Abstract
RNA oligonucleotides having triazole linkages between uridine and adenosine nucleosides have been prepared and studied using spectroscopic techniques. UV melting and CD showed that triazole strongly destabilized RNA duplex (7 to 14 °C per modification). NMR data suggested that, despite relative flexibility around the modified linkage, all base pairs were formed.
Keywords: RNA, Modified nucleoside, Triazole internucleoside linkage, Huisgen dipolar cycloaddition, Click reaction
RNA interference may become a promising alternative to conventional chemotherapy of genetic disorders, cancer, viral infections and other diseases for which the target mRNA can be identified.1 However, for short interfering RNAs (siRNAs) to be efficient therapeutic agents they need to be chemically modified to optimize nuclease resistance, crossing of cell membranes, biodistribution and pharmacokinetics.2 We are interested in developing RNA analogues that have the phosphate linkages replaced by non-ionic and hydrophobic modifications, which we believe may improve the above properties of siRNAs. We have found that amide3 (3′-CH2-CO-NH-5′) and formacetal4 (3′-O-CH2-O-5′) linkages have surprisingly little effect on the global A-type structure, thermal stability and hydration of RNA and appear to be excellent mimics of phosphodiester linkages. Meanwhile, preparation of RNA bearing these modifications requires extensive synthetic effort, which hinders rapid testing and development of modified siRNAs. In a search for a non-ionic modification that would be easy to synthesize both at the modified nucleoside and siRNA level, we became interested in triazole linkages that could be made via the Huisgen [3+2] dipolar cycloaddition (the “click” reaction).5,6
Von Matt et al.7 were the first to report synthesis and thermal stability of triazole-linked DNA, where the triazole was derived from 3′-azidothymidine. They found that the triazole modification of the DNA strand destabilized DNA-RNA heteroduplexes by about 1 to 2 °C per modification.7b Recently, Isobe et al.8 synthesized triazole-linked dT10, using Huisgen cycloaddition of 3′-azido and 5′-ethynyl thymidine. The triazole in Isobe’s work was isomeric to von Matt’s structure and led to an exceptionally stable duplex with complementary DNA.8a Similar triazole linkage was later used by El-Sagheer and Brown to ligate synthetic DNA fragments and to amplify the resulting construct using PCR.9 Dondoni and co-workers10 also described preparation of trizole-linked DNA trinucleosides, but did not report on biophysical properties of these compounds.
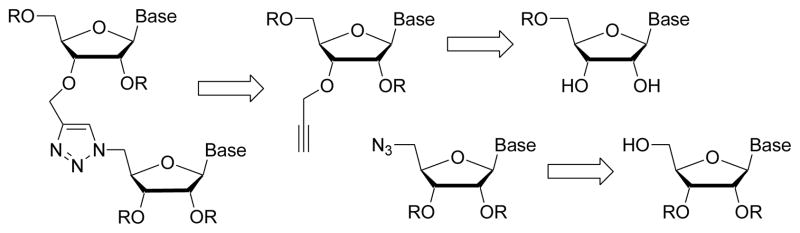
Although triazole linkages prepared from 3′-azidothymidine have shown great promise in DNA, we expected that the presence of 2′-hydroxyl would complicate both preparation of 3′-azidouridine and synthesis of the corresponding triazole linked RNA. We envisioned that the easiest approach to triazole linked RNA would be a coupling of 3′-O-propargyl and 5′-azidoribonucleosides (Figure 1).
Figure 1.
Retrosynthetic analysis of triazole-linked RNA.
Zerrouki and co-workers11 incorporated the trizole linkage shown in Figure 1 in DNA but did not report on biophysical properties of the modified oligonucleotides. El-Sagheer and Brown used the “click” reaction between 3′-O-propargyl-5-methyldeoxycytidine and 5′-azidouridine to incorporate a single triazole linkage at the active site of a hammerhead ribozyme.12 The modified ribozyme was fully functional suggesting that the triazole linkage in Figure 1 may be well tolerated in RNA despite being two atoms longer than the phosphate. Our own recent results, suggesting that RNA may tolerate hydrophobic non-ionic modifications better than DNA,3,4 inspired us to test the properties of triazole-linked RNA. We found that the triazole linkage strongly destabilized RNA double helix by about 7 to 14 °C per modification depending on the sequence and location of the modifications. Although satisfactory melting curves could be recorded for all model systems, NMR and CD spectra suggested that the double helical structure is relatively flexible.
The key consideration in our design was minimizing the synthetic effort. While preparation of 5′-azidoribonucleosides was straightforward,13 synthesis of 3′-O-propargyl ribonucleosides involved alkylation of the 2′,3′-cis-diol system, which we expected to proceed with low selectivity and require separation of isomeric products.14 However, we envisioned that the quick access of the alkyne coupling partner (only three to four steps from ribonucleosides) would offset the difficulties and low yields of the alkylation step.
Treatment of the known 5′-O-methoxytrityluridine 1 with dibutyltin oxide followed by propargyl bromide gave the expected mixture of 3′- and 2′-O-propargyluridine (2 and 3, in an approximate ratio of 1:1.3, respectively) from which the target compound was isolated in 28% yield (Scheme 1). To complete the synthesis of the alkyne coupling partner 4, 2′-OH was protected with acetyl group.
Scheme 1.
Propargylation of 5′-O-methoxytrityluridine. Steps: (a) Bu4NBr, Bu2SnO, DMF 100 °C, 2 h, then propargyl bromide, 45 °C, 24 h, 28%; (b) acetic anhydride, pyridine, rt, 5 h 98%.
We recently developed synthesis of selectively protected 5′-azido-2′-O-TOM adenosine 5,3a which in the present study served well as the coupling partner for 4 in the Huisgen cycloaddition reaction to prepare the triazole linked r(UTRA) dimer 6 (Scheme 2). The structure of the triazole linkage was confirmed using 2D NOESY NMR spectroscopy. In particular, the NOEs of adenosine H5′, triazole aromatic H and 3′-OCH2 of uridine confirmed the assigned structure of 6 (for details see Supplementary Information, Figures S1 and S2). Standard phosphoramidite synthesis gave dimer 7 suitable for solid phase synthesis (Expedite 8909) of RNA sequences bearing the triazole modification.
Scheme 2.
Synthesis of triazole-linked r(UTRA) phosphoramidite. Steps: (a) CuSO4, sodium ascorbate, THF/ethanol/water, 60 °C, 12 h, 79%; (b) iPr2NEt, ClP(OCH2CH2CN)N(iPr)2, CH2Cl2, rt, 6 h, 47 %.
To evaluate the effect of triazole linkage on thermal stability of RNA we synthesized several self-complementary sequences (Table 1) that had performed well in our previous thermodynamic and structural studies.3a,4a UV thermal melting (Figure S7) was done as previously reported.3a,4a Results in Table 1 show that triazole linkage strongly destabilized RNA helices. The effect was more dramatic for RNA3 and RNA4 (Δtm >10 °C per modification) where the triazole linkages were placed opposite to each other in the double helix than for RNA1 and RNA2 (Δtm ca 7 °C per modification) where the triazoles were shifted relative to each other.
Table 1.
Thermal stability of triazole modified RNA.
| Entry | Sequence | tm TRa | tm RNAb | Δ tm |
|---|---|---|---|---|
| RNA1 | GCGUAUTRACGC | 39.5 | 53.7 | −7.1 |
| RNA2 | GCGUTRAUACGC | 38.7 | 53.7 | −7.5 |
| RNA3 | GUGUTRACAC | 16.0 | 40.8 | −12.4 |
| RNA4 | GCGUTRACGC | 29.5 | 58.3 | −14.4 |
tm (°C) of triazole-modified RNA, TR denotes position of the triazole linkage;
tm (°C) of unmodified RNA control.
To gain insights into how the triazole linkage affected the structure of modified RNA we compared the circular dichroism (CD) spectra of RNA1-RNA4 with the spectra of the corresponding unmodified RNAs (Figure 2). RNA2 gave the expected CD spectrum (Figure 2A) similar to the unmodified RNA. Despite the similar tm and melting curves (Figure S7) RNA1 gave almost no CD signal (Figure 2A). A similar result was obtained with RNA3 (Figure S8). RNA4 gave CD spectrum similar to but less intense than the unmodified control (Figure 2B). Overall, the CD data suggested that triazole modification made RNA duplexes more flexible and that RNA1 and RNA3 might be significantly disordered even at temperatures well below the tm obtained by UV melting (20 and 0 °C, respectively).
Figure 2.
Comparison of CD spectra of triazole-linked RNAs (broken lines) and the unmodified RNA controls (solid lines). Recorded at A 20 °C, B 0 °C.
To gain insight into base pair formation in the triazole modified RNA we studied RNA4 using NMR techniques.15 Spectra in 95% H2O/5% D2O exhibit three clear imino proton signals (Figure 3, top), one of which is clear only at low temperature where exchange of the imino proton with water is sufficiently slowed.
Figure 3.
NMR spectra and 2D water-NOESY spectrum of RNA4 duplex (0.1 mM). Upper left: 1D spectra of the imino proton region of the duplex (upper right with numbering) showing that U4 imino is only observed at low temperature. Bottom: 2D water-NOESY acquired at 1°C with mixing time of 150 msec. Red circles indicate A5 H2 NOEs to U4 imino, C6 H1′, and cross-strand A5 H1′. Cytosine intramolecular amino-amino cross-peaks typical in Watson-Crick GC pairs are labeled, including C8 in the terminal base pair for which no G imino is observed. Strong cross-peaks from G3 and G7 iminos to C6 and C2 aminos, respectively, indicate GC Watson-Crick pairs.
Strong NOEs to cytosine amino protons allow assignment of the two temperature-stable imino protons to G3 and G7 in GC base pairs. An NOE to the H2 proton of the lone adenine, despite the high rate of exchange with water, identifies the third imino peak as belonging to uracil in an AU base pair. Further evidence of this AU pair is contained in typical NOEs from adenine H2 to the cross-strand adenine H1′ and to the intrastrand H1′ of C6. Finally, the chemical shift of the adenine H2 proton (7.14 ppm) is nearly identical to that observed for the amide-modified and the unmodified RNA in our previous study (7.08 ppm).3a While no imino proton signal was observed for the terminal guanosine (G1), formation of the closing GC base pair was suggested by the chemical shifts of C8 amino protons. One of these was shifted greater than 1 ppm downfield relative to the other, suggesting involvement in a hydrogen bond of one proton but not the other. This pattern, similar to that observed for C2 and C6 amino protons, is typical of Watson-Crick GC pairs. Thus, the NMR evidence indicates formation of a duplex that includes the central AU pair, although that pair may be slightly distorted or dynamic allowing some water access. The relative flexibility of the central AU pair is consistent with reduced intensity of the CD signal for RNA4 (Figure 2B).
In summary, we found that the triazole 3′-O-CH2-C=CH-N-5′ linkage strongly destabilized RNA double helix (Δtm −7 to −14 °C per modification) and is not a good mimic of the phosphate linkage in RNA. Our results on the triazole linkage that has four bonds (3′-O-CH2-C=CH-N-5′) instead of the two bonds long phosphate (3′-O-P(O)2-O-5′) linkage are in contrast to results reported by Isobe et al.8 who found that the two bonds long triazole (3′-N-CH=CH-5′) linkage strongly stabilized DNA. The most likely explanation for these differences is that the significantly longer linkage adopts conformation that is less than optimal for RNA double helix and causes the flexibility of the UA base pairs as suggested by the NMR and CD spectra of RNA4. Depending on the sequence the duplex may become disordered, as suggested by the lack of CD signal for RNA1 and RNA3. The striking observation that modified duplexes with similar melting curves gave different CD spectra underlines the need for careful and comprehensive biophysical studies before conclusions can be made about the properties of a nucleic acid modification.
Supplementary Material
Acknowledgments
We thank National Institutes of Health (R01 GM071461) for financial support of this research and Dr. Pradeep S. Pallan (Vanderbilt University) for help with MALDI-TOF MS.
Footnotes
Supplementary material available: experimental procedures, copies of HPLC traces, UV melting curves, CD and NMR spectra.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and notes
- 1.Watts JK, Corey DR. Bioorg Med Chem Lett. 2010;20:3203. doi: 10.1016/j.bmcl.2010.03.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.(a) Watts JK, Deleavey GF, Damha MJ. Drug Discovery Today. 2008;13:842. doi: 10.1016/j.drudis.2008.05.007. [DOI] [PubMed] [Google Scholar]; (b) Corey DR. J Clin Invest. 2007;117:3615. doi: 10.1172/JCI33483. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Rozners E. Curr Org Chem. 2006;10:675. [Google Scholar]; (d) Manoharan M. Curr Opin Chem Biol. 2004;8:570. doi: 10.1016/j.cbpa.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 3.(a) Chelliah S, Thomas S, Abbott J, Kennedy SD, Rozners E. Angew Chem, Int Ed. 2011;50:2068. doi: 10.1002/anie.201007012. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Rozners E, Katkevica D, Bizdena E, Strömberg R. J Am Chem Soc. 2003;125:12125. doi: 10.1021/ja0360900. [DOI] [PubMed] [Google Scholar]
- 4.(a) Kolarovic A, Schweizer E, Greene E, Gironda M, Pallan PS, Egli M, Rozners E. J Am Chem Soc. 2009;131:14932. doi: 10.1021/ja904926e. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Rozners E, Katkevica D, Strömberg R. Chem BioChem. 2007;8:537. doi: 10.1002/cbic.200600515. [DOI] [PubMed] [Google Scholar]; (c) Rozners E, Strömberg R. J Org Chem. 1997;62:1846. [Google Scholar]
- 5.(a) Rostovtsev VV, Green LG, Fokin VV, Sharpless KB. Angew Chem Int Ed. 2002;41:2596. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]; (b) Tornoe CW, Christensen C, Meldal M. J Org Chem. 2002;67:3057. doi: 10.1021/jo011148j. [DOI] [PubMed] [Google Scholar]
- 6.For a recent review, see El-Sagheer AH, Brown T. Chem Soc Rev. 2010;39:1388. doi: 10.1039/b901971p.
- 7.(a) Von Matt P, Lochmann T, Altmann KH. Bioorg Med Chem Lett. 1997;7:1549. [Google Scholar]; (b) Von Matt P, Altmann KH. Bioorg Med Chem Lett. 1997;7:1553. [Google Scholar]
- 8.(a) Isobe H, Fujino T, Yamazaki N, Guillot-Nieckowski M, Nakamura E. Org Lett. 2008;10:3729. doi: 10.1021/ol801230k. [DOI] [PubMed] [Google Scholar]; (b) Fujino T, Yamazaki N, Isobe H. Tetrahedron Lett. 2009;50:4101. [Google Scholar]
- 9.El-Sagheer AH, Brown T. J Am Chem Soc. 2009;131:3958. doi: 10.1021/ja8065896. [DOI] [PubMed] [Google Scholar]
- 10.Nuzzi A, Massi A, Dondoni A. QSAR Comb Sci. 2007:1191. [Google Scholar]
- 11.(a) Lucas R, Zerrouki R, Granet R, Krausz P, Champavier Y. Tetrahedron. 2008;64:5467. [Google Scholar]; (b) Lucas R, Neto V, Hadj Bouazza A, Zerrouki R, Granet R, Krausz P, Champavier Y. Tetrahedron Lett. 2008;49:1004. [Google Scholar]; (c) Lucas R, Elchinger PH, Faugeras PA, Zerrouki R. Nucleosides, Nucleotides Nucleic Acids. 2010;29:168. doi: 10.1080/15257771003708579. [DOI] [PubMed] [Google Scholar]
- 12.El-Sagheer AH, Brown T. Proc Natl Acad Sci U S A. 2010;107:15329. doi: 10.1073/pnas.1006447107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamamoto I, Sekine M, Hata T. J Chem Soc, Perkin Trans. 1980;1:306. [Google Scholar]
- 14.Srivastava SC, Raza SK. 5 744 595. US Patent US. 1998
- 15.Hammond NB, Tolbert BS, Kierzek R, Turner DH, Kennedy SD. Biochemistry. 2010;49:5817. doi: 10.1021/bi100332r. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.