Abstract
Because elevated ubiquitin ligase atrogin-1/MAFbx and MuRF1 mediate skeletal muscle wasting associated with various catabolic conditions, the signaling pathways involved in the upregulation of these genes under pathological conditions are considered therapeutic targets. AKT and NF-κB have been previously shown to regulate the expression of atrogin-1/MAFbx or MuRF1, respectively. In addition, we recently found that p38 MAPK mediates TNF-α upregulation of atrogin-1/MAFbx expression, suggesting that multiple signaling pathways mediate muscle wasting in inflammatory diseases. To date, however, these advances have not resulted in a practical clinical intervention for disease-induced muscle wasting. In the present study, we tested the effect of curcumin—a non-toxic anti-inflammatory reagent that inhibits p38 and NF-κB—on lipopolysaccharide (LPS)-induced muscle wasting in mice. Daily intraperitoneal (i.p.) injection of curcumin (10–60 μg/kg) for 4 days inhibited, in a dose-dependent manner, the LPS-stimulated (1 mg/kg, i.p.) increase of atrogin-1/MAFbx expression in gastrocnemius and extensor digitorum longus (EDL) muscles, resulting in the attenuation of muscle protein loss. It should also be noted that curcumin administration did not alter the basal expression of atrogin-1/MAFbx, nor did it affect LPS-stimulated MuRF1 and polyubiquitin expression. LPS activated p38 and NF-κB, while inhibiting AKT; whereas, curcumin administration inhibited LPS-stimulated p38 activation, without altering the effect of LPS on NF-κB and AKT. These results indicate that curcumin is effective in blocking LPS-induced loss of muscle mass through the inhibition of p38-mediated upregulation of atrogin-1/MAFbx.
Keywords: endotoxemia, ubiquitin–proteasome pathway, p38, AKT, NF-κB
Muscle wasting is a major feature of the cachexia associated with such diseases as sepsis, cancer, AIDS, diabetes, uremia, congestive heart failure, and chronic obstructive pulmonary disease (COPD) [Hasselgren, 1995; Tisdale, 1997]. Muscle wasting increases patient morbidity and mortality through disability, injury, osteoporosis, impairment of respiratory function, and depletion of the free amino acid pool for enzyme and antibody production. The progressive loss of body protein becomes lethal when it depletes approximately 40% of lean body mass [Winick, 1979]. However, to date, no drug has been approved for muscle wasting.
Muscle wasting is primarily caused by accelerated muscle protein breakdown via the ubiquitin–proteasome pathway [Jagoe and Goldberg, 2001]. Proteins degraded by this pathway are first covalently linked to a chain of ubiquitin molecules that marks them for rapid breakdown by the 26S proteasome. The selectivity of ubiquitin targeting is primarily a function of ubiquitin ligase (E3 protein) [Hershko and Ciechanover, 1998]. Two muscle-enriched ubiquitin ligases, atrogin-1/MAFbx and MuRF1, are rate limiting for muscle protein loss in various catabolic conditions. Expression of these two ubiquitin ligases is upregulated in the muscle of various animal models of muscle atrophy, including fasting, disuse, denervation, cancer, diabetes, uremia, and sepsis [Bodine et al., 2001; Gomes et al., 2001; Dehoux et al., 2003; Wray et al., 2003; Lecker et al., 2004], and in humans suffering from immobilization [Jones et al., 2004], acute quadriplegic myopathy and neurogenic atrophy [Di Giovanni et al., 2004]. Individual knockout of the atrogin-1/MAFbx or MuRF1 gene results in the attenuation of denervation-induced muscle atrophy [Bodine et al., 2001], suggesting that atrogin-1/MAFbx and MuRF1 are critical to muscle protein loss. Consequently, these genes are considered important targets for therapeutic intervention.
Intense research has been devoted to the elucidation of signaling mechanisms that control expression of these two ubiquitin ligases. AKT-regulated FOXO transcription factors have been shown to control atrogin-1/MAFbx and MuRF1 expression. Anabolic signals such as IGF-I activate AKT, resulting in the suppression of atrogin-1/MAFbx and MuRF1 expression. Conversely, catabolic signals such as glucocorticoids inhibit AKT activity, resulting in the upregulation of atrogin-1/MAFbx and MuRF1 expression [Sandri et al., 2004; Stitt et al., 2004; Latres et al., 2005]. In addition, MuRF1 expression is stimulated by NF-κB [Cai et al., 2004], an inflammatory stimulus-activated transcription factor that mediates muscle protein loss via multiple components of the ubiquitin proteasome pathway [Li and Reid, 2000; Li et al., 2003b]. We recently found that atrogin-1/MAFbx expression is upregulated by TNF-α via a p38 MAPK-dependent mechanism in C2C12 myotubes [Li et al., 2005]. Although the p38, ERK1/2, and JNK MAPKs are all activated by TNF-α, only the inhibition of p38 blocks atrogin-1/MAFbx upregulation [Li et al., 2005]. This suggests the existence of a specific signaling mechanism that upregulates atrogin-1/MAFbx expression via p38 in response to inflammatory stimuli. In fact, increased muscle p38 activity has been reported in a number of pro-catabolic states, including limb immobilization [Childs et al., 2003; O’keefe et al., 2004], type 2 diabetes [Koistinen et al., 2003], acute quadriplegic myopathy [Di Giovanni et al., 2004], neurogenic atrophy [Di Giovanni et al., 2004], and aging [Williamson et al., 2003]. Considering that p38 and NF-κB are activated by cytokines TNF-α and IL-1 [Zarubin and Han, 2005]—two recognized mediators of muscle catabolism [Jackman and Kandarian, 2004]—it is likely that these molecules mediate the excessive muscle protein breakdown often associated with inflammatory diseases. Hence, we hypothesized that inflammation-stimulated muscle wasting can be prevented by inhibiting p38 and NF-κB activation.
Sepsis is an inflammatory condition that causes a severe and rapid loss of body protein, much of which originates from skeletal muscle [Rosenblatt et al., 1983]. Many studies have provided evidence that muscle atrophy in sepsis is primarily the result of increased protein breakdown [Hasselgren et al., 1986, 1989; Ash and Griffin, 1989] via the ubiquitin–proteasome pathway [Tiao et al., 1994, 1997]. Indeed, both atrogin-1/MAFbx and MuRF1 are upregulated in the muscle of septic models induced by either cecal ligation and puncture [Wray et al., 2003] or lipopolysaccharide (LPS) administration [Dehoux et al., 2003]. Endotoxemia, which induces a number of catabolic factors in sepsis (e.g., TNF-α, IL-1, glucocorticoids, and glucocorticoid receptors), also suppresses anabolic factor IGF-I [Vary, 1998]. These catabolic factors contribute to the ensuing muscle catabolism by activating p38 and NF-κB (TNF-α and IL-1), while inhibiting AKT (glucocorticoid secretion and IGF-I suppression).
As a widely used muscle catabolism model with a major inflammation component, LPS-induced septic mice were chosen to test our hypothesis; curcumin, a non-toxic inhibitor of p38 and NF-κB, was employed for the intervention. Similar to the selective p38 inhibitor, SB203580, curcumin (diferuloylmethane)—the main biologically active phytochemical compound extracted from turmeric—has a p38-inhibiting property [Carter et al., 2003], and also obstructs p38-mediated TNF-α upregulation of atrogin-1/MAFbx in C2C12 myotubes [Li et al., 2005]. Moreover, curcumin is reportedly capable of inhibiting NF-κB [Jobin et al., 1999], and has no known toxicity to humans, even at very high doses [Chainani-Wu, 2003]. These attributes indicated curcumin would be suitable for testing our hypothesis in mice—and, if successful, for future use in humans. In the current study, we have demonstrated that curcumin obstructs the action of p38 induced by LPS, effectively inhibits LPS-induced atrogin-1/MAFbx upregulation, and averts the loss of muscle mass.
MATERIALS AND METHODS
Materials
Curcumin was purchased from LKT Laboratories, Inc. (St. Paul, MN). LPS (derived from Pseudomonas aeruginosa) and the antibody against β-actin were obtained from Sigma-Aldrich Corporation (St. Louis, MO). Antibodies for p38, phospho-p38, AKT, and phospho-AKT were obtained from Cell Signaling Technology, Inc. (Danvers, MA).
Animals
Experimental protocols were approved in advance by the Institutional Animal Care and Use Committee (IACUC) at Baylor College of Medicine. Adult male ICR mice (~30 g) purchased from Harlan (Indianapolis, IN) were preconditioned by intraperitoneal (i.p.) injection of 100 μl curcumin (10–60 μg/kg) or an equal volume of vehicle (PBS containing 0.1% DMSO) for 4 consecutive days. After the fourth injection of curcumin, one injection (i.p.) of LPS (1 mg/kg in 50 μl) or an equal volume of vehicle (PBS) was administered. At the indicated times, gastrocnemius and extensor digitorum longus (EDL) muscles were collected from the mice immediately after rapid euthanization. In the experiment testing prolonged exposure to LPS, curcumin and LPS were concomitantly administered (daily) over the course of 3 days.
Northern Blot Analysis
Northern blot analysis was conducted, as previously described [Li et al., 2003a]. Levels of mRNA were quantified by determining the optical density of bands through the use of densitometry software (ImageQuant, GE Healthcare).
Western Blot Analysis
Muscle proteins were extracted by homogenization using a glass homogenizer, followed by three cycles of freezing-thawing in a buffer solution containing 20 mM HEPES-KOH (pH 7.9), 25% glycerol, 420 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 1 mM dithiothreitol, 0.5 mM PMSF, 1 μg/ml leupeptin, 2 μg/ml aprotinin, 2 mM Na3VO4, 10 mM Na4P2O7, and 10 mM NaF. Protein concentrations were determined with the Bio-Rad protein assay kit (Bio-Rad Laboratories, Hercules, CA). Western blotting was conducted, as previously described [Li et al., 2005].
Electrophoresis Mobility Shift Assay (EMSA)
Gastrocnemius muscle was homogenized in a glass homogenizer; nuclear proteins were extracted and EMSA was performed, as previously described [Li et al., 2003a]. The DNA fragment used for probing NF-κB binding was 5′-AGTTGAGGGGACTTTCCCAGGC-3′.
Statistical Analysis
The data were analyzed with SigmaStat software (Chicago, IL), using a one-way analysis of variance (ANOVA) combined with Tukey’s multiple comparison test, or the Student’s t-test, when appropriate. Differences between groups were considered significant at P < 0.05; values are reported as means ± SE.
RESULTS
Curcumin Inhibits LPS Stimulation of Atrogin-1/MAFbx Gene Expression in a Dose-Dependent Manner
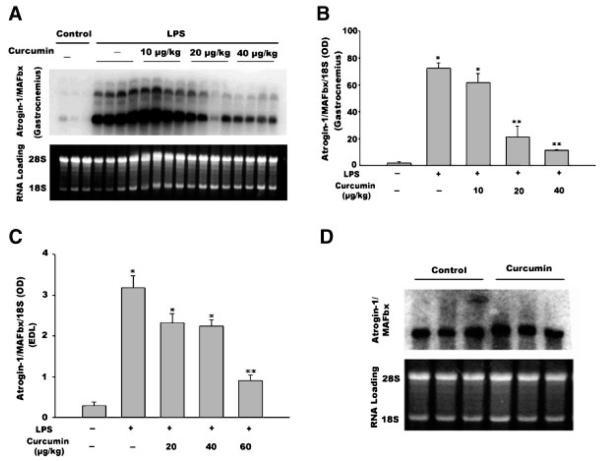
Northern blot analysis was performed to evaluate atrogin-1/MAFbx mRNA levels. Multiple atrogin-1/MAFbx mRNA transcripts ranging from ~6.5 to ~ 2.4 kb were detected, which is consistent with data from previous studies [Gomes et al., 2001; Li et al., 2005]. Similar to a prior report of data in rats [Dehoux et al., 2003], at 12 h, LPS administration (1 mg/kg, i.p.) induced a 36-fold increase in atrogin-1/MAFbx mRNA levels in the gastrocnemius muscle of adult mice (Fig. 1A and By). The administration of 10, 20, or 40 μg/kg of curcumin daily for 4 consecutive days prior to LPS injection blunted LPS-stimulation of atrogin1-/MAFbx mRNA expression to 30-, 11-, and 6-fold, respectively. To evaluate the effect of curcumin on atrogin-1/MAFbx expression during longer term LPS exposure, mice were injected daily with LPS and curcumin for an additional 3-day period. A similar dose-dependent inhibition of LPS-induced atrogin-1/MAFbx upregulation by curcumin was observed in gastrocnemius upon termination of the 3-day exposure to LPS (data not shown). Since the inhibitory effect of 40 μg/kg curcumin appeared still well within the linear range, we increased the dose range to 60 μg/kg and observed a dose-dependent inhibition of LPS-stimulated atrogin-1/MAFbx upregulation in the EDL muscle—although the inhibitory effect appeared less potent than in gastrocnemius (Fig. 1C). Due to the fact that atrogin-1/MAFbx is constitutively expressed in muscle, and may have a role in normal protein turnover [Bodine et al., 2001; Gomes et al., 2001], we examined the effect of curcumin administration (60 μg/kg i.p. daily for 4 consecutive days) on the basal expression of the atrogin-1/MAFbx gene. As shown in Figure 1D, curcumin did not alter the basal levels of atrogin-1/MAFbx mRNA in gastrocnemius muscle (P > 0.05).
Fig. 1.
Curcumin inhibits LPS upregulation of atrogin-1/MAFbx gene expression in a dose-dependent manner. Mice receiving curcumin or vehicle for 4 consecutive days were i.p. injected with LPS (1 mg/kg). Gastrocnemius and EDL were collected 12 h after LPS injection. Northern blot analysis was conducted to determine the atrogin-1/MAFbx mRNA isolated from gastrocnemius (panel A). Four mice were used in each group, excluding the control group, which contained three mice. The autoradiograph shown in panel A was quantified via densitometry analysis and normalized to 18S (panel B). Densitometry data (OD) from the Northern analysis of atrogin-1/MAFbx mRNA isolated from EDL are shown in panel C. Three mice were used in each group; however, the control group contained two mice. Panel D shows densitometry data from Northern blot analysis of the effect of curcumin on the basal levels of atrogin-1/MAFbx mRNA in gastrocnemius. Three mice were used in each group. Data were analyzed by ANOVA and *(P < 0.05) or **(P < 0.01) indicates difference from the control group determined by Tukey’s multiple comparison test.
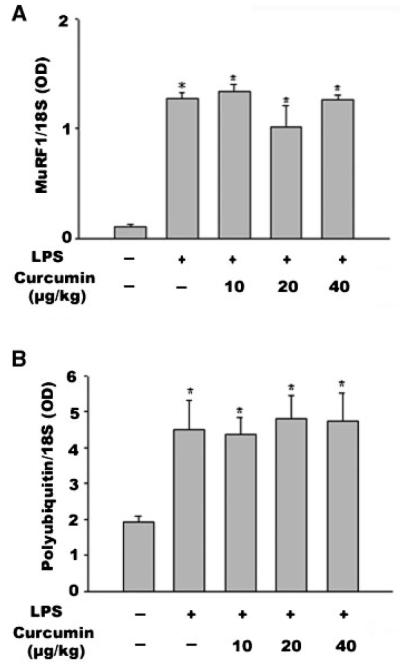
Since LPS has been shown to upregulate MuRF1 and polyubiquitin [Dehoux et al., 2003], we employed Northern blot analysis to evaluate whether curcumin also affects LPS upregulation of these two genes. We observed the upregulation of MuRF1 gene expression by LPS at 12 h; however, curcumin did not inhibit this action (Fig. 2A). Moreover, LPS stimulation of polyubiquitin expression was not inhibited by curcumin (Fig. 2B). These results demonstrate that curcumin selectively inhibits LPS stimulation of atrogin-1/MAFbx expression.
Fig. 2.
Curcumin does not affect LPS upregulation of the MuRF1 and polyubiquitin genes. Mice receiving curcumin or vehicle for 4 consecutive days were i.p. injected with LPS (1 mg/kg). Gastrocnemius was collected 12 h after LPS injection. Northern blot analysis was performed for MuRF1 mRNA (panel A) or polyubiquitin mRNA (panel B). Data were analyzed by ANOVA (three mice per group), and * indicates difference (P < 0.05) from the control group determined by Tukey’s multiple comparison test.
Curcumin Inhibits LPS-Induced Muscle Protein Loss
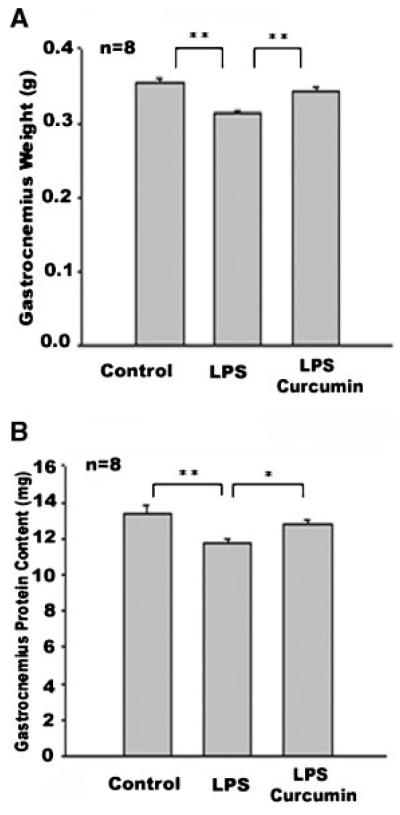
LPS is known to rapidly induce muscle protein loss within 24 h [Premer et al., 2002]. This is consistent with LPS upregulation of atrogin-1/MAFbx and MuRF1 expression within 12 h, as shown above, and by others [Dehoux et al., 2003]. To determine whether curcumin inhibition of atrogin-1/MAFbx upregulation by LPS results in a blockade of muscle protein loss, mice with essentially the same average body weight were divided into three groups (control, LPS, and curcumin plus LPS); subsequently, 18 h after LPS injection, the effect of curcumin on gastrocnemius mass and protein content was measured. The weight of gastrocnemius from mice receiving LPS was 11.7% less than that of the control mice (Fig. 3A); however, in mice receiving curcumin, LPS-induced muscle mass loss was largely prevented (Fig. 3A). Total protein content quantified from gastrocnemius confirmed a 12.3% loss of muscle protein stimulated by LPS—and this loss was blocked by curcumin (Fig. 3B). These measurements indicate that curcumin is effective in preventing LPS-induced muscle protein loss. It should also be noted that we observed no effect of curcumin administration on body and gastrocnemius weight in the control mice; thus, it appears curcumin does not affect muscle mass in normal mice.
Fig. 3.
Curcumin inhibits LPS-induced muscle mass/protein loss in gastrocnemius. Following curcumin (60 μg/kg, i.p., daily) or vehicle administration for 4 consecutive days, mice were i.p. injected with LPS (1 mg/kg). Gastrocnemius was collected 18 h after LPS injection. A: Muscle mass was assessed by weighing gastrocnemius from both legs. B: Protein content in the gastrocnemius was measured. Data were analyzed by ANOVA and *(P < 0.05) or **(P < 0.01) indicates difference from the control group determined by Tukey’s multiple comparison test.
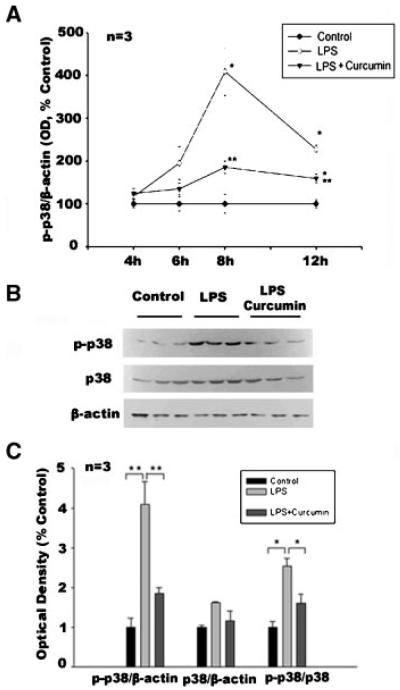
Curcumin Blocks LPS-Induced Activation of p38 MAPK in Muscle
Although we previously demonstrated that TNF-α upregulates atrogin-1/MAFbx via p38 in C2C12 myotubes [Li et al., 2005], it was unknown whether LPS activates p38 in skeletal muscle. To determine this, adult mice were injected with LPS (1 mg/kg, i.p.), and levels of phosphorylated p38 in gastrocnemius extracts were analyzed by Western blotting at 4, 6, 8, and 12 h (post-injection). As shown in Figure 4A, we observed increased phosphorylated p38 in gastrocnemius, which peaked at 8 h (4.1-fold) and continued to be elevated for at least 12 h (2.3-fold). The time course of LPS-stimulated p38 activity was consistent with LPS upregulation of atrogin-1/MAFbx expression at 12 h. Four-day curcumin administration (60 μg/kg, i.p. daily) inhibited LPS stimulation of p38 activity. There was also an increase in total p38 levels induced by LPS, which peaked at 6 h, but this was unaffected by curcumin (data not shown). As indicated in Figure 4C, at the peak of p38 activation (8 h), p38 expression (p38/β-actin) was not increased significantly by LPS, while total p38 activity (p-p38/β-actin) and the ratio of active p38 to total p38 (p-p38/p38) were both increased significantly by LPS; curcumin, however, inhibited this LPS action. Therefore, curcumin serves to impede the stimulation of p38 by LPS, primarily through inhibiting the activation of p38; on the other hand, curcumin had no effect on the basal activity of p38 in control mice (data not shown). In sum, these findings demonstrate, for the first time, that: (1) LPS activates p38 in skeletal muscle and (2) curcumin effectively inhibits this action.
Fig. 4.
LPS activates p38 in gastrocnemius muscle and curcumin inhibits this action. Following curcumin (60 μg/kg, i.p., daily) or vehicle administration for 4 consecutive days, mice were i.p. injected with LPS (1 mg/kg). At the indicated times, gastrocnemius was collected for analysis. Panel A shows optical density data (OD) (derived from Western blot analysis) on the activation (phosphorylated form) of p38 (p-p38); for loading control p-p38 level was normalized to β-actin. Data were analyzed by ANOVA, combined with Tukey’s multiple comparison test. A statistically significant difference (P < 0.05) from the control is indicated by *, while a difference from the LPS-treated group is indicated by **. Panel B shows Western blot analysis of p38, phosphorylated p38 and β-actin from samples collected at 8h. Panel C shows optical density data derived from panel B. Data were analyzed by ANOVA and *(P < 0.05) or **(P < 0.01) indicates difference from the control group determined by Tukey’s multiple comparison test.
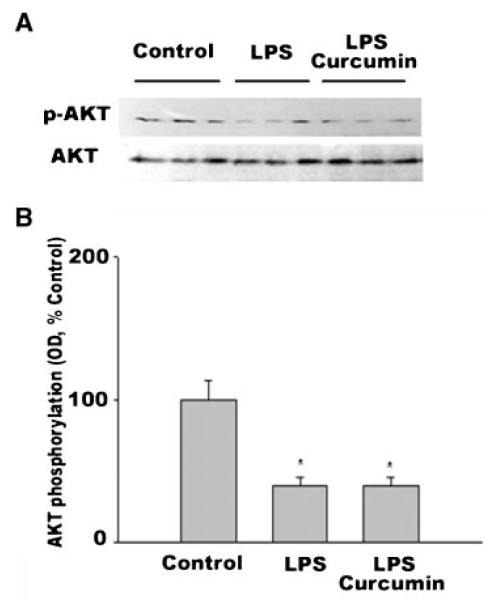
LPS is known to increase glucocorticoid activity and decrease IGF-I expression [Vary, 1998]. These events lead to the upregulation of atrogin-1/MAFbx and MuRF1 via downregulation of AKT activity [Stitt et al., 2004; Latres et al., 2005]. Therefore, it was necessary to determine whether curcumin inhibits LPS stimulation of atrogin-1/MAFbx expression by altering the effect of LPS on AKT activity. Although we observed that LPS downregulated AKT activity in gastrocnemius—as indicated by a 61% decrease in phosphorylated AKT (P < 0.05) at 8 h post-LPS administration (Fig. 5)—curcumin treatment did not alter the LPS effect. These data show that curcumin does not inhibit LPS-stimulated atrogin-1/MAFbx expression through mediating AKT activity.
Fig. 5.
Curcumin does not alter LPS downregulation of AKT activity in gastrocnemius. Mice were injected with LPS following curcumin or vehicle administration, as described in Figure 3. Gastrocnemius was collected 8 h after LPS injection. A: Gastrocnemius protein extracts were analyzed with Western blotting using an antibody against AKT or phosphorylated AKT (p-AKT). Three mice were used in each group. B: Optical density data (OD) of p-AKT normalized to AKT were analyzed by ANOVA combined with Tukey’s multiple comparison test. A statistically significant difference (P < 0.05) from the control is indicated by *.
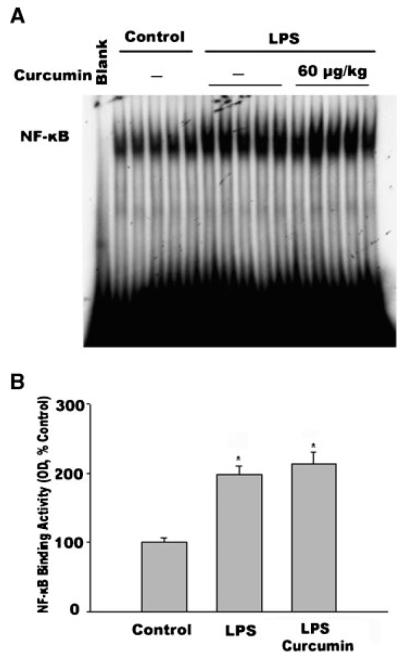
Curcumin is reported to inhibit NF-κB [Jobin et al., 1999], a transcription factor that has been shown to upregulate MuRF1 gene expression [Cai et al., 2004]. Utilizing EMSA, we examined the effect of curcumin on the LPS-induced activation of NF-κB; our results showed an NF-κB binding pattern similar to that found during our previous study, in which supershift analysis identified p50 and p65 bands as components of the NF-κB complex [Durham et al., 2004]. In the current study, LPS administration resulted in increased NF-κB binding activity in the nuclear protein extracts prepared from gastrocnemius, after 4 h (Fig. 6)—the time point at which NF-κB activation was previously shown to peak in septic muscle [Penner et al., 2001]. Curcumin, however, did not alter the action of LPS on NF-κB (Fig. 6). This result is consistent with our observation above that curcumin does not affect LPS-stimulated MuRF1 expression.
Fig. 6.
Curcumin does not inhibit LPS stimulation of NF-κB binding activity in gastrocnemius. Mice were injected with LPS following curcumin administration, as described in Figure 1. Gastrocnemius was collected 4 h after LPS injection. A: NF-κB DNA-binding activity in nuclear extracts prepared from gastrocnemius was determined by EMSA. Five mice were used in each group. B: Optical density data (OD) derived from panel A were analyzed using ANOVA combined with Tukey’s multiple comparison test; * indicates a statistically significant difference (P < 0.05) from the control.
DISCUSSION
Despite recent progress in our understanding of the mechanism of muscle protein degradation mediated by the ubiquitin–proteasome pathway, no effective intervention has been established to reverse catabolic disease-induced muscle wasting. It is currently believed that specific ubiquitin ligases such as atrogin-1/MAFbx and MuRF1 are critical to the elevated activity of the ubiquitin–proteasome pathway in various catabolic diseases. Yet, it is equally probable that these constitutively expressed enzymes play a role in the normal turnover of cellular proteins. Thus, direct inhibition of the ubiquitin ligases is likely to have unwanted side effects, independent of any anti-muscle wasting effect. Consequently, a more desirable therapeutic strategy is to target the signaling mechanisms that mediate the upregulation of relevant ubiquitin ligases induced under pathological conditions. The present study in mice demonstrates the feasibility of this strategy and the usefulness of curcuminasapotentialremedy for sepsis-induced muscle wasting in humans. We have shown that: (1) curcumin selectively blocks the elevation of LPS-stimulated atrogin-1/MAFbx expression, in a dose-dependent manner; (2) curcumin does not affect the basal expression of atrogin-1/MAFbx; (3) curcumin effectively inhibits LPS-stimulated muscle protein loss; and (4) the inhibition of p38 activity is implicated in the action of curcumin.
A remarkable feature of curcumin is that the dose which effectively blocks LPS-stimulated atrogin-1/MAFbx upregulation and muscle protein loss is very low (60 μg/kg), indicating a highly efficient interaction between curcumin and the relevant target(s). In addition, curcumin is non-toxic [Chainani-Wu, 2003] and readily available as a diet supplement; these features make it an attractive drug candidate in the treatment of muscle wasting associated with sepsis—and possibly, other inflammatory conditions.
Our analysis of the potential mechanism of curcumin to inhibit LPS-stimulated atrogin-1/MAFbx expression also yielded interesting information. Although higher p38 activity in muscle has been observed in a number of procatabolic states, including acute quadriplegic myopathy, neurogenic atrophy, Type 2 diabetes, limb immobilization, and aging [Childs et al., 2003; Koistinen et al., 2003; Williamson et al., 2003; Di Giovanni et al., 2004; O’keefe et al., 2004], the present study reveals, for the first time, that LPS—a potent catabolic stimulus—activates p38 in skeletal muscle while stimulating atrogin-1/MAFbx expression. Moreover, LPS-stimulated atrogin-1/MAFbx expression is blocked when p38 activation is inhibited by curcumin. We recently observed a positive correlation between p38 activity and atrogin-1/MAFbx expression in C2C12 myotubes [Li et al., 2005]; this finding suggested a role for p38 in mediating the intensified muscle protein breakdown associated with inflammatory stimuli. The current study provides in vivo data that show a similar relationship between atrogin-1/MAFbx expression levels and p38 activity.
It is noteworthy that p38 mediates diverse cellular events, some of which appear to be in functional opposition. For example, p38 mediates both insulin stimulation of glucose uptake [Tremblay et al., 2003] and TNF-α stimulation of insulin resistance in muscle [de Alvaro et al., 2004]. Additionally, p38 stimulates muscle satellite cell proliferation, as well as differentiation [Jones et al., 2005]. Also, in muscle protein homeostasis, p38 appears to respond to both catabolic (LPS, TNF-α, and IL-1, etc.) and anabolic (insulin and exercise) stimuli [Long et al., 2004]. These discrepancies may result from the presence of p38 subtypes [Raingeaud et al., 1995; Wang et al., 1997; Enslen et al., 1998; Yang et al., 1999] and the subcellular compartmentalization of p38 [Yu et al., 1998; Chevalier et al., 2000], through which a variety of extracellular signals can be transduced into distinct nuclear responses. Interestingly, it has been shown that p38 phosphorylates different protein substrates, depending on the cellular environment [Jones et al., 2005].
While we observed that LPS downregulates AKT activity, which is known to promote atrogin-1/MAFbx expression [Sandri et al., 2004; Stitt et al., 2004], curcumin does not alter the effect of LPS on AKT. Notwithstanding, although curcumin blocks LPS stimulation of atrogin-1/MAFbx expression by inhibiting p38, our data do not distinguish whether, in mediating atrogin-1/MAFbx upregulation, p38 acts in a pathway that is independent of AKT or downstream of AKT.
Another unresolved issue is that we do not yet know why curcumin does not exert an inhibitory effect on NF-κB activation, as previously reported. It does not appear that this is due to poor bioavailability, since curcumin effectively blocks p38 activation. In a prior study, we observed that curcumin supplementation in the diet inhibited NF-κB activity in the peripheral tissues of ambulatory mice, but not in unloaded soleus muscle [Farid et al., 2005]. As LPS activates NF-κB through both classical and alternative pathways [Mordmuller et al., 2003], it is possible that curcumin only inhibits the classical [Jobin et al., 1999], but not the alternative, pathway of NF-κB activation. Due to the ineffectiveness of curcumin in inhibiting NF-κB activation, it is not surprising that curcumin does not prevent LPS-induced upregulation of MuRF1 expression. Considering that curcumin does not affect LPS stimulation of MuRF1 expression, atrogin-1/MAFbx upregulation appears to be responsible for a significant portion of muscle protein loss caused by LPS.
Independent of its p38 inhibitory effect, curcumin has multiple pharmacological effects [Carter et al., 2003], including an antioxidant effect [Araujo and Leon, 2001]. Reactive oxygen species (ROS) is implicated in muscle wasting, and LPS is known to induce oxidative stress [Reid and Li, 2001]. However, our finding that activation of the redox-sensitive transcription factor NF-κB [Li et al., 1999] by LPS in muscle is not affected by curcumin suggests that curcumin does not inhibit atrogin-1/MAFbx upregulation by functioning as a general antioxidant. In addition, we previously observed that the general antioxidant N-acetylcysteine did not inhibit the effect of curcumin on p38-dependent atrogin-1/MAFbx upregulation by TNF-α in C2C12 myotubes (unpublished data). Cumulatively, these observations suggest that the general antioxidant effect of curcumin is not responsible for its inhibition of atrogin-1/MAFbx upregulation.
Although we concede that the inhibitory action of curcumin on atrogin-1/MAFbx upregulation may involve currently unidentified mechanisms unrelated to its inhibition of p38, our current findings indicate that the pathway for this effect is the curcumin-mediated blockade of atrogin-1/MAFbx gene induction, via inhibition of p38. Nevertheless, the remarkable effectiveness of curcumin in blocking LPS-induced muscle protein loss suggests its potential usefulness in the clinical intervention of LPS-induced muscle wasting.
ACKNOWLEDGMENTS
This work has been supported by the Curtis Hankamer Basic Research Fund and National Institutes of Health Grant AR049022 to Y-P Li. We thank Eric D. Rabinovsky, Baylor College of Medicine, for his insightful comments and J.E. Young, for editing this manuscript.
Grant sponsor: Curtis Hankamer Basic Research Fund; Grant sponsor: National Institutes of Health; Grant number: AR049022.
REFERENCES
- Araujo CC, Leon LL. Biological activities of Curcuma longa L. Mem Inst Oswaldo Cruz. 2001;96:723–728. doi: 10.1590/s0074-02762001000500026. [DOI] [PubMed] [Google Scholar]
- Ash SA, Griffin GE. Effect of parenteral nutrition on protein turnover in endotoxaemic rats. Clin Sci (Lond) 1989;76:659–666. doi: 10.1042/cs0760659. [DOI] [PubMed] [Google Scholar]
- Bodine SC, Latres E, Baumhueter S, Lai VK, Nunez L, Clarke BA, Poueymirou WT, Panaro FJ, Na E, Dharmarajan K, Pan ZQ, Valenzuela DM, DeChiara TM, Stitt TN, Yancopoulos GD, Glass DJ. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science. 2001;294:1704–1708. doi: 10.1126/science.1065874. [DOI] [PubMed] [Google Scholar]
- Cai D, Frantz JD, Tawa NE, Jr., Melendez PA, Oh BC, Lidov HG, Hasselgren PO, Frontera WR, Lee J, Glass DJ, Shoelson SE. IKKbeta/NF-kappaB activation causes severe muscle wasting in mice. Cell. 2004;119:285–298. doi: 10.1016/j.cell.2004.09.027. [DOI] [PubMed] [Google Scholar]
- Carter Y, Liu G, Yang J, Fier A, Mendez C. Sublethal hemorrhage induces tolerance in animals exposed to cecal ligation and puncture by altering p38, p44/42, and SAPK/JNK MAP kinase activation. Surg Infect (Larchmt) 2003;4:17–27. doi: 10.1089/109629603764655245. [DOI] [PubMed] [Google Scholar]
- Chainani-Wu N. Safety and anti-inflammatory activity of curcumin: A component of tumeric (Curcuma longa) J Altern Complement Med. 2003;9:161–168. doi: 10.1089/107555303321223035. [DOI] [PubMed] [Google Scholar]
- Chevalier D, Thorin E, Allen BG. Simultaneous measurement of ERK, p38, and JNK MAP kinase cascades in vascular smooth muscle cells. J Pharmacol Toxicol Methods. 2000;44:429–439. doi: 10.1016/s1056-8719(00)00118-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs TE, Spangenburg EE, Vyas DR, Booth FW. Temporal alterations in protein signaling cascades during recovery from muscle atrophy. Am J Physiol Cell Physiol. 2003;285:C391–C398. doi: 10.1152/ajpcell.00478.2002. [DOI] [PubMed] [Google Scholar]
- de Alvaro C, Teruel T, Hernandez R, Lorenzo M. Tumor necrosis factor alpha produces insulin resistance in skeletal muscle by activation of inhibitor kappaB kinase in a p38 MAPK-dependent manner. J Biol Chem. 2004;279:17070–17078. doi: 10.1074/jbc.M312021200. [DOI] [PubMed] [Google Scholar]
- Dehoux MJ, van Beneden RP, Fernandez-Celemin L, Lause PL, Thissen JP. Induction of MafBx and Murf ubiquitin ligase mRNAs in rat skeletal muscle after LPS injection. FEBS Lett. 2003;544:214–217. doi: 10.1016/s0014-5793(03)00505-2. [DOI] [PubMed] [Google Scholar]
- Di Giovanni S, Molon A, Broccolini A, Melcon G, Mirabella M, Hoffman EP, Servidei S. Constitutive activation of MAPK cascade in acute quadriplegic myopathy. Ann Neurol. 2004;55:195–206. doi: 10.1002/ana.10811. [DOI] [PubMed] [Google Scholar]
- Durham WJ, Li YP, Gerken E, Farid M, Arbogast S, Wolfe RR, Reid MB. Fatiguing exercise reduces DNA binding activity of NF-kappaB in skeletal muscle nuclei. J Appl Physiol. 2004;97:1740–1745. doi: 10.1152/japplphysiol.00088.2004. [DOI] [PubMed] [Google Scholar]
- Enslen H, Raingeaud J, Davis RJ. Selective activation of p38 mitogen-activated protein (MAP) kinase isoforms by the MAP kinase kinases MKK3 and MKK6. J Biol Chem. 1998;273:1741–1748. doi: 10.1074/jbc.273.3.1741. [DOI] [PubMed] [Google Scholar]
- Farid M, Reid MB, Li YP, Gerken E, Durham WJ. Effects of dietary curcumin or N-acetylcysteine on NF-kappaB activity and contractile performance in ambulatory and unloaded murine soleus. Nutr Metab (Lond) 2005;2:20. doi: 10.1186/1743-7075-2-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes MD, Lecker SH, Jagoe RT, Navon A, Goldberg AL. Atrogin-1, a muscle-specific F-box protein highly expressed during muscle atrophy. Proc Natl Acad Sci USA. 2001;98:14440–14445. doi: 10.1073/pnas.251541198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasselgren PO. Muscle protein metabolism during sepsis. Biochem Soc Trans. 1995;23:1019–1025. doi: 10.1042/bst0231019. [DOI] [PubMed] [Google Scholar]
- Hasselgren PO, Talamini M, James JH, Fischer JE. Protein metabolism in different types of skeletal muscle during early and late sepsis in rats. Arch Surg. 1986;121:918–923. doi: 10.1001/archsurg.1986.01400080064011. [DOI] [PubMed] [Google Scholar]
- Hasselgren PO, James JH, Benson DW, Hall-Angeras M, Angeras U, Hiyama DT, Li S, Fischer JE. Total and myofibrillar protein breakdown in different types of rat skeletal muscle: Effects of sepsis and regulation by insulin. Metabolism. 1989;38:634–640. doi: 10.1016/0026-0495(89)90100-5. [DOI] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- Jackman RW, Kandarian SC. The molecular basis of skeletal muscle atrophy. Am J Physiol Cell Physiol. 2004;287:C834–C843. doi: 10.1152/ajpcell.00579.2003. [DOI] [PubMed] [Google Scholar]
- Jagoe RT, Goldberg AL. What do we really know about the ubiquitin-proteasome pathway in muscle atrophy? Curr Opin Clin Nutr Metab Care. 2001;4:183–190. doi: 10.1097/00075197-200105000-00003. [DOI] [PubMed] [Google Scholar]
- Jobin C, Bradham CA, Russo MP, Juma B, Narula AS, Brenner DA, Sartor RB. Curcumin blocks cytokine-mediated NF-kappa B activation and proinflammatory gene expression by inhibiting inhibitory factor I-kappa B kinase activity. J Immunol. 1999;163:3474–3483. [PubMed] [Google Scholar]
- Jones SW, Hill RJ, Krasney PA, O’Conner B, Peirce N, Greenhaff PL. Disuse atrophy and exercise rehabilitation in humans profoundly affects the expression of genes associated with the regulation of skeletal muscle mass. FASEB J. 2004;18:1025–1027. doi: 10.1096/fj.03-1228fje. [DOI] [PubMed] [Google Scholar]
- Jones NC, Tyner KJ, Nibarger L, Stanley HM, Cornelison DD, Fedorov YV, Olwin BB. The p38{alpha}/{beta} MAPK functions as a molecular switch to activate the quiescent satellite cell. J Cell Biol. 2005;169:105–116. doi: 10.1083/jcb.200408066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koistinen HA, Chibalin AV, Zierath JR. Aberrant p38 mitogen-activated protein kinase signalling in skeletal muscle from Type 2 diabetic patients. Diabetologia. 2003;46:1324–1328. doi: 10.1007/s00125-003-1196-3. [DOI] [PubMed] [Google Scholar]
- Latres E, Amini AR, Amini AA, Griffiths J, Martin FJ, Wei Y, Lin HC, Yancopoulos GD, Glass DJ. Insulin-like growth factor-1 (IGF-1) inversely regulates atrophy-induced genes via the phosphatidylinositol 3-kinase/Akt/mammalian target of rapamycin (PI3K/Akt/mTOR) pathway. J Biol Chem. 2005;280:2737–2744. doi: 10.1074/jbc.M407517200. [DOI] [PubMed] [Google Scholar]
- Lecker SH, Jagoe RT, Gilbert A, Gomes M, Baracos V, Bailey J, Price SR, Mitch WE, Goldberg AL. Multiple types of skeletal muscle atrophy involve a common program of changes in gene expression. FASEB J. 2004;18:39–51. doi: 10.1096/fj.03-0610com. [DOI] [PubMed] [Google Scholar]
- Li YP, Reid MB. NF-kB mediates the protein loss induced by TNF-alpha in differentiated skeletal muscle myotubes. Am J Physiol. 2000;279:R1165–R1170. doi: 10.1152/ajpregu.2000.279.4.R1165. [DOI] [PubMed] [Google Scholar]
- Li YP, Atkins CM, Sweatt JD, Reid BR. Mitochondria mediate tumor necrosis factor-alpha/NF-keppa B signaling in skeletal muscle myotubes. Antioxidants & Redox Signaling. 1999;1:97–104. doi: 10.1089/ars.1999.1.1-97. [DOI] [PubMed] [Google Scholar]
- Li YP, Chen Y, Li AS, Reid MB. Hydrogen peroxide stimulates ubiquitin-conjugating activity and expression of genes for specific E2 and E3 proteins in skeletal muscle myotubes. Am J Physiol Cell Physiol. 2003a;285:C806–C812. doi: 10.1152/ajpcell.00129.2003. [DOI] [PubMed] [Google Scholar]
- Li YP, Lecker SH, Chen Y, Waddell ID, Goldberg AL, Reid MB. TNF-alpha increases ubiquitin-conjugating activity in skeletal muscle by up-regulating UbcH2/E220k. FASEB J. 2003b;17:1048–1057. doi: 10.1096/fj.02-0759com. [DOI] [PubMed] [Google Scholar]
- Li YP, Chen Y, John J, Moylan J, Jin B, Mann DL, Reid MB. TNF-alpha acts via p38 MAPK to stimulate expression of the ubiquitin ligase atrogin1/MAFbx in skeletal muscle. FASEB J. 2005;19:362–370. doi: 10.1096/fj.04-2364com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long YC, Widegren U, Zierath JR. Exercise-induced mitogen-activated protein kinase signalling in skeletal muscle. Proc Nutr Soc. 2004;63:227–232. doi: 10.1079/PNS2004346. [DOI] [PubMed] [Google Scholar]
- Mordmuller B, Krappmann D, Esen M, Wegener E, Scheidereit C. Lymphotoxin and lipopolysaccharide induce NF-kappaB-p52 generation by a co-translational mechanism. EMBO Rep. 2003;4:82–87. doi: 10.1038/sj.embor.embor710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’keefe MP, Perez FR, Kinnick TR, Tischler ME, Henriksen EJ. Development of whole-body and skeletal muscle insulin resistance after one day of hindlimb suspension. Metabolism. 2004;53:1215–1222. doi: 10.1016/j.metabol.2004.02.025. [DOI] [PubMed] [Google Scholar]
- Penner CG, Gang G, Wray C, Fischer JE, Hasselgren PO. The transcription factors NF-kappab and AP-1 are differentially regulated in skeletal muscle during sepsis. Biochem Biophys Res Commun. 2001;281:1331–1336. doi: 10.1006/bbrc.2001.4497. [DOI] [PubMed] [Google Scholar]
- Premer DM, Goertz R, Georgieff MK, Mammel MC, Schwarzenberg SJ. Muscle proteolysis and weight loss in a neonatal rat model of sepsis syndrome. Inflammation. 2002;26:97–101. doi: 10.1023/a:1014840412552. [DOI] [PubMed] [Google Scholar]
- Raingeaud J, Gupta S, Rogers JS, Dickens M, Han J, Ulevitch RJ, Davis RJ. Pro-inflammatory cytokines and environmental stress cause p38 mitogen-activated protein kinase activation by dual phosphorylation on tyrosine and threonine. J Biol Chem. 1995;270:7420–7426. doi: 10.1074/jbc.270.13.7420. [DOI] [PubMed] [Google Scholar]
- Reid MB, Li Y-P. Cytokines and oxidative signaling in skeletal muscle cells. Acta Physiol Scand. 2001;171:225–232. doi: 10.1046/j.1365-201x.2001.00824.x. [DOI] [PubMed] [Google Scholar]
- Rosenblatt S, Clowes GH, Jr., George BC, Hirsch E, Lindberg B. Exchange of amino acids by muscle and liver in sepsis. Arch Surg. 1983;118:167–175. doi: 10.1001/archsurg.1983.01390020023004. [DOI] [PubMed] [Google Scholar]
- Sandri M, Sandri C, Gilbert A, Skurk C, Calabria E, Picard A, Walsh K, Schiaffino S, Lecker SH, Goldberg AL. Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell. 2004;117:399–412. doi: 10.1016/s0092-8674(04)00400-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stitt TN, Drujan D, Clarke BA, Panaro F, Timofeyva Y, Kline WO, Gonzalez M, Yancopoulos GD, Glass DJ. The IGF-1/PI3K/Akt pathway prevents expression of muscle atrophy-induced ubiquitin ligases by inhibiting FOXO transcription factors. Mol Cell. 2004;14:395–403. doi: 10.1016/s1097-2765(04)00211-4. [DOI] [PubMed] [Google Scholar]
- Tiao G, Fagan JM, Samuels N, James JH, Hudson K, Lieberman M, Fischer JE, Hasselgren PO. Sepsis stimulates nonlysosomal, energy-dependent proteolysis and increases ubiquitin mRNA levels in rat skeletal muscle. J Clin Invest. 1994;94:2255–2264. doi: 10.1172/JCI117588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiao G, Hobler S, Wang JJ, Meyer TA, Luchette FA, Fischer JE, Hasselgren PO. Sepsis is associated with increased mRNAs of the ubiquitin-proteasome proteolytic pathway in human skeletal muscle. J Clin Invest. 1997;99:163–168. doi: 10.1172/JCI119143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tisdale MJ. Biology of cachexia. J Natl Cancer Inst. 1997;89:1763–1773. doi: 10.1093/jnci/89.23.1763. [DOI] [PubMed] [Google Scholar]
- Tremblay F, Dubois MJ, Marette A. Regulation of GLUT4 traffic and function by insulin and contraction in skeletal muscle. Front Biosci. 2003;8:d1072–d1084. doi: 10.2741/1137. [DOI] [PubMed] [Google Scholar]
- Vary TC. Regulation of skeletal muscle protein turnover during sepsis. Curr Opin Clin Nutr Metab Care. 1998;1:217–224. doi: 10.1097/00075197-199803000-00013. [DOI] [PubMed] [Google Scholar]
- Wang XS, Diener K, Manthey CL, Wang S, Rosenzweig B, Bray J, Delaney J, Cole CN, Chan-Hui PY, Mantlo N, Lichenstein HS, Zukowski M, Yao Z. Molecular cloning and characterization of a novel p38 mitogen-activated protein kinase. J Biol Chem. 1997;272:23668–23674. doi: 10.1074/jbc.272.38.23668. [DOI] [PubMed] [Google Scholar]
- Williamson D, Gallagher P, Harber M, Hollon C, Trappe S. Mitogen-activated protein kinase (MAPK) pathway activation: Effects of age and acute exercise on human skeletal muscle. J Physiol. 2003;547:977–987. doi: 10.1113/jphysiol.2002.036673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winick M. Hunger Disease—Studies by Jewish Physicians in the Warsaw Ghetto. John Wiley & Sons; New York: 1979. [Google Scholar]
- Wray CJ, Mammen JM, Hershko DD, Hasselgren PO. Sepsis upregulates the gene expression of multiple ubiquitin ligases in skeletal muscle. Int J Biochem Cell Biol. 2003;35:698–705. doi: 10.1016/s1357-2725(02)00341-2. [DOI] [PubMed] [Google Scholar]
- Yang SH, Galanis A, Sharrocks AD. Targeting of p38 mitogen-activated protein kinases to MEF2 transcription factors. Mol Cell Biol. 1999;19:4028–4038. doi: 10.1128/mcb.19.6.4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Bozza PT, Tzizik DM, Gray JP, Cassara J, Dvorak AM, Weller PF. Co-compartmentalization of MAP kinases and cytosolic phospholipase A2 at cytoplasmic arachidonate-rich lipid bodies. Am J Pathol. 1998;152:759–769. [PMC free article] [PubMed] [Google Scholar]
- Zarubin T, Han J. Activation and signaling of the p38 MAP kinase pathway. Cell Res. 2005;15:11–18. doi: 10.1038/sj.cr.7290257. [DOI] [PubMed] [Google Scholar]