Abstract
Two types of necrosis-inducing lipodepsipeptide toxins, called syringomycin and syringopeptin, are major virulence factors of Pseudomonas syringae pv. syringae strain B301D. A previous study showed that a locus, called syrA, was required for both syringomycin production and plant pathogenicity, and the syrA locus was speculated to encode a regulator of toxin production. In this study, sequence analysis of the 8-kb genomic DNA fragment that complements the syrA phenotype revealed high conservation among a broad spectrum of fluorescent pseudomonads. The putative protein encoded by open reading frame 4 (ORF4) (1,299 bp) in the syrA locus region exhibited 85% identity to ArgA, which is involved in arginine biosynthesis in Pseudomonas aeruginosa. Growth of strain W4S2545, the syrA mutant, required supplementation of N minimal medium with arginine. Similarly, syringomycin production of syrA mutant W4S2545 was restored by the addition of arginine to culture media. Furthermore, the insertion of Tn5 in the genome of the syrA mutant W4S2545 was localized between nucleotides 146 and 147 in ORF4, and syringomycin production was complemented in trans with the wild-type DNA fragment containing intact ORF4. These results demonstrate that the syrA locus is the argA gene of P. syringae pv. syringae and that argA is directly involved in arginine biosynthesis and therefore indirectly affects syringomycin production because of arginine deficiency.
A notable feature of Pseudomonas syringae pv. syringae, an economically important pathogen causing necrotic infections of a broad range of plant species, is the production of two lipopeptide phytotoxins, syringomycin and syringopeptin (6). The toxins cause necrosis in host plants and are major virulence factors (17, 32). Syringomycin is a cyclic lipodepsinonapeptide composed of a 3-hydroxy fatty acid tail attached to a polar peptide head that contains nine amino acid residues, including arginine (6). Three forms of syringomycin that differ by the length of the 3-hydroxy fatty acid tail are produced (5). Syringomycin E (SRE), which contains a 3-hydroxy dodecanoic acid tail, is the major form produced by strain B301D of P. syringae pv. syringae (5). Syringomycin is synthesized via a nonribosomal peptide synthetase system (18, 43). The syringomycin (syr) gene cluster is located in a 55-kb DNA region in the strain B301D genome, and the syr gene cluster is composed of genes required for biosynthesis, regulation, and secretion of syringomycin (18, 23, 28, 43, 44).
Xu and Gross (40) described a gene called syrA that is an essential locus for syringomycin production and the pathogenicity of P. syringae pv. syringae B301D. The syrA mutant strain W4S2545 was generated by random Tn5 mutagenesis of the B301D genome and was selected due to the loss of syringomycin production and pathogenicity in pear seedlings and cherry fruits. Although the syrA mutant was unchanged with regard to its ability to elicit a hypersensitive reaction on tobacco leaves, the syrA mutant was significantly attenuated in its ability to grow in planta (41). Syringomycin production of strain W4S2545 was restored both in trans and in cis by the introduction of cosmid pGX183 (41). The syrA gene was localized on an 8-kb EcoRI-KpnI DNA fragment in the B301D genome and was estimated to be 2.3 to 2.8 kb in size based on restriction enzyme mapping and Tn5 mutagenesis. Consequently, the syrA locus was predicted to be a regulatory gene that might control several pathogenicity genes (41). Recent sequence analysis found that the syrA locus was not located in the syr gene cluster (23, 33). The function of the syrA gene in syringomycin production was unclear prior to this work.
To define the function of the syrA locus in relation to syringomycin production and the pathogenicity of P. syringae pv. syringae strain B301D, the syrA gene region was cloned from pGX183 (41) and from the chromosome of syrA mutant W4S2545. Sequence analysis revealed significant nucleotide identity of the syrA gene locus to the argA gene of Pseudomonas aeruginosa, which is involved in arginine biosynthesis (9). The Tn5 transposon insertion in the strain W4S2545 genome was mapped and confirmed by sequencing. The results of this report demonstrate that strain W4S2545 is an arginine-auxotrophic mutant of strain B301D and that the syrA locus, renamed argA, encodes an N-acetylglutamate synthetase required for arginine biosynthesis. The conservation of the argA gene region among fluorescent pseudomonads is also discussed.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and growth conditions.
The bacterial strains and plasmids used in this study are described in Table 1. Escherichia coli strain DH10B (14) was used as a host for cloning and was cultured in Luria-Bertani medium (31) at 37°C. Strains of P. syringae pv. syringae were cultured at 25°C in nutrient broth-yeast extract (NBY) medium (38). Potato dextrose agar (PDA) supplemented with 0.4% Casamino Acids (16) was used in plate bioassays of P. syringae pv. syringae strains for syringomycin production. N minimal (NM) agar medium (38) was used for the investigation of arginine auxotrophy. Antibiotics (Sigma Chemical Co., St. Louis, Mo.) were added to media as required in the following concentrations: 100 μg/ml for ampicillin, 100 μg/ml for kanamycin, and 25 μg/ml for tetracycline.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Relevant characteristicsa | Source or reference |
|---|---|---|
| Strains | ||
| E. coli DH10B | F−mcrA ΔlacX74 (φ80dlacZΔM15) Δ(mrr-hsdRMS-mcrB) deoR recA1 endA1 araD139 Δ(ara leu)7697 galU galK λ−rpsL nupG | 14 |
| P. syringae pv. syringae | ||
| B301D | Wild-type strain from pear | 8 |
| W4S2545 | B301D-R::Tn5; Rifr Kmr | 40 |
| B301DSL22 | ORF5::nptII derivative of B301D; Kmr | This study |
| B301DSL23 | ORF6::nptII derivative of B301D; Kmr | This study |
| BR132 | syrB1::Tn3HoHo1 derivative of B301DR; Pipr | 43 |
| Plasmids | ||
| pBlueScript II SK | Cloning vector; Apr | Strategene |
| pUC18 | Cloning vector; Apr | 42 |
| pUCP26 | Broad-host range vector; Tcr | 39 |
| pBR325 | Cloning vector; Cmr Tcr Apr | 27 |
| pGX15 | pLAFR3 containing the 12-kb B301D genomic DNA fragment; Tcr | 41 |
| pJS100 | pBlueScript II SK containing the 8-kb EcoRI-KpnI DNA fragment from pGX15; Apr | This study |
| pSL101 | pBlueScript II SK containing the 14-kb EcoRI-KpnI DNA fragment of the W4S2545 genome; Kmr Apr | This study |
| pSL102 | pBlueScript II SK containing the 5-kb BamHI-KpnI DNA fragment from pGX15; Apr | This study |
| pSL108 | pUC18 containing the 3.6-kb SalI-kpnI fragment from pJS100; Apr | This study |
| pSL113 | pSL108 with the nptII insertion at the MscI site of ORF5; Kmr Apr | This study |
| pSL114 | pSL108 with the nptII insertion at the EcoRV site of ORF6; Kmr Apr | This study |
| pSL115 | pBR325 containing the disrupted argE gene with the nptII insertion from pSL113; Kmr Tcr | This study |
| pSL116 | pBR325 containing the disrupted ORF6 with the nptII insertion from pSL114; Kmr Tcr | This study |
| pSL117 | pUCP26 containing the 8-kb EcoRI-KpnI DNA fragment from pGX15; Tcr | This study |
Kmr, Tcr, Apr, Pip,r and Rifr, resistance to kanamycin, tetracycline, ampicillin, piperacillin, and rifampin, respectively.
DNA manipulations.
Routine procedures were used for preparing plasmid DNA and for subcloning DNA fragments into designated vectors (31). Genomic DNA was prepared with the cetyltrimethylammonium bromide protocol (10). A QIAEX II gel extraction kit (QIAGEN Inc., Valencia, Calif.) was used to recover DNA fragments for cloning and for use as hybridization probes. Electroporation of plasmids into cells of E. coli and P. syringae was completed with a Gene Pulser II (Bio-Rad Laboratories, Hercules, Calif.) as described by Scholz-Schroeder et al. (32).
Sequence analysis.
To sequence the syrA gene region, plasmid pJS100 (Table 1) was generated from pGX15 (41) and mutagenized by using an EZ::TN <Kan-2> insertion kit according to the manufacturer's instructions (Epicentre Technologies Corp., Madison, Wis.). Based on mapping with restriction endonucleases, the plasmids with nptII insertions at different positions were selected for sequencing. Forward and reverse primers supplied with the EZ::TN <Kan-2> insertion kit were used for initial sequence reactions off each plasmid carrying an EZ::TN transposon. Sequencing was completed with primer walking when noncontiguous sequences existed. Primers were designed for primer walking with the PRIME program within the Wisconsin Sequence Analysis programs of Genetics Computer Group (GCG) package version 10.0 (12) and were synthesized by Operon Technologies, Inc. (Alameda, Calif.). Sequence reactions were completed by using fluorescence-based dideoxy terminators and Ampli-Taq polymerase and were run on an ABI PRISM 377 DNA sequencer (PerkinElmer Applied Biosystems, Inc., Norwalk, Conn.). At least triple coverage of sequencing reactions was achieved to generate consensus DNA sequence.
Sequence analysis was accomplished using the GCG package and Lasergene expert sequence analysis software version 5.0 (DNASTAR, Inc., Madison, Wis.). The following GCG programs were used: GELASSEMBLE for the assembly of nucleotide sequences, FINDPATTERNS for the identification of Shine-Dalgarno sequences, TERMINATOR for the prediction of rho-independent transcriptional terminators, and BESTFIT for the comparison of two sequences. Database searches for genes and proteins homologous to the predicted open reading frames (ORFs) were conducted by using the BLAST server at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov). To evaluate the significance of the protein sequence similarities, the GAP program of GCG was run with the randomization option of 100. The Z scores were calculated by using the actual quality score minus the mean quality value of the randomized scores divided by the standard deviation of randomized score distribution. Protein homology was significant if the Z score value was larger than 6. The MEGALIGN program within the Lasergene software package was used for multiple sequence analysis.
Complementation assays.
The growth of P. syringae pv. syringae syrA mutant strain W4S2545 (40) was evaluated on NM medium supplemented with a 1-μg/ml concentration of l-glutamate, N-acetyl-l-glutamate, N-acetyl-l-ornithine, or l-arginine (Sigma Chemical Co.). Bacterial cells were grown with shaking in 4 ml of NBY medium for 6 h at 25°C, harvested by centrifugation, and diluted to a cell density of ∼2 × 108 CFU/ml. NM agar medium was supplemented with 1 μg of l-arginine/ml or selected intermediates in the arginine biosynthesis pathway. NM plates were inoculated with a 5-μl droplet containing 106, 104, or 102 cells per spot. Wild-type strain B301D of P. syringae pv. syringae was used as the reference strain for growth. The ability to produce visible colonies was recorded after the inoculated plates were incubated at 25°C for 96 h. Each experiment was repeated independently three times with three plates per replicate.
Mutagenesis of the ORFs upstream of the syrA gene.
ORF5 and ORF6 were disrupted by the insertion of the nptII gene to generate nonpolar mutations. A 1.4-kb fragment containing the nptII gene from pBSL15 (1) was cloned into pSL108 at the MscI site to disrupt translation of ORF5, resulting in plasmid pSL113. Likewise, the nptII fragment was inserted in pSL108 at the EcoRV site to disrupt the translation of ORF6 and to generate plasmid pSL114. The nptII insertions in the two ORFs were confirmed by restriction endonuclease mapping and sequencing. The disrupted gene fragments from plasmids pSL113 and pSL114 were cloned into pBR325 at the EcoRI site that was previously blunted by T4 DNA polymerase to construct plasmids pSL115 and pSL116, respectively. Plasmids pSL115 and pSL116 were electroporated into P. syringae pv. syringae strain B301D for marker exchange mutagenesis as previously described by Scholz-Schroeder et al. (32). Homologous recombination of the disrupted gene fragments in the genome of P. syringae pv. syringae B301D was verified by Southern analysis (31).
Assays for syringomycin production.
Syringomycin production by P. syringae pv. syringae strains was evaluated by using bioassays as previously described by Gross and DeVay (16). In brief, bacterial cells were grown overnight in NBY medium at 25°C. The cells were harvested by centrifugation, washed with sterile deionized water, and adjusted to a final cell density of ∼2 × 108 CFU/ml. To test the effects of additions of l-arginine and selected intermediates of the arginine biosynthesis pathway on the restoration of syringomycin production of the syrA mutant, each PDA plate was supplemented with a 1-μg/ml concentration of either l-glutamate, N-acetyl-glutamate, N-acetyl-l-ornithine, or l-arginine. The PDA plates were inoculated with 5 μl of the bacterial suspension and incubated at 25°C for 4 days. The inoculated plates were oversprayed with Geotrichum candidum F-260 as an indicator organism and were further incubated for 24 h at 25°C. Zones inhibitory to the growth of G. candidum indicated syringomycin production. P. syringae pv. syringae wild-type strain B301D and syrB1 mutant strain BR132 (43) were used as positive and negative controls, respectively, for syringomycin production. Each assay was repeated independently three times with three plates per replicate.
Nucleotide sequence accession number.
The nucleotide sequence of the 8,080-bp DNA region (Fig. 1) is available in the GenBank database under accession number AY374326.
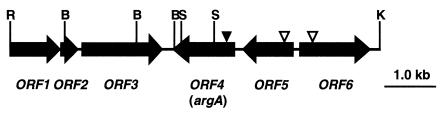
FIG. 1.
Physicalmap of the 8,080-bp syrA gene region of P. syringae pv. syringae B301D. The positions and orientations of the potential ORFs are shown as horizontal arrows. The solid-black triangle indicates the position of the Tn5 insertion in strain W4S2545, and the open triangles mark the positions of the nptII insertions. Restriction enzyme sites are abbreviated as follows: R, EcoRI; B, BglII; S, SalI; and K, KpnI.
RESULTS
Sequence analysis of the syrA gene region.
To characterize the genomic DNA region that restored the phenotype of the syrA mutation, an 8,080-bp DNA EcoRI-KpnI fragment of pGX15 was sequenced from pJS100 (Table 1). Sequence analysis revealed six potential ORFs in the region (Fig. 1). A striking feature of the 8,080-bp region was the presence of ORF4 and ORF5, which exhibited significant homology, respectively, to the argA and argE genes involved in arginine biosynthesis in prokaryotes (9). The other four ORFs in this region were also conserved among pseudomonads (25, 36).
ORF4 shared the highest identity to the argA gene, encoding N-acetylglutamate synthetase in bacteria (9). This ORF, 1,299 bp in length, was preceded by a potential Shine-Dalgarno sequence (ACAGG) 11 bp upstream of the start codon (ATG) of the ORF, and a rho-independent transcriptional terminator was identified 58 bp downstream of the stop codon (TAA). Nucleotide sequence analysis revealed that ORF4 exhibited 84 and 80% identity to the argA genes of Pseudomonas putida KT2440 (25) and P. aeruginosa PAO1 (36), respectively. ORF4 was predicted to code for a protein 432 amino acids in size. Database searches demonstrated that the putative protein from ORF4 shared the highest identity (88%; Z score, 364) to the ArgA protein of P. putida KT2440 (25). In addition, this protein encoded by ORF4 showed identities of 85, 52, and 49% to the putative ArgA proteins of P. aeruginosa PAO1 (36), Yersinia pestis KIM (11), and E. coli K-12 (29), respectively. The N-acetylglutamate synthetase encoded by argA is the first enzyme of the arginine biosynthesis pathway in bacteria and catalyzes the acetylation of l-glutamate with acetyl coenzyme A (Fig. 2).
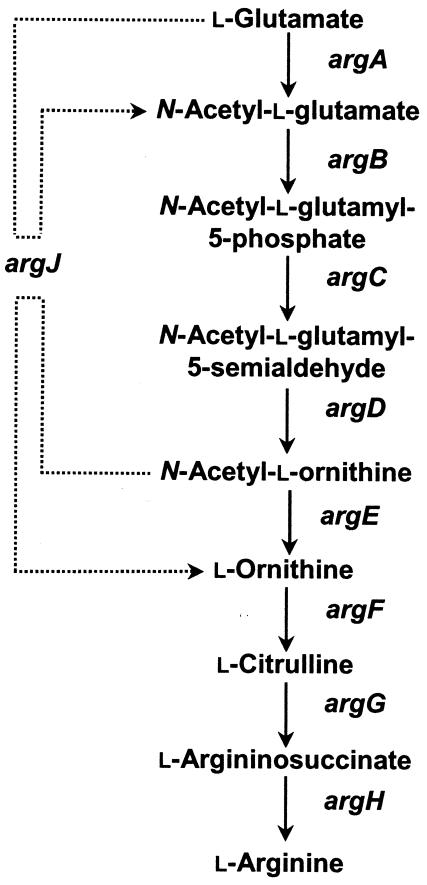
FIG. 2.
Pathwayof arginine biosynthesis in bacteria modified from the work of Cunin et al. (9). Gene designations in the arginine biosynthesis pathway are argA (N-acetylglutamate synthetase), argB (N-acetylglutamate 5-phosphotransferase), argC (N-acetylglutamate 5-semialdehydrogenase), argD (N-acetylornithine aminotransferase), argE (acetylornithine deacetylase), argF (anabolic ornithine carbamoyltransferase), argG (argininosuccinate synthetase), argH (argininosuccinase), and argJ (ornithine acetyltransferase). The dashed line indicates that the acetyl group of N-acetyl-l-ornithine is recycled to form N-acetylglutamate by ArgJ in the cyclic pathway.
ORF5 exhibited significant homology to the argE gene, predicted to code for acetylornithine deacetylase in bacteria (7, 9). This ORF was 1,149 bp in length and had a conserved Shine-Dalgarno sequence (AGGAAG) 5 bp upstream of the start codon (ATG) of the ORF. A potential rho-independent transcriptional terminator was predicted 79 bp downstream of the stop codon (TGA). Likewise, ORF5 exhibited 78 and 77% identity to genes designated argE in the genomes of P. aeruginosa PAO1 (36) and P. putida KT2440 (25), respectively. ORF5 was predicted to encode a protein 382 amino acids in size which shares 81 and 78% identity to the putative ArgE proteins of P. putida KT2440 (25) and P. aeruginosa PAO1 (36), respectively. In addition, the putative protein encoded by ORF5 exhibited 49% identity to the well-characterized acetylornithine deacetylase (ArgE) of E. coli (7, 21). The acetylornithine deacetylase encoded by argE is known to catalyze deacetylation of N-acetylornithine into ornithine in the arginine biosynthesis pathway of E. coli (Fig. 2) (9).
Four additional ORFs (ORF1, ORF2, ORF3, and ORF6) in the syrA gene region were predicted to code for conserved proteins in pseudomonads (Fig. 1; Table 2). These four potential ORFs had the same transcriptional orientation, which was opposite to that of ORF4 and ORF5. ORF1 was 1,032 bp in length, and the putative protein product exhibited 90% identity to the C terminus of the S1 RNA binding domain protein (PP0245) of P. putida KT2440 (25) and 85% identity to the C terminus of a hypothetical protein (PA5201) of P. aeruginosa (36). ORF2 was 384 bp in size, and the predicted protein from this ORF showed 76 and 72% identity to proteins PP0244 of P. putida and PA5202 of P. aeruginosa, respectively, whose functions are unknown. ORF3, 1,608 bp in size, was predicted to code for a protein that shared the highest identity (78%) to a putative glutamate-cysteine ligase (PP0243) of P. putida KT2440 and 76% identity to the glutamate-cysteine ligase (PA5203) of P. aeruginosa. ORF6, which was 1,455 bp in length, was localized upstream of ORF5, and the putative protein from ORF6 shared 75 and 67% identity to proteins of P. putida KT2440 (PP5187) and P. aeruginosa PAO1 (PA5209), respectively, whose functions are unknown.
TABLE 2.
Genes and ORFs identified in the argA gene region of strain B301D
| ORF | Homologya
|
Predicted function | ||
|---|---|---|---|---|
| Homolog | Identity (%) | Z score | ||
| ORF1 | Tex (part) | 91 | 262 | S1 RNA binding domain protein |
| ORF2 | PP0244 | 76 | 86 | Unknown |
| ORF3 | GshA | 74 | 392 | Glutamate-cysteine ligase |
| ORF4 | ArgA | 88 | 364 | N-Acetylglutamate synthetase |
| ORF5 | ArgE | 81 | 328 | Acetylornithine deacetylase |
| ORF6 | PP5187 | 75 | 255 | Unknown |
Homology to genes or ORFs of P. putida KT2440 (26).
ORF4 was disrupted by the insertion of a Tn5 transposon in the syrA mutant genome.
An EcoRI-KpnI DNA fragment 14 kb in length and containing a Tn5 transposon was cloned from the genome of the syrA mutant W4S2545 to localize the transposon in the genome. Sequence analysis of the 14-kb DNA fragment carrying a Tn5 transposon (5.8 kb) demonstrated that the sequence from this fragment was identical to the 8.1-kb EcoRI-KpnI DNA fragment of pGX183 (41). Mapping and further sequence analysis revealed that the Tn5 transposon was inserted within ORF4 between nucleotides 146 and 147 relative to the start codon of ORF4 (Fig. 1).
Mutation of ORF4 resulted in a lack of syringomycin production of P. syringae pv. syringae B301D. The syrA mutant W4S2545 failed to produce syringomycin (40, 41) on PDA medium. As expected, cells of strain W4S2545 containing plasmid pSL117, which is pUCP26 (39) carrying the entire ORF4 DNA fragment, produced a zone of inhibition to G. candidum that was comparable to that of the wild-type strain B301D (data not shown). These results show that a functional ORF4 is critical for syringomycin production by P. syringae pv. syringae.
To evaluate the effect of mutations in ORF5 and ORF6 on syringomycin production, insertions of the nptII gene fragment at the unique EcoRV and SmaI sites of the two respective ORFs constructed mutations in both ORF5 and ORF6. The mutations in the two ORFs are nonpolar due to the lack of a transcriptional terminator in the nptII gene (1). As a result, the risk of polar effects of mutation on downstream ORFs was eliminated. The nptII-disrupted ORFs were individually recombined into the genome of P. syringae pv. syringae B301D by marker exchange mutagenesis, generating ORF5 mutant B301DSL22 and ORF6 mutant B301DSL23 (Table 1). Southern analysis verified the recombination of the nptII disruption of each ORF in the genomes of resultant mutants. Plate bioassays of syringomycin production on PDA showed that both strains B301DSL22 (ORF5::nptII) and B301DSL23 (ORF6::nptII) produced zones of inhibition approximately 10 mm in radius, which was not significantly different from that produced by the wild-type strain B301D (data not shown). These data show that neither ORF5 nor ORF6 is required for syringomycin production by P. syringae pv. syringae.
Growth restoration of strain W4S2545 with supplementation of l-arginine and its biosynthetic intermediates.
Growth of syrA mutant W4S2545 was tested on NM agar medium after 96 h of inoculation. No visible colony growth was observed on plates inoculated with a droplet containing 106, 104, or 102 cells of strain W4S2545, unlike the normal growth observed for wild-type strain B301D. The results show that strain W4S2545 is an auxotrophic mutant of P. syringae pv. syringae. In contrast, both strains B301DSL22 (ORF5::nptII) and B301DSL23 (ORF6::nptII) grew on NM plates as well as did wild-type strain B301D (data not shown), which demonstrated that disruption of either ORF5 or ORF6 did not result in an auxotrophic phenotype.
Supplementation of the NM agar medium with l-arginine restored the growth of strain W4S2545. Based on sequence analysis of the syrA gene region and the location of the Tn5 insert in the W4S2545 genome, this strain was predicted to be an arginine-auxotrophic mutant. Growth measurements were performed on NM agar plates supplemented with l-arginine or one of the intermediates in the arginine biosynthesis pathway (Fig. 2). The results demonstrated that addition of l-arginine restored the growth of the syrA mutant. Similarly, colonies of the syrA mutant were observed after incubation for 96 h on the NM agar medium supplemented with N-acetyl-l-ornithine. As expected, supplementation of l-glutamate could not restore the growth of strain W4S2545. However, the growth of strain W4S2545 could not be stimulated by supplementation of N-acetyl-glutamate, which was also reported for argA mutants of P. aeruginosa (19) and Salmonella enterica serovar Typhimurium (13).
Syringomycin production of strain W4S2545 was restored by arginine supplementation.
The effects of supplementation of l-glutamate, N-acetyl-glutamate, N-acetyl-l-ornithine, and l-arginine on syringomycin production were measured by using the standard procedure of plate assays (Fig. 3). The cells of strain W4S2545 growing on the PDA plates supplemented with either l-arginine or N-acetyl-l-ornithine produced zones (8 to 10 mm in radius) inhibitory to G. candidum that were comparable to those of the wild type (12 mm in radius) (Fig. 3). In contrast, no inhibition zone was observed when the cells were cultured on PDA plates supplemented with either l-glutamate or N-acetyl-l-glutamate (Fig. 3). These results showed that the addition of l-arginine or its intermediates, except N-acetyl-l-glutamate, restored syringomycin production by strain W4S2545 on PDA medium. In addition, it was found that the radius of the inhibition zone produced by strain B301D on PDA plates supplemented with 1 μg of l-arginine/ml was increased approximately 20% compared to that without the addition of l-arginine (data not shown).
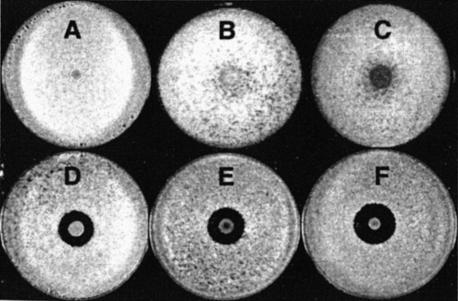
FIG. 3.
Plate bioassays for syringomycin production by P. syringae pv. syringae strain W4S2545. The PDA plates were supplemented to a final concentration of 1 μg/ml with l-glutamate (B), N-acetyl-l-glutamate (C), N-acetyl-l-ornithine (D), or l-arginine (E). As controls, plates A and F were supplemented with sterile water. Plates A to E were inoculated with strain W4S2545, and plate F was inoculated with strain B301D as a control. The inoculated plates were incubated for 4 days at 25°C and oversprayed with G. candidum. Syringomycin production is observed by zones of inhibition of G. candidum. It was observed that the indicator fungus grew on colonies of strain W4S2545 on the PDA plates supplemented with water, l-glutamate, and N-acetyl-l-glutamate but not with the other compounds tested due to syringomycin production.
Conservation of the argA gene region in pseudomonads.
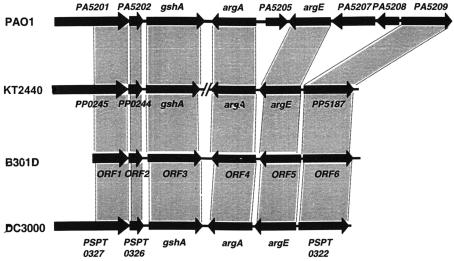
Comparative analyses revealed that the argA gene regions of related strains of P. syringae are conserved. The sequence of the argA gene region of a bean strain of P. syringae pv. syringae, B728a, was nearly identical to that of strain B301D, with an overall nucleotide substitution rate of ∼0.7%, and all of the putative proteins from this region of strain B301D were identical to that of strain B728a (http://www.jgi.doe.gov), which reveals conservation of the argA regions in closely related strains that differ in host specificity (16). More variation in the argA gene region was observed between P. syringae pv. syringae B301D and P. syringae pv. tomato DC3000. The argA gene region was identified in the genome sequence of strain DC3000 approximately 350 kb downstream of dnaA, the chromosomal replication initiator gene (http://www.tigr.org; GenBank accession number, AE016853). The gene organization of the argA region in strain DC3000 was the same as that in strain B301D; however, putative proteins of strain DC3000 exhibited ∼93% identity to those of strain B301D on average. In addition, an overall nucleotide substitution rate of ∼13.3% was observed for strains DC3000 and B301D in the corresponding argA (syrA) gene regions. Deletions also were identified in the argA (syrA) gene region of strain DC3000, compared to that of strain B301D. Most of the deletions occurred within noncoding regions, and the biggest deletion, 96 bp in length, was found between gshA and argA in DC3000. All deletions within an ORF did not result in a translational frameshift.
Further analyses revealed the presence of gene reorganizations among the related species of pseudomonads in the syrA (argA) gene region (Fig. 4). The argA region was identified in the genome sequence of P. aeruginosa strain PAO1 approximately 400 kb upstream of the dnaA gene (http://www.pseudomonas.com; GenBank accession number, AE004091). Compared with the syrA gene region of strain B301D, strain PAO1 carried an extra region, ∼1.2 kb in size (between ORF4 and ORF5 of strain B301D) (Fig. 4), encoding PA5202. The other extra region, ∼2.0 kb in size (between ORF5 and ORF6 of strain B301D), encoded hypothetical proteins PA5207 and PA5208. Interestingly, gene rearrangement in the argA gene region was observed when P. syringae pv. syringae B301D and P. putida KT2440 were compared (http://www.tigr.org) (Fig. 4). The argA gene region of strain KT2440 was divided into two parts. The location of three ORFs, which corresponds to the strain B301D chromosome region containing ORF1, ORF2, and ORF3, was identified in the genome sequence of strain KT2440 approximately 300 kb downstream of the dnaA gene (http://www.tigr.org; GenBank accession number, AE015451). The other region, carrying argA, argE, and a homolog of ORF6 of B301D, was observed approximately 270 kb upstream of the dnaA gene. As a result, the two parts of the argA gene regions observed in strain B301D flank both sides of the origin of replication of the strain KT2440 chromosome.
FIG. 4.
Comparisonof the gene organization in the argA (syrA) gene region of strain B301D with the corresponding argA region of related pseudomonads. The following strains were included in the comparative analysis: P. syringae pv. syringae B301D, P. syringae pv. tomato DC3000, P. putida KT2440, and P. aeruginosa PAO1. Gray boxes connect homologous ORFs. For strain KT2440, the “//” symbolizes an ∼560-kb DNA fragment that separates the locus into two parts in the genome of strain KT2440. Gene regions and designations were obtained from the following websites: http://www.tigr.org for strain DC3000, http://www.pseudomonas.com for strain PAO1, and http://www.tigr.org for strain KT2440.
DISCUSSION
Phenotypic and genetic analyses demonstrated that the syrA locus is equivalent to the argA gene, which encodes N-acetylglutamate synthetase in bacteria. The putative protein from the syrA gene shared a high similarity to the known ArgA proteins of P. putida KT2440 (25), P. aeruginosa PAO1 (36), Y. pestis Kim (11), and E. coli K-12 (29). The ArgA protein, an N-acetylglutamate synthetase, catalyzes the first step of the arginine biosynthesis pathway and converts l-glutamate to N-acetyl-l-glutamate (9, 19). Mutation in the syrA locus resulted in the failure of P. syringae pv. syringae to grow on minimal medium and to produce syringomycin. The phenotype of W4S2545 was recovered after supplementation of the media with arginine and its intermediates and by complementation in trans with the wild-type DNA fragment containing the entire syrA gene. N-Acetyl-l-glutamate, one of the intermediates of arginine biosynthesis, was not able to restore the phenotype of the syrA mutant (W4S2545) in this study. The same results were observed for an argA mutant of P. aeruginosa, and it was predicted that N-acetyl-l-glutamate is deacetylated outside the cytoplasm to facilitate uptake into the cell (19). Finally, the syrA gene was the best-fit hit in the genomic DNA sequence draft of strain B728a (http://www.jgi.doe.gov) when the known argA sequences were used to search a genomic sequence database. These results demonstrated that strain W4S2545 is an arginine auxotrophic mutant and that the syrA phenotype resulted from a mutation of the argA gene. Consequently, the syrA gene is renamed the argA gene.
A functional argA gene is critical for syringomycin production of P. syringae pv. syringae. Xu and Gross (40, 41) demonstrated that a mutation in the syrA (argA) gene eliminated syringomycin production by P. syringae pv. syringae. Subsequently, Grgurina et al. (14) confirmed that reverse-phase high-performance liquid chromatographyanalysis of the syrA mutant W3S2545 failed to produce syringomycin (15). This phenotype was recovered in trans and in cis from strain W4S2545 (argA::Tn5) by the introduction of the wild-type syrA gene, which was further confirmed in trans in this study. Although strain W4S2545 was able to grow as well as its wild-type strain on PDA medium, no measurable syringomycin was produced. The apparent explanation is that PDA medium contained a limited amount of arginine that supports the growth of the argA mutant but not syringomycin production. Because arginine is one of nine amino acids in the syringomycin molecule produced by strain B301D (5), mutation of the argA gene resulted in a corresponding failure to produce syringomycin. This explanation was supported by the observation that syringomycin production by wild-type strain B301D was increased approximately 20% with the addition of arginine to PDA medium.
Arginine biosynthesis as a primary metabolic system is notable for its complexity and variability at the genetic level among prokaryotes (9) (Fig. 2). Two patterns for arginine biosynthesis were proposed for prokaryotes: a linear pathway and a cyclic pathway based on whether argJ is involved in the biosynthesis pathways (4, 9). The argJ gene is absent and the argE gene is required for arginine biosynthesis in the linear pathway, which was found in Enterobacteriaceae (9) and Sulfolobus solfataricus (37). In contrast, all other prokaryotes, including P. aeruginosa, were shown or predicted to employ the cyclic pathway, in which a functional argJ gene is able to complement the argE mutant (4, 9). In addition, the ArgJ proteins are classified into two categories: bifunctional and monofunctional (4, 9). A bifunctional ArgJ protein is capable of complementing both argA and argE auxotrophs as observed for Bacillus stearothermophilus (30), Bacillus subtilis (26), and Neisseria gonorrhoeae (24). In contrast, the monofunctional argJ gene product is unable to restore arginine production to an argA mutant as observed for Streptomyces coelicolor (20). An intact argJ gene was found in the draft genomic sequence of P. syringae pv. syringae strain B728a (http://www.jgi.doe.gov). Therefore, as in P. aeruginosa (19), it is predicted that argJ is monofunctional in P. syringae pv. syringae based on the fact that argA is required for arginine biosynthesis. However, more extensive mutagenesis and biochemical analyses are needed to characterize the function of the predicted argJ gene in P. syringae.
The argA regions in pseudomonads are conserved (Fig. 4). All six ORFs identified in the strain B301D genome were highly conserved, with identities ranging from 67 to 91% among a broad spectrum of fluorescent pseudomonads. The 8-kb region of strains B301D and B728a of P. syringae pv. syringae shared more than 99% identity at the nucleotide level, which was similar to the nucleotide conservation among different strains of P. aeruginosa (35). In comparison, P. syringae pv. tomato strain DC3000 showed higher variability in the argA region, with a 13.5% nucleotide substitution rate, than strains B301D and B728a of P. syringae pv. syringae. Another interesting observation about the argA region was that gene rearrangements occurred in the genome of P. putida KT2440. Consequently, the 8-kb region was split into two parts that flanked both sides of the putative replication origin (i.e., dnaA) of the strain KT2440 chromosome.
The ability to synthesize arginine is critical for pathogenicity of P. syringae pv. syringae B301D. Xu and Gross (41) demonstrated that mutant strain W4S2545 of P. syringae pv. syringae developed significantly smaller populations, peaking at approximately 104 CFU per fruit 3 days after inoculation (108 CFU per fruit for wild-type strain B301D) and failed to cause disease on sweet cherry fruits even though the cells of this strain still possessed the ability to cause a hypersensitive reaction on tobacco leaves. In contrast, BR132 (a syrB1 mutant), defective in syringomycin production, grew in planta to levels comparable to those of the wild-type strain yet was reduced in virulence by approximately 40% (43). Results of this study reveal that strain W4S2545 was an arginine auxotroph of P. syringae pv. syringae. Considering all of the above observations together, the loss of pathogenicity of strain W4S2545 results primarily from an inability for growth in planta due to arginine deficiency. In fact, it has long been known that auxotrophic mutants of pathogenic bacteria are oftentimes reduced in virulence or lose plant pathogenicity altogether. In the early 1960s, for example, Garber (as reported in reference 22) observed that mutants of P. syringae pv. tabaci that were unable to synthesize certain amino acids lost virulence. Similarly, effects of virulence and pathogenicity were observed for certain auxotrophic mutants of P. syringae pv. syringae strains, namely, PS9020 (2) and B728a (3) and P. syringae pv. phaseolicola strains PP7010 (2) and S2 (34). However, the effects of various auxotrophic mutants requiring different amino acids on pathogenesis varied greatly. They ranged from being indistinguishable from those of the wild-type strain to a complete loss of pathogenicity. Results of this study show that the syrA (argA) mutant W4S2545 was nonpathogenic due to arginine auxotrophy.
Acknowledgments
We are grateful to Brenda K. Scholz-Schroeder for helpful discussions and critical review of the manuscript.
This work was supported by grant 2001-35319-10400 from the National Research Competitive Grants Program of the Science and Education Administration, U.S. Department of Agriculture.
REFERENCES
- 1.Alexeyev, M. F. 1995. Three kanamycin resistance gene cassettes with different polylinkers. BioTechniques 18:52-56. [PubMed] [Google Scholar]
- 2.Anderson, D. M., and D. Mills. 1985. The use of transposon mutagenesis in the isolation of nutritional and virulence mutants in two pathovars of Pseudomonas syringae. Phytopathology 75:104-108. [Google Scholar]
- 3.Anderson, G. L., G. A. Beattie, and S. E. Lindow. 1998. Molecular characterization and sequence of a methionine biosynthetic locus from Pseudomonas syringae. J. Bacteriol. 180:4497-4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baetens, M., C. Legrain, A. Boyen, and N. Glansdorff. 1998. Genes and enzymes of the acetyl cycle of arginine biosynthesis in the extreme thermophilic bacterium Thermus thermophilus HB27. Microbiology 144:479-492. [DOI] [PubMed] [Google Scholar]
- 5.Ballio, A., D. Barra, F. Bossa, J. E. DeVay, I. Grgurina, N. S. Iacobellis, S. Marino, P. Pucci, M. Simmaco, and G. Surico. 1988. Multiple forms of syringomycin. Physiol. Mol. Plant Pathol. 33:493-496. [Google Scholar]
- 6.Bender, C. L., F. Alarcón-Chaidez, and D. C. Gross. 1999. Pseudomonas syringae phytotoxins: mode of action, regulation, and biosynthesis by peptide and polyketide synthetases. Microbiol. Mol. Biol. Rev. 63:266-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boyen, A., D. Charlier, J. Charlier, V. Sakanyan, I. Mett, and N. Glansdorff. 1992. Acetylornithine deacetylase, succinyldiaminopimelate desuccinylase and carboxypeptidase G2 are evolutionarily related. Gene 116:1-6. [DOI] [PubMed] [Google Scholar]
- 8.Cody, Y. S., D. C. Gross, E. L. Proebsting, Jr., and R. A. Spotts. 1987. Suppression of ice nucleation-active Pseudomonas syringe by antagonistic bacteria in fruit tree orchards and evaluations of frost control. Phytopathology 77:1036-1044. [Google Scholar]
- 9.Cunin, R., N. Glansdorff, A. Piérard, and V. Stalon. 1986. Biosynthesis and metabolism of arginine in bacteria. Microbiol. Rev. 50:314-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Del Sal, G., G. Manfioletti, and C. Schneider. 1989. The CTAB-DNA precipitation method: a common mini-scale preparation of template DNA from phagemids, phages or plasmids suitable for sequencing. BioTechniques 7:514-520. [PubMed] [Google Scholar]
- 11.Deng, W., V. Burland, G. Plunkett III, A. Boutin, G. F. Mayhew, P. Liss, N. T. Perna, D. J. Rose, B. Mau, S. Zhou, D. C. Schwartz, J. D. Fetherston, L. E. Lindler, R. R. Brubaker, G. V. Plano, S. C. Straley, K. A. McDonough, M. L. Nilles, J. S. Matson, F. R. Blattner, and R. D. Perry. 2002. Genome sequence of Yersinia pestis KIM. J. Bacteriol. 184:4601-4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Devereux, J., P. Haeberli, and O. Smithies. 1984. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 12:387-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Emmett, M., and W. E. Kloos. 1979. The nature of arginine auxotrophy in cutaneous populations of staphylococci. J. Gen. Microbiol. 110:305-314. [DOI] [PubMed] [Google Scholar]
- 14.Grant, S. G., J. Jessee, F. R. Bloom, and D. Hanahan. 1990. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc. Natl. Acad. Sci. USA 87:4645-4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grgurina, I., D. C. Gross, N. S. Iacobellis, P. Lavermicocca, J. Y. Takemoto, and M. Benincasa. 1996. Phytotoxin production by Pseudomonas syringae pv. syringae: syringopeptin production by syr mutants defective in biosynthesis or secretion of syringomycin. FEMS Microbiol. Lett. 138:35-39. [Google Scholar]
- 16.Gross, D. C., and J. E. DeVay. 1977. Population dynamics and pathogenesis of Pseudomonas syringae in maize and cowpea in relation to the in vitro production of syringomycin. Phytopathology 67:475-483. [Google Scholar]
- 17.Gross, D. C. 1991. Molecular and genetic analysis of toxin production by pathovars of Pseudomonas syringae. Annu. Rev. Phytopathol. 29:247-278. [Google Scholar]
- 18.Guenzi, E., G. Galli, I. Grgurina, D. C. Gross, and G. Grandi. 1998. Characterization of the syringomycin synthetase gene cluster—a link between prokaryotic and eukaryotic peptide synthetases. J. Biol. Chem. 273:32857-32863. [DOI] [PubMed] [Google Scholar]
- 19.Haas, D., B. W. Holloway, A. Schambock, and T. Leisinger. 1977. The genetic organization of arginine biosynthesis in Pseudomonas aeruginosa. Mol. Gen. Genet. 154:7-22. [DOI] [PubMed] [Google Scholar]
- 20.Hindle, Z., R. Callis, S. Dowden, B. A. Rudd, and S. Baumberg. 1994. Cloning and expression in Escherichia coli of a Streptomyces coelicolor A3(2) argCJB gene cluster. Microbiology 140:311-320. [DOI] [PubMed] [Google Scholar]
- 21.Javid-Majd, F., and J. S. Blanchard. 2000. Mechanistic analysis of the argE-encoded N-acetylornithine deacetylase. Biochemistry 39:1285-1293. [DOI] [PubMed] [Google Scholar]
- 22.Klement, Z., and R. N. Goodman. 1967. The hypersensitive reaction to infection by bacterial plant pathogens. Annu. Rev. Phytopathol. 5:17-44. [Google Scholar]
- 23.Lu, S. E., B. K. Scholz-Schroeder, and D. C. Gross. 2002. Characterization of the salA, syrF, and syrG regulatory genes located at the right border of the syringomycin gene cluster of Pseudomonas syringae pv. syringae. Mol. Plant-Microbe Interact. 15:43-53. [DOI] [PubMed] [Google Scholar]
- 24.Martin, P. R., and M. H. Mulks. 1992. Sequence analysis and complementation studies of the argJ gene encoding ornithine acetyltransferase from Neisseria gonorrhoeae. J. Bacteriol. 174:2694-2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nelson, K. E., C. Weinel, I. T. Paulsen, R. J. Dodson, H. Hilbert, V. A. Martins dos Santos, D. E. Fouts, S. R. Gill, M. Pop, M. Holmes, L. Brinkac, M. Beanan, R. T. DeBoy, S. Daugherty, J. Kolonay, R. Madupu, W. Nelson, O. White, J. Peterson, H. Khouri, I. Hance, L. P. Chris, E. Holtzapple, D. Scanlan, K. Tran, A. Moazzez, T. Utterback, M. Rizzo, K. Lee, D. Kosack, D. Moestl, H. Wedler, J. Lauber, D. Stjepandic, J. Hoheisel, M. Straetz, S. Heim, C. Kiewitz, J. Eisen, K. N. Timmis, A. Dusterhoft, B. Tummler, and C. M. Fraser. 2002. Complete genome sequence and comparative analysis of the metabolically versatile Pseudomonas putida KT2440. Environ. Microbiol. 4:799-808. [DOI] [PubMed] [Google Scholar]
- 26.O'Reilly, M., and K. M. Devine. 1994. Sequence and analysis of the citrulline biosynthetic operon argC-F from Bacillus subtilis. Microbiology 140:1023-1025. [DOI] [PubMed] [Google Scholar]
- 27.Prentki, P., F. Karch, S. Iida, and J. Meyer. 1981. The plasmid cloning vector pBR325 contains a 482 base-pair-long inverted duplication. Gene 14:289-299. [DOI] [PubMed] [Google Scholar]
- 28.Quigley, N. B., Y. Y. Mo, and D. C. Gross. 1993. SyrD is required for syringomycin production by Pseudomonas syringae pathovar syringae and is related to a family of ATP-binding secretion proteins. Mol. Microbiol. 9:787-801. [DOI] [PubMed] [Google Scholar]
- 29.Rajagopal, B. S., J. DePonte III, M. Tuchman, and M. H. Malamy. 1998. Use of inducible feedback-resistant N-acetylglutamate synthetase (argA) genes for enhanced arginine biosynthesis by genetically engineered Escherichia coli K-12 strains. Appl. Environ. Microbiol. 64:1805-1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sakanyan, V., D. Charlier, C. Legrain, A. Kochikyan, I. Mett, A. Pierard, and N. Glansdorff. 1993. Primary structure, partial purification and regulation of key enzymes of the acetyl cycle of arginine biosynthesis in Bacillus stearothermophilus: dual function of ornithine acetyltransferase. J. Gen. Microbiol. 139:393-402. [DOI] [PubMed] [Google Scholar]
- 31.Sambrook, J., E. F. Frisch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 32.Scholz-Schroeder, B. K., M. L. Hutchison, I. Grgurina, and D. C. Gross. 2001. The contribution of syringopeptin and syringomycin to virulence of Pseudomonas syringae pv. syringae strain B301D on the basis of sypA and syrB1 biosynthesis mutant analysis. Mol. Plant-Microbe Interact. 14:336-348. [DOI] [PubMed] [Google Scholar]
- 33.Scholz-Schroeder, B. K., J. D. Soule, S. E. Lu, I. Grgurina, and D. C. Gross. 2001. A physical map of the syringomycin and syringopeptin gene clusters localized to an approximately 145-kb DNA region of Pseudomonas syringae pv. syringae strain B301D. Mol. Plant-Microbe Interact. 14:1426-1435. [DOI] [PubMed] [Google Scholar]
- 34.Somlyai, G., M. Hevesi, Z. Banfalvi, Z. Klement, and A. Kondorosi. 1986. Isolation and characterization of non-pathogenic and reduced virulence mutants of Pseudomonas syringae pv. phaseolicola induced by Tn5 transposon insertions. Physiol. Mol. Plant Pathol. 29:369-380. [Google Scholar]
- 35.Spencer, D. H., A. Kas, E. E. Smith, C. K. Raymond, E. H. Sims, M. Hastings, J. L. Burns, R. Kaul, and M. V. Olson. 2003. Whole-genome sequence variation among multiple isolates of Pseudomonas aeruginosa. J. Bacteriol. 185:1316-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 37.Van de Casteele, M., M. Demarez, C. Legrain, N. Glansdorff, and A. Pierard. 1990. Pathways of arginine biosynthesis in the extreme thermophilic archaeo- and eubacteria. J. Gen. Microbiol. 136:1177-1183. [Google Scholar]
- 38.Vidaver, A. K. 1967. Synthetic and complex media for the rapid detection of fluorescence of phytopathogenic pseudomonads: effect of the carbon source. Appl. Microbiol. 15:1523-1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.West, S. E., H. P. Schweizer, C. Dall, A. K. Sample, and L. J. Runyen-Janecky. 1994. Construction of improved Escherichia-Pseudomonas shuttle vectors derived from pUC18/19 and sequence of the region required for their replication in Pseudomonas aeruginosa. Gene 148:81-86. [DOI] [PubMed] [Google Scholar]
- 40.Xu, G.-W., and D. C. Gross. 1988. Evaluation of the role of syringomycin in plant pathogenesis by using Tn5 mutants of Pseudomonas syringae pv. syringae defective in syringomycin production. Appl. Environ. Microbiol. 54:1345-1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu, G.-W., and D. C. Gross. 1988. Physical and functional analyses of the syrA and syrB genes involved in syringomycin production by Pseudomonas syringae pv. syringae. J. Bacteriol. 170:5680-5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 43.Zhang, J.-H., N. B. Quigley, and D. C. Gross. 1995. Analysis of the syrB and syrC genes of Pseudomonas syringae pv. syringae indicates that syringomycin is synthesized by a thiotemplate mechanism. J. Bacteriol. 177:4009-4020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang, J.-H., N. B. Quigley, and D. C. Gross. 1997. Analysis of the syrP gene, which regulates syringomycin synthesis by Pseudomonas syringae pv. syringae. Appl. Environ. Microbiol. 63:2771-2778. [DOI] [PMC free article] [PubMed] [Google Scholar]