.
RESULTS
Enzyme assay.
A highly sensitive, rapid, and reproducible HPLC spectrofluorometric detection method was established for the measurement of GTPCH enzyme reactions. The optimum times for the I2 and KI oxidation of dihydroneopterin triphosphate—the immediate product of GTPCH—to neopterin triphosphate and for the alkaline phosphatase-mediated cleavage of the phosphate moieties from neopterin triphosphate were 15 and 45 min, respectively. Standard curves for neopterin, which eluted at a retention volume of 6.7 ml by this method, ranged from 1 to 100 ng per sample with reproducibilities of 0.999. This sensitive assay tool permitted the ready analysis of GTPCH enzyme activity in crude and purified enzyme fractions. Product increased in a linear fashion over a 2-h enzyme incubation period.
Enzyme purification.
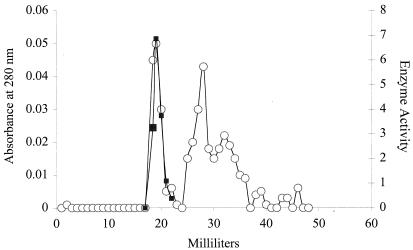
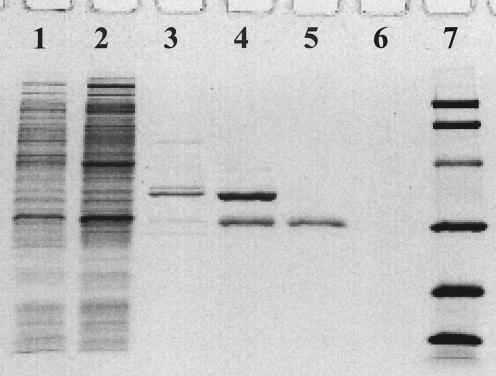
GTPCH was purified many times, and the results of a typical four-step purification of Nocardia GTPCH are summarized in Table 1. The specific activity of GTPCH in crude Nocardia cell extracts obtained from cells growing on standard medium was generally about 66 pmol · h−1 · mg of protein−1. The addition of phenylalanine slightly increased GTPCH levels in crude extracts to 71 pmol · h−1 · mg of protein−1. The purity of the 100,000 × g-soluble enzyme was greatly enhanced (290-fold) by the GTP-agarose affinity chromatography step. Single symmetrical peaks of enzyme activity were observed in elution profiles from each of the chromatographic columns. As shown in Fig. 2 for the last step, Ultragel AcA 34 column chromatography, enzyme activity and protein coeluted to afford pure GTPCH. The enzyme from Nocardia cells was purified 5,246-fold by this process with a specific activity of 373 U · mg of protein−1 and an overall recovery of nearly 10%. Soluble Nocardia GTPCH represents approximately 0.02% of total protein in crude supernatants. The pure enzyme was homogeneous by SDS-PAGE (Fig. 3).
FIG. 2.
Elution profile of Nocardia GTPCH obtained by Ultragel AcA 34 column chromatography. ○, absorbance at 280 nm; ▪, enzyme activity.
FIG. 3.
SDS-PAGE of samples from the purification of Nocardia GTPCH. Lane 1, crude extract; lane 2, dialyzed ammonium sulfate precipitate; lane 3, pooled fractions from GTP-agarose; lane 4, pooled fractions from DEAE Sepharose; lane 5, pooled fractions from Ultragel AcA 34; lane 6, control with sample buffer; lane 7, molecular mass markers (phosphorylase b [97.4 kDa], serum albumin [66.2 kDa], ovalbumin [45 kDa], carbonic anhydrase [31 kDa], trypsin inhibitor [21.5 kDa], and lysozyme [14.4 kDa]).
Properties of Nocardia GTPCH.
By SDS-PAGE, the purified enzyme gave a single band of an apparent molecular mass of 32 kDa. The molecular mass of the native enzyme, estimated by AcA 34 gel permeation chromatography, was 253 kDa, suggesting that the active enzyme was a homo-octameric protein complex.
Two separately purified Nocardia GTPCH samples gave identical N-terminal amino acid sequences of H2N-Ser-Ala-Asn-Asn-His-Val-Gly-Gly-His-Ala-Leu-Ala. The BLASTP and TBLASTN programs (1) used for searching the updated Swissport, Genpept, GenBank, and EMBL databases gave no matching amino acid sequences.
Nocardia GTPCH specifically used GTP as a substrate. No products were detected by HPLC when GTP was replaced by GDP, GMP, ATP, UTP, or CTP. The Km values for GTP were estimated from double-reciprocal Lineweaver-Burk plots to be 6.5 ± 0.53 μM. The range of GTP used for Km determination was 1 to 100 μM. Maximum enzyme activity was at pH 7.8 in 0.05 M sodium phosphate buffer, and the temperature optimum was 56°C. Nocardia GTPCH was heat stable, withstanding temperatures as high as 60°C for 10 min while still retaining 50% of its original activity. Enzyme activity remained essentially unchanged when it was incubated for 1 h at 37°C. For this reason, the standard incubation temperature selected was 37°C.
The effects of other nucleotide triphosphates versus GTP on purified GTPCH activity are summarized in Table 2. At a concentration of 2 mM, GDP and GMP caused about 20 and 10% decreases in GTPCH activity, respectively. ATP, TTP, CTP, and UTP caused reductions in GTPCH activity ranging from 50 to 33%, with ATP showing nearly 50% inhibition. Purified Nocardia GTPCH was significantly inhibited by 5 mM concentrations of a variety of divalent cations, including calcium (82% inhibition), copper (9% inhibition), zinc (90% inhibition), and mercury (87% inhibition at 0.1 mM). Magnesium had little effect on enzyme activity (Table 3). Enzyme activity was increased 30% by the addition of 0.1 M KCl, similar to all other known GTPCHs. At 100 μM, BH4 inhibited Nocardia GTPCH activity by 25%. On the other hand, folate, dihydrobiopterin, biopterin, and sepiapterin had no significant effects on GTPCH activity. DAHP was an apparent competitive inhibitor of Nocardia GTPCH, with a Ki of 0.23 ± 0.1 mM similar to that from rat aortic small muscle cells (0.76 mM) (43).
TABLE 2.
Effects of nucleotides on Nocardia GTPCH activity
| Nucleotide | Remaining activity (%)a
|
|
|---|---|---|
| 1 mM | 2 mM | |
| None | 100 | 100 |
| UTP | 71.7 | 67.1 |
| TTP | 65.6 | 52.6 |
| CTP | 67.7 | 59.1 |
| ATP | 64.3 | 50.7 |
| GDP | 82.7 | 81.2 |
| GMP | 95.4 | 89.4 |
The activity of controls (None) containing no nucleotide additive was 0.02 nmol−1·h−1 as 100%.
TABLE 3.
Effects of metal ions on Nocardia GTPCH activity
| Salt added | Remaining activity (%)a
|
|
|---|---|---|
| 1 mM | 5 mM | |
| None | 100 | 100 |
| CaCl2 | 100 | 18.8 |
| CoCl2 | 110 | 87.4 |
| CuCl2 | 95.1 | 9.1 |
| FeCl2 | 94.2 | 51.9 |
| HgCl2 | 13 | 58.1 (0.1 mM) |
| MgCl2 | 100 | 90.7 |
| MnCl2 | 102 | 65.5 |
| ZnCl2 | 100 | 9.5 |
The activity of controls with no added metal salts was 0.02 nmol−1·h−1 as 100%.
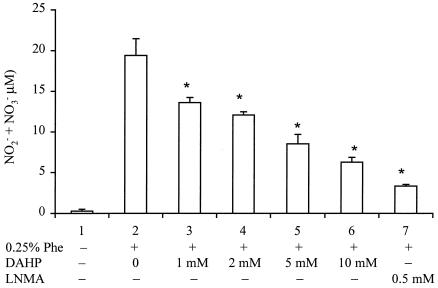
Effects of DAHP and LNMA on NO2− plus NO3− synthesis.
Measurements of NO2− plus NO3− were difficult with medium samples that were subjected only to microfiltration. We discovered that substances interfering with the Griess reaction causing high blank values were effectively removed by ultrafiltration through a 3-kDa cutoff membrane to give very low blank values. Figure 4 shows that Nocardia cultures grown in phenylalanine-free medium produced a base level of 0.26 ± 0.22 μM NO2− plus NO3− (lane 1). Filtrates from cultures grown in medium containing 0.25% phenylalanine contained 19.41 ± 2.04 μM NO (lane 2). Cultures grown in phenylalanine medium plus DAHP gave NO2− plus NO3− concentrations decreasing in a dose-dependent fashion from a high of 19.41 ± 2.04 μM to 6.28 ± 0.58 μM (lanes 2 to 6). Cultures grown in phenylalanine medium plus 0.5 mM LNMA, a competitive inhibitor of NOSNoc, reduced NO2− plus NO3− levels to 3.33 ± 0.22 μM (lane 7).
FIG. 4.
Effects of DAHP and LNMA on NO2− plus NO3− formation in Nocardia. Controls were cells grown in phenylalanine-free medium (lane 1). Cells grown in Phe-containing medium (lane 2) were incubated with various concentrations of DAHP or LNMA for 24 h. The resulting supernatants were assayed for NO2− plus NO3−, as described in Materials and Methods. Data represent means ± standard deviations (n = 3). *, P < 0.05 versus the respective value obtained from cells without DAHP or LNMA treatment.
DISCUSSION
This work describes the first GTPCH enzyme isolated from an actinomycete. GTPCHs are zinc-containing enzymes (2) that catalyze the oxidative cleavage of the imidazole ring of GTP by initial C8 hydroxylation, deformylation of the initial product, and cyclization of the first two carbons of the attached and isomerized ribose to afford dihydroneopterin triphosphate. Nocardia GTPCH appears to be a constitutive enzyme because the addition of phenylalanine only caused a reproducible small increase (10%) in specific activity of the enzyme in crude extracts. Purification of Nocardia GTPCH posed significant analytical challenges brought about by the very low level of GTPCH activity, the inherently low reaction rate (32) measured in nanomoles · hour−1 · milligram of protein−1, and the instability of the immediate GTPCH reaction product, dihydroneopterin triphosphate (10, 31). A rapid, highly specific, and extremely sensitive enzyme assay was based on optimized methods for oxidation of dihydroneopterin triphosphate to the completely aromatic equivalent, i.e., enzymatic hydrolysis of the triphosphate moiety to liberate neopterin that was readily detected by a modification of the HPLC method of Viveros et al. (40). We believe that the analytical methods described herein are superior to the radiometry-based assays (4, 44) used by others for GTPCH purification and analysis.
Initial attempts to purify GTPCH used a sequence of ammonium sulfate fractionation followed sequentially by DEAE Sepharose, GTP-agarose, and Ultragel AcA 34 chromatographies. This process was slow, requiring nearly 6 days of work to obtain an enzyme preparation that was nearly inactive. The final sequence of steps (Table 1) was much faster, requiring 3 days to obtain highly active and relatively stable GTPCH.
GTP-agarose has been effectively used for the purification of other GTPCHs (10, 20, 21, 44). GTP-agarose affinity chromatography was the most effective step, giving a 290-fold improvement in Nocardia GTPCH purity.
Interestingly, relatively large amounts of dihydroneopterin triphosphate were detected in active enzyme fractions eluting from the GTP-agarose affinity column. This was due to the presence of GTP, a substrate for GTPCH, in the eluting buffer. The presence of significant amounts of the GTPCH product was surprising because at 4°C, the specific activity of Nocardia GTPCH was only about 2% of that obtained under standard assay conditions. Dihydroneopterin triphosphate was removed from active enzyme samples during the subsequent ion-exchange chromatography step.
All known GTPCHs show considerable variability both in amino acid sequence and in the sizes of the active multimeric forms into which they assemble. By SDS-PAGE and gel filtration, the molecular masses of the GTPCH-denatured subunit and intact native forms were 32 and 253 kDa, respectively. Thus, active Nocardia GTPCH appears to be a homo-octamer. GTPCHs (molecular mass of monomeric subunit, molecular mass of mass active multimer) from B. subtilis (21 kDa, 180 kDa), S. indica (25 kDa, 200 kDa), and E. coli (25.5 kDa, 210 kDa) indicate active homomultimers similar to that found in Nocardia. GTPCH molecular mass values from the rat (30 kDa, 300 kDa) and human (50 kDa, 440 kDa) samples suggest active homodecameric forms for the active enzymes. X-ray crystal studies confirmed that GTPCHs from E. coli (28) and the rat (35) have decameric active structures consisting of two pentameric subunit rings associated face to face. However, GTPCHs from these species share only 34% amino acid identity.
One other actinomycete enzyme that cleaves the diazole ring of GTP has been identified as GTP-8-formylhydrolase (11, 46). GTP-8-formylhydrolase cleaved GTP in an early step in the biosynthesis of pyrrolopyrimidine nucleoside antibiotics, toyocamycin, sangivamycin, and tubercidin. The enzyme purified from S. tubercidicus (46) cleaved the GTP imidazole ring, but the product was converted to pyrrolopyrimidine, not pteridines (11). The Streptomyces enzyme subunit size is 58 kDa (46), twice that of Nocardia and other bacterial GTPCHs.
Influences of divalent cations, nucleotides, biopterin biosynthetic intermediates, folate, and DAHP on Nocardia GTPCH were generally similar to those observed by others. While 12 mM MgCl2 inhibits B. subtilis GTPCH activity by 80% (10), 20 mM MgCl2 was only slightly inhibitory to Nocardia GTPCH, as was found previously for E. coli (31). The reduction in activity with nucleotide triphosphates and diphosphates shows that GTPCH activity and pteridine biosynthesis (10) are subject to fluctuations in intracellular nucleotide levels. The inhibitory effects of ATP on Nocardia GTPCH were similar to those observed with GTPCHs from E. coli (44) and D. melanogaster (41). ATP showed no inhibitory effect on B. subtilis GTPCH (10). However, UTP competitively inhibited B. subtilis (10) GTPCH by interfering with the binding of GTP to the enzyme and therefore increasing the apparent Km for GTP. The slight (25%) inhibition of GTPCH activity by BH4, a biopterin produced by Nocardia (34), could suggest possible end-product control in biopterin biosynthesis. However, the lack of inhibition by other intermediates in BH4 biosynthesis and the typically low intracellular concentrations of BH4 (1.6 ± 0.4 μg per gram [wet weight] of rat liver [12, 38]) indicate that feedback control of GTPCH activity is physiologically unimportant. A complex GTPCH feedback regulatory protein known in mammals has not been observed in bacteria (38). Influences of calcium, iron, mercury, and zinc are similar to the effects of these cations on other GTPCHs, except that 5 mM CaCl2 caused 5% inhibition on S. indica GTPCH (21). Concentrations greater than 1 mM for Zn2+ inhibited purified GTPCHs from bacteria (4, 21) and mammals (20, 33), even though zinc is required for catalytic activity (2).
In eukaryotes, stimulation of GTPCH activity occurs when oligomeric GTPCH associates with pentameric regulatory proteins in the presence of phenylalanine (38, 45). Although no such process is yet known in bacteria, addition of 0.25% phenylalanine to Nocardia culture media dramatically increased levels of NO2− plus NO3− from 0.26 to 19.41 μM. LNMA, a specific inhibitor of NOSNoc (7), dramatically reduced NO2− plus NO3− levels in culture media, thus relating the source of NO2− plus NO3− to NO, the product of nitric oxide synthase. The GTP inhibitor DAHP caused dose-related decreases in the amounts of NO2− plus NO3− in media from treated cells. These results confirmed our earlier observations that BH4 was essential for NO synthesis by NOSNoc (34). The present work provides direct biochemical evidence for the pathway by which BH4, a relatively rare pteridine cofactor in bacteria, is synthesized by Nocardia. We recently discovered that Nocardia sp. strain NRRL 5646 contains another BH4-dependent enzyme, phenylalanine hydroxylase (to be reported elsewhere). Thus, in Nocardia spp., GTPCH plays an essential role in providing BH4 as a cofactor in at least two enzyme reactions: nitric oxide synthesis and phenylalanine hydroxylation.
Nocardia sp. strain NRRL 5646 is a unique organism requiring BH4 for its well-characterized nitric oxide synthase, NOSNoc (7). The physiological roles of NOS and NO in Nocardia remain unclear. Our first observation of a NOS enzyme system in prokaryotes initially raised many new questions about the presence of supporting biochemical pathways that may serve to produce essential cofactors for the NOS reaction and that may yield functionally active products that may play roles in cellular physiology, metabolism, and possibly pathogenicity (7, 34). The occurrence and properties of Nocardia GTPCH, the first enzyme in BH4 biosynthesis, have now been clearly elaborated. In earlier work from our laboratory, the use of inhibitors with Nocardia cells was highly successful in demonstrating a putative novel signaling role for NO, the product of NOSNoc in this organism (34). GC activity was eliminated when cultures were incubated with either the NOS inhibitor Nω-nitro-l-arginineand LNMA or the GC inhibitor ODQ. Previous work (34) has also pointed to a centrally important role for GTP metabolism in Nocardia.
Finally, our previous discovery of the occurrence of a NO-dependent, cyclic GMP-mediated second messenger system in Nocardia raised the interesting prospect of similar systems in other actinomycetes. In Streptomycetes, such systems could play regulatory roles in antibiotic and other secondary metabolite biosyntheses. In related pathogens, such as Mycobacterium, second messenger systems could be involved in virulence or pathogenesis. With the present study, our work with Nocardia has now identified at least three possible target enzymes, the selective inhibition or stimulation of which could subvert key microbial metabolic processes involved in secondary metabolite control or pathogenesis. The possibility of achieving selective inhibition of bacterial NOSNoc (7), GC (34), or GTPCH may provide substantial motivation for the design of new and useful antibacterial agents, for example, for use against tuberculosis.
Acknowledgments
Aimin He is grateful for financial support through a University of Iowa, Center for Biocatalysis and Bioprocessing, fellowship.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-41410. [DOI] [PubMed] [Google Scholar]
- 2.Auerbach, G., A. Herrmann, A. Bracher, G. Bader, M. Gütlich, M. Fischer, M. Garrido-Franco, J. Richardson, H. Nar, and R. Huber. 2000. Zinc plays a key role in human and bacterial GTP cyclohydrolase I. Proc. Natl. Acad. Sci. USA 97:13567-13572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Betts, R. E., D. E. Walters, and J. P. N. Rosazza. 1974. Microbial transformation of antitumor compounds. I. Conversion of acronycine to 9-hydroxyacronycine by Cunninghamella echinulata. J. Med. Chem. 17:599-602. [DOI] [PubMed] [Google Scholar]
- 4.Blau, N., and A. Niederwieser. 1985. GTP-cyclohydrolases: a review. J. Clin. Chem. Clin. Biochem. 23:169-176. [PubMed] [Google Scholar]
- 5.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 6.Burg, A. W., and G. M. Brown. 1968. The biosynthesis of folic acid. VIII: Purification and properties of the enzyme that catalyzes the production of formate from carbon atom 8 of guanosine triphosphate. J. Biol. Chem. 243:2349-2358. [PubMed] [Google Scholar]
- 7.Chen, Y., and J. P. N. Rosazza. 1995. Purification and characterization of nitric oxide synthase (NOSNoc) from a Nocardia species. J. Bacteriol. 177:5122-5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cho, S. H., J. U. Na, H. Youn, C. S. Hwang, C. H. Lee, and S. O. Kang. 1999. Sepiapterin reductase producing l-threo-dihydrobiopterin from Chlorobium tepidum. Biochem. J. 340:497-503. [PMC free article] [PubMed] [Google Scholar]
- 9.Cleland, W. W. 1979. Statistical analysis of enzyme kinetic data. Methods Enzymol. 63:103-138. [DOI] [PubMed] [Google Scholar]
- 10.De Saizieu, A., P. Vankan, and A. P. G. M. van Loon. 1995. Enzymic characterization of Bacillus subtilis GTP cyclohydrolase I. Biochem. J. 306:371-377. [PMC free article] [PubMed] [Google Scholar]
- 11.Elstner, E. F., and R. J. Suhadolnik. 1971. The biosynthesis of the nucleoside antibiotics. J. Biol. Chem. 246:6973-6981. [PubMed] [Google Scholar]
- 12.Fukushima, T., and J. C. Nixon. 1980. Analysis of reduced forms of biopterin in biological tissues and fluids. Anal. Biochem. 102:176-188. [DOI] [PubMed] [Google Scholar]
- 13.Giovannoni, G., J. M. Land, G. Keir, E. J. Thompson, and S. J. R. Heales. 1997. Adaptation of the nitrate reductase and Griess reaction methods for the measurement of serum nitrate plus nitrite levels. Ann. Clin. Biochem. 34:193-198. [DOI] [PubMed] [Google Scholar]
- 14.Green, L. C., D. A. Wagner, J. Glogowski, P. L. Skipper, J. S. Wishnok, and S. R. Tannenbaum. 1982. Analysis of nitrate, nitrite, and [15N] nitrite in biological fluids. Anal. Biochem. 126:131-138. [DOI] [PubMed] [Google Scholar]
- 15.Hatakeyama, K., T. Harada, S. Suzuki, Y. Watanabe, and H. Kagamiyama. 1989. Purification and characterization of rat liver GTP cyclohydrolase I. Cooperative binding of GTP to the enzyme. J. Biol. Chem. 264:21660-21664. [PubMed] [Google Scholar]
- 16.Hesslinger, C., E. Kremmer, L. Hültner, M. Ueffing, and I. Ziegler. 1998. Phosphorylation of GTP cyclohydrolase I and modulation of its activity in rodent mast cells. J. Biol. Chem. 273:21616-21622. [DOI] [PubMed] [Google Scholar]
- 17.Jackson, R. J., and T. Shiota. 1975. The nature of the multiple forms of d-erythrodihydroneopterin triphosphate synthetase. Biochim. Biophys. Acta 403:232-244. [DOI] [PubMed] [Google Scholar]
- 18.Kaneko, T., S. Sato, H. Kotani, A. Tanaka, E. Asamizu, Y. Nakamura, N. Miyajima, M. Hirosawa, M. Sugiura, S. Sasamoto, T. Kimura, T. Hosouchi, A. Matsuno, A. Muraki, N. Nakazaki, K. Naruo, S. Okumura, S. Shimpo, C. Takeuchi, T. Wada, A. Watanabe, M. Yamada, M. Yasuda, and S. Tabata. 1996. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis strain PCC 6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 3:109-136. [DOI] [PubMed] [Google Scholar]
- 19.Kapatos, G., S. L. Stegenga, and K. Hirayama. 2000. Identification and characterization of basal and cyclic AMP response elements in the promoter of the rat GTP cyclohydrolase I gene. J. Biol. Chem. 275:5947-5957. [DOI] [PubMed] [Google Scholar]
- 20.Kia, K. W., K. B. Jacobson, and J. J. Yim. 1991. Isolation and characterization of GTP cyclohydrolase I from mouse liver. Comparison of normal and the hph-1 mutant. J. Biol. Chem. 266:12294-12300. [PubMed] [Google Scholar]
- 21.Kohashi, M., T. Itadani, and K. Iwai. 1980. Purification and characterization of guanosine triphosphate cyclohydrolase I from Serratia indica. Agric. Biol. Chem. 44:271-278. [Google Scholar]
- 22.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, et al. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 23.Kwon, N. S., C. F. Nathan, and D. J. Stuehr. 1989. Reduced biopterin as a cofactor in the generation of nitrogen oxides by murine macrophages. J. Biol. Chem. 264:20496-20501. [PubMed] [Google Scholar]
- 24.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 25.Lee, S. W., H. W. Lee, H. J. Chung, Y. A. Kim, Y. J. Kim, Y. Hahn, J. H. Chung, and Y. S. Park. 1999. Identification of the genes encoding enzymes involved in the early biosynthetic pathway of pteridines in Synechocystis sp. PCC 6803. FEMS Microbiol. Lett. 176:169-176. [DOI] [PubMed] [Google Scholar]
- 26.Maden, B. E. H. 2000. Tetrahydrofolate and tetrahydromethanopterin compared: functionally distinct carrier in C1 metabolism. Biochem. J. 350:609-629. [PMC free article] [PubMed] [Google Scholar]
- 27.Nar, H., R. Huber, G. Auerbach, M. Fischer, C. Hösl, H. Ritz, A. Bracher, W. Meining, S. Eberhardt, and A. Bacher. 1995. Active site topology and reaction mechanism of GTP cyclohydrolase I. Proc. Natl. Acad. Sci. USA 92:12120-12125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nar, H., R. Huber, W. Meining, C. Schmid, S. Weinkauf, and A. Bacher. 1995. Atomic structure of GTP cyclohydrolase I. Structure 3:459-466. [DOI] [PubMed] [Google Scholar]
- 29.Nichol, C. A., G. K. Smith, and D. S. Duch. 1985. Biosynthesis and metabolism of tetrahydrobiopterin and molybdopterin. Annu. Rev. Biochem. 54:729-764. [DOI] [PubMed] [Google Scholar]
- 30.Rebelo, J., G. Auerbach, G. Bader, A. Bracher, H. Nar, C. Hosl, N. Schramek, J. Kaiser, A. Bacher, R. Huber, and M. Fischer. 2003. Biosynthesis of pteridine. Reaction mechanism of GTP cyclohydrolase I. J. Mol. Biol. 326:503-516. [DOI] [PubMed] [Google Scholar]
- 31.Schoedon, G., U. Redweik, G. Frank, R. G. H. Cotton, and N. Blau. 1992. Allosteric characteristics of GTP cyclohydrolase I from Escherichia coli. Eur. J. Biochem. 210:561-568. [DOI] [PubMed] [Google Scholar]
- 32.Schramek, N., A. Bracher, M. Fischer, G. Auerbach, H. Nar, R. Huber, and A. Bacher. 2002. Reaction mechanism of GTP cyclohydrolase I: single turnover experiments using a kinetically competent reaction intermediate. J. Mol. Biol. 282:829-837. [DOI] [PubMed] [Google Scholar]
- 33.Shen, R. S., A. Alam, and Y. Zhang. 1989. Human liver GTP cyclohydrolase I: purification and some properties. Biochimie 71:343-349. [DOI] [PubMed] [Google Scholar]
- 34.Son, J. K., and J. P. N. Rosazza. 2000. Cyclic guanosine-3′,5′-monophosphate and biopteridine biosynthesis in Nocardia sp. J. Bacteriol. 182:3644-3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Steinmetz, M. O., C. Plüss, U. Christen, B. Wolpensinger, A. Lustig, E. R. Werner, H. Wachter, A. Engel, U. Aebi, J. Pfeilschifter, and R. A. Kammerer. 1998. Rat GTP cyclohydrolase I is a homodecameric protein complex containing high-affinity calcium-binding sites. J. Mol. Biol. 279:189-199. [DOI] [PubMed] [Google Scholar]
- 36.Tayeh, M. A., and M. A. Marletta. 1989. Macrophage oxidation of l-arginine to nitric oxide, nitrite, and nitrate. J. Biol. Chem. 264:19654-19658. [PubMed] [Google Scholar]
- 37.Thöny, B., G. Auerbach, and N. Blau. 2000. Tetrahydrobiopterin biosynthesis, regulation and function. Biochem. J. 347:1-16. [PMC free article] [PubMed] [Google Scholar]
- 38.Toshie, H., H. Kagamiyama, and K. Hatakeyama. 1993. Feedback regulation mechanisms for the control of GTP cyclohydrolase I activity. Science 260:1507-1510. [DOI] [PubMed] [Google Scholar]
- 39.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 76:4350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Viveros, O. H., C. L. Lee, M. M Abou-Donia, J. C. Nixon, and C. A. Nichol. 1981. Biopterin cofactor biosynthesis: independent regulation of GTP cyclohydrolase in adrenal medulla and cortex. Science 213:349-350. [DOI] [PubMed] [Google Scholar]
- 41.Weisberg, E., and J. M. O'Donnell. 1986. Purification and characterization of GTP cyclohydrolase I from Drosophila melanogaster. J. Biol. Chem. 261:1453-1458. [PubMed] [Google Scholar]
- 42.Werner, E. R., G. Werner-Felmayer, and B. Mayer. 1998. Tetrahydrobiopterin, cytokines, and nitric oxide synthases. Proc. Soc. Exp. Biol. Med. 219:171-182. [DOI] [PubMed] [Google Scholar]
- 43.Xie, L., J. A. Smith, and S. S. Gross. 1998. GTP cyclohydrolase I inhibition by the prototypic inhibitor 2,4-diamino-6-hydroxypyrimidine. J. Biol. Chem. 273:21091-21098. [DOI] [PubMed] [Google Scholar]
- 44.Yim, J. J., and G. M. Brown. 1976. Characterization of guanosine triphosphate cyclohydrolase I purified from Escherichia coli. J. Biol. Chem. 251:5087-5094. [PubMed] [Google Scholar]
- 45.Yoneyama, T., and K. Hatakeyama. 1998. Decameric GTP cyclohydrolase I forms complexes with two pentameric GTPCH feedback regulatory proteins in the presence of phenylalanine or of a combination of BH4 and GTP. J. Biol. Chem. 273:20102-20108. [DOI] [PubMed] [Google Scholar]
- 46.Yoo, J. C., J. M. Han, O. H. Ko, and H. J. Bang. 1998. Purification and characterization of GTP cyclohydrolase I from Streptomyces tubercidicus, a producer of tubercidin. Arch. Pharm. Res. 21:692-697. [DOI] [PubMed] [Google Scholar]