Abstract
We report the identification and chemical characterization of four antifungal substances, 3-(R)-hydroxydecanoic acid, 3-hydroxy-5-cis-dodecenoic acid, 3-(R)-hydroxydodecanoic acid and 3-(R)-hydroxytetradecanoic acid, from Lactobacillus plantarum MiLAB 14. The concentrations of the 3-hydroxy fatty acids in the supernatant followed the bacterial growth. Racemic mixtures of the saturated 3-hydroxy fatty acids showed antifungal activity against different molds and yeasts with MICs between 10 and 100 μg ml−1.
Lactic acid bacteria (LAB) have a long history of use as biopreservatives for food and feed storage. The general preserving ability of lactic acid and other fermentation end products and the antibacterial effects of LAB proteinaceous bacteriocins are well documented (14, 24). Recent research has revealed that LAB can produce low-molecular-weight antifungal substances, e.g., phenyllactic acid, p-hydroxyphenyllactic acid (12, 25), cyclic dipeptides such as cyclo(Gly-l-Leu), cyclo(l-Phe-l-Pro), and cyclo(l-Phe-trans-4-OH-l-Pro) (18, 25), benzoic acid, methylhydantoin, mevalonolactone (18), and short-chain fatty acids (FAs) (4).
Previously, a large number of LAB strains with antifungal effects have been isolated in vitro from plant material stored under anaerobic conditions (16). In a continuous study of LAB strains with antifungal effects, procedures for isolating antifungal compounds among a background of high concentrations of lactic acid were devised (25). In this study, the characterization of four antifungal 3-hydroxy FAs (3-OH-FAs) from Lactobacillus plantarum MiLAB 14 is reported.
Strain MiLAB 14, isolated from lilac flowers (16), was identified as L. plantarum from both the fermentation pattern and the 16S rRNA gene sequence. The API 50 CHL test (bioMérieux, Marcy L'Etoile, France) was used for identification by fermentation pattern. Chromosomal DNA isolation and PCR amplification were performed as previously described (25). Approximately 1,400 bp of the 16S rRNA gene were sequenced as previously described (25) but with additional customized primers covering the whole fragment.
The strain MiLAB 14 was grown on MRS agar and stored as previously described (15). The molds Aspergillus fumigatus J9, Aspergillus nidulans J283 (FSGC A4 wt), Penicillium roqueforti J268 (IBT 6754), and Penicillium commune J238 (IBT 12400) and the yeasts Kluyveromyces marxianus J137 (CBS 6556), Pichia anomala J121, and Rhodotorula mucilaginosa J350 (CFSQE 63) were used as target organisms for assay of antifungal activity. The target fungi were chosen to represent potential spoilage fungi in silage and dairy products (20). All fungi are kept in the culture collection at the Department of Microbiology, Swedish University of Agricultural Sciences. Molds and yeasts were prepared as previously described (15). A microtiter plate assay (15) was used for bioassay-guided fractionation. A. fumigatus was used as the target organism, as it has been shown to be sensitive to antifungal strains of LAB (16) as well as being a serious pathogen of animals and humans (6).
The cell-free supernatant of L. plantarum MiLAB 14 from a 48-h still culture at 30°C was obtained and fractionated by the same method with solid-phase extraction (SPE) and high-performance liquid chromatography, as previously described (25). As a negative control, noninoculated MRS broth was fractionated and evaluated in the bioassay by the procedure used for the cell-free supernatant. The structures of the antifungal compounds were determined by nuclear magnetic resonance (NMR) spectroscopy, electrospray ionization mass spectrometry (ESI-MS), and gas chromatography-mass spectrometry (GC-MS). Absolute configuration was determined by preparation of 3-O-methyl N-(S)-phenylethylamide derivates of the 3-OH-FAs followed by GC-MS analysis (7).
Three aliquots of 800-ml cultures in MRS broth were inoculated with 104 bacteria ml−1, and concentrations of 3-OH-FAs, growth (number of CFU on MRS agar plates), and pH were monitored for 78 h. Each sample of cell-free supernatant (10 ml) was fractionated by SPE (Isolute, C18 end capped, 1 g). The fraction eluted with 4 ml of aqueous 95% acetonitrile, after a wash with 3 ml of aqueous 30% acetonitrile, was dried under vacuum. The material was dissolved in 100 μl of hexane in 1.5-ml Eppendorf tubes and derivatized with 50 μl of N,O-bis(trimethylsilyl)trifluoroacetamide (Supelco, Stainheim, Germany) and 10 μl of pyridine at 80°C for 1 h. GC-MS was performed on a fused-silica capillary column using a temperature gradient (70°C for 3 min; 70 to 240°C at 10°C min−1; injector, 240°C; interface, 260°C; carrier gas, He, 1 ml min−1). The reference was a mixture of trimethylsilyl derivatives of 3-hydroxydecanoic acid, 3-hydroxydodecanoic acid, and 3-hydroxytetradecanoic acid. The concentrations of the 3-OH-FAs were determined by using 3-hydroxyundecanoic acid as an internal standard (2 μg ml of supernatant−1 added before preparation of cell-free supernatant) and assuming similar response factors for the different 3-OH-FAs.
The MIC was determined in duplicate as the lowest concentration where total inhibition of spore germination was observed. MICs were determined for decanoic acid, 2-hydroxydodecanoic acid, 3-hydroxydecanoic acid, 3-hydroxyundecanoic acid, 3-hydroxydodecanoic acid, and 3-hydroxytetradecanoic acid (Larodan Fine Chemicals, Malmö, Sweden), and all OH-FAs were racemic. FAs were dissolved in methanol and diluted with 10 mM acetic acid. The molds A. fumigatus, A. nidulans, P. roqueforti, and P. commune and the yeasts K. marxianus, P. anomala, and R. mucilaginosa were used as target organisms. MIC determinations were performed as serial twofold dilutions by a microtiter plate method (15) with malt extract broth (2%) instead of MRS.
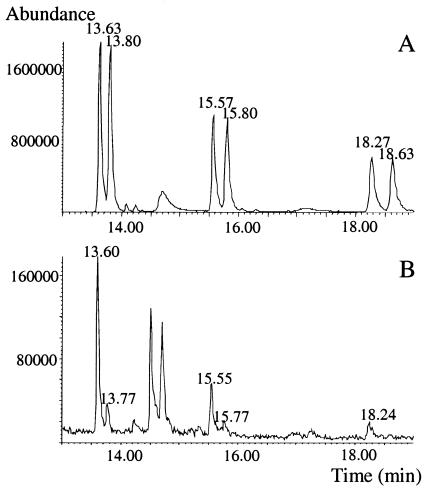
Two active compounds, 3-hydroxydecanoic acid and 3-hydroxy-5-cis-dodecenoic acid, were isolated from cell-free supernatant by bioassay-guided fractionation. The former compound was identified by comparing data from NMR, ESI-MS, and high-performance liquid chromatography with data from commercial racemic 3-hydroxydecanoic acid, whereas the latter compound was identified by comparison of experimental and literature NMR data (9, 11) together with the molecular mass from ESI-MS. No activity was observed from the corresponding fractions isolated from noninoculated MRS broth. Further, two active compounds, 3-hydroxydodecanoic acid and 3-hydroxytetradecanoic acid, were identified in cell-free supernatant by GC-MS. The saturated 3-OH-FAs were determined to have the (R) configuration (Fig. 1), whereas the absolute configuration of 3-hydroxy-5-cis-dodecenoic acid could not be determined due to the lack of reference compounds. The presence of small peaks corresponding to 3-OH-FAs in the (S) configuration in Fig. 1 is due to racemization during the derivatization procedure. This racemization was detected by using methanol-d4/D2O instead of methanol/water as the solvent in the hydrolysis step (data not shown). All saturated 3-OH-FAs (17, 19, 22) in this work as well as 3-hydroxy-5-cis-dodecenoic acid (3, 11) have previously been isolated. Hydroxy FAs are commonly found in animals, plants, and fungi (26), and their antifungal properties have been reported (8, 10). In bacteria, 3-OH-FAs are present in lipopolysaccharides (1) or in poly-hydroxyalkanoic acids (23). On the other hand, gram-positive bacteria, such as LAB, have no lipopolysaccharides, and there are no reports on poly-hydroxyalkanoic acids from LAB. The cellular FAs that have been used for classification of different LAB are mainly saturated and monounsaturated FAs containing 12 to 20 carbons (5, 21) and representing more than 90% of all cellular FAs in LAB (5). However, Lee et al. (13) identified 2-hydroxyhexadecanoic acid and 3-hydroxyheptadecanoic acid from different Leuconostoc strains. LAB can metabolize unsaturated FAs to OH-FAs (27, 28), indicating metabolic pathways for hydroxylation of FAs, but the exact role of 3-OH-FAs in LAB metabolism remains to be elucidated.
FIG. 1.
Extracted ion chromatogram of m/z 120 for 3-O-methyl N-(S)-phenylethylamide derivatives of a standard solution with racemic 3-hydroxydecanoic acid (R = 13.63 min and S = 13.80 min), 3-hydroxydodecanoic acid (R = 15.57 min and S = 15.80 min), and 3-hydroxytetradecanoic acid (R = 18.27 min and S = 18.63 min) (A) and samples from L. plantarum MiLAB 14 after SPE of cell-free supernatant, 3-hydroxydecanoic acid (R = 13.60 min and S = 13.77 min [the (S) form is formed by racemization of the sample during derivatization]), 3-hydroxydodecanoic acid (R = 15.55 min and S = 15.77 min [the (S) form is formed by racemization of the sample during derivatization]), and 3-hydroxytetradecanoic acid (R = 18.24 min) (B) (see the text for conditions).
The concentration of 3-(R)-hydroxydecanoic acid in the culture supernatant increased during the logarithmic growth phase of L. plantarum MiLAB 14 and reached a maximum of 1.7 μg ml−1 after 38 h (Fig. 2). The concentration of 3-(R)-hydroxydecanoic acid increased only during the exponential phase and not when the cells reached the stationary phase. This indicates that 3-OH-FAs do not originate from disrupted cell membranes of dead bacteria but instead are excreted to the culture broth by living bacterial cells. The concentrations of 3-hydroxy-5-cis-dodecenoic acid (1.0 μg ml−1), 3-(R)-hydroxydodecanoic acid (0.5 μg ml−1), and 3-(R)-hydroxytetradecanoic acid (0.2 μg ml−1) were lower than the concentration of 3-(R)-hydroxydecanoic acid (1.6 μg ml−1) after 78 h of growth, but all concentrations varied over time in a similar way (data not shown). None of the acids were detected at the time of inoculation.
FIG. 2.
Concentration of 3-hydroxydecanoic acid (▴), number of CFU (•), and pH (▪) during still-culture growth of L. plantarum MiLAB 14 in MRS broth at 30°C. Bars indicate standard deviations (n = 3).
For both yeasts and molds, the MICs for total growth inhibition were between 10 and 100 μg ml−1 for the racemic forms of the 3-OH-FAs (Table 1). Yeasts appeared to be more sensitive than filamentous fungi to the different 3-OH-FAs. Among the filamentous fungi, P. roqueforti was the most sensitive (5 to 50 μg ml−1), whereas A. fumigatus, previously found to be highly sensitive to LAB strains with antifungal properties (16), was among the least sensitive fungi (25 to 100 μg ml−1). The concentrations of the 3-OH-FAs found in MiLAB 14 supernatant are about 10 to 200 times lower than the MICs. However, the 3-OH-FAs could still contribute to the antifungal activity of L. plantarum MiLAB 14, since higher local concentrations of the 3-OH-FAs are expected to be found close to bacterial colonies and there could also be synergistic effects.
TABLE 1.
MIC of racemic mixtures of fatty acids for different yeasts and molds in a microtiter plate assay
| Organism | MIC (μg ml−1) ofa:
|
|||||
|---|---|---|---|---|---|---|
| C10 | 2-OH-C12 | 3-OH-C10b | 3-OH-C11 | 3-OH-C12b | 3-OHC14b | |
| P. roqueforti | 25 | 5 | 25 | 10 | 25 | 50 |
| P. commune | 50 | 25 | 100 | 50 | 50 | >100c |
| A. nidulans | 100 | 25 | 50 | 50 | 25 | >100c |
| A. fumigatus | 100 | 25 | 100 | 50 | 25 | >100c |
| K. marxianus | 25 | 25 | 50 | 25 | 25 | 10 |
| R. mucilaginosa | 5 | 5 | 10 | 25 | 10 | 10 |
| P. anomala | 25 | 25 | 50 | 25 | 25 | 50 |
C10, decanoic acid; 2-OH-C12, 2-hydroxydecanoic acid; 3-OH-C10, 3-hydroxydecanoic acid; 3-OH-C11, 3-hydroxyundecanoic acid; 3-OH-C12, 3-hydroxydodecanoic acid; 3-OH-C14, 3-hydroxytetradecanoic acid.
Detected in L. plantarum MiLAB 14 culture supernatant.
Not determined due to low solubility.
The mechanisms behind the antifungal effect of the 3-OH-FAs are not known, but the MICs for all FAs and target organisms are within a fairly narrow range. This suggests that all the 3-OH-FAs from L. plantarum MiLAB 14, as well as 2-hydroxydodecanoic acid and decanoic acid, would have similar modes of action. As enantiomerically pure 3-OH-FAs were not available, possible differences in MICs between (R) and (S) forms could not be established. One general mechanism that has been proposed for antifungal FAs is that the activity is due to detergent-like properties of the compounds, affecting the structure of cell membranes of the target organisms. Indeed, cis-9-heptadecenoic acid, a compound similar to the 3-OH-FAs identified here, readily partitions into the lipid bilayers of fungal membranes (2). This increases membrane permeability and the release of intracellular electrolytes and proteins and, eventually, leads to cytoplasmic disintegration of fungal cells.
Future studies on lactic acid bacteria with antifungal properties could lead to useful biopreservation systems, preventing fungal spoilage and mycotoxin formation in both food and animal feed.
Nucleotide sequence accession number.
The sequence determined in this study has been deposited in GenBank with accession number AY383631.
Acknowledgments
The financial support of the Foundation for Strategic Environmental Research (MISTRA) is gratefully acknowledged.
Stefan Roos assisted in confirming bacterial species identity.
REFERENCES
- 1.Alexander, C., and U. Zähringer. 2002. Chemical structure of lipid A—the primary immunomodulatory center of bacterial lipopolysaccharides. Trends Glycosci. Glyc. 14:69-86. [Google Scholar]
- 2.Avis, T. J., and R. R. Bélanger. 2001. Specificity and mode of action of the antifungal fatty acid cis-9-heptadecenoic acid produced by Pseudozyma flocculosa. Appl. Environ. Microbiol. 67:956-960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bishop, D. G., and J. L. Still. 1962. Fatty acid metabolism in Serratia marcescens. II. The occurrence of hydroxy acids. Biochem. Biophys. Res. Commun. 7:337-341. [DOI] [PubMed] [Google Scholar]
- 4.Corsetti, A., M. Gobbetti, J. Rossi, and P. Damiani. 1998. Antimould activity of sourdough lactic acid bacteria: identification of a mixture of organic acids produced by Lactobacillus sanfrancisco CB1. Appl. Microbiol. Biotechnol. 50:253-256. [DOI] [PubMed] [Google Scholar]
- 5.Decallonne, J., M. Delmee, P. Wauthoz, M. El Lioui, and R. Lambert. 1991. A rapid procedure for the identification of lactic acid bacteria based on the gas chromatographic analysis of the cellular fatty acids. J. Food Prot. 54:217-224. [DOI] [PubMed] [Google Scholar]
- 6.de Hoog, G. S. 1996. Risk assessment of fungi reported from humans and animals. Mycoses 39:407-417. [DOI] [PubMed] [Google Scholar]
- 7.Gradowska, W., and L. Larsson. 1994. Determination of absolute configurations of 2-and 3-hydroxy fatty acids in organic dust by gas chromatography-mass spectrometry. J. Microbiol. Methods. 20:55-67. [Google Scholar]
- 8.Granér, G., M. Hamberg, and J. Meijer. 2003. Screening of oxylipins for control of oilseed rape (Brassica napus) fungal pathogens. Phytochemistry 63:89-95. [DOI] [PubMed] [Google Scholar]
- 9.Gunstone, F. D., M. R. Pollard, C. M. Scrimgeour, and H. S. Vedanayagam. 1977. Fatty acids. Part 50. 13C nuclear magnetic resonance studies of olefinic fatty acids and esters. Chem. Phys. Lipids 18:115-129. [DOI] [PubMed] [Google Scholar]
- 10.Hou, C. T., and R. J. Forman. 2000. Growth inhibition of plant pathogenic fungi by hydroxy fatty acids. J. Ind. Microbiol. Biotechnol. 24:275-276. [Google Scholar]
- 11.Ichihara, A., M. Hashimoto, and S. Sakamura. 1985. (3R, 5Z)-(−)-3-Hydroxy-5-dodecenoic acid, a phytotoxic metabolite of Pythium ultimum. Agric. Biol. Chem. 49:2207-2209. [Google Scholar]
- 12.Lavermicocca, P., F. Valerio, A. Evidente, S. Lazzaroni, A. Corsetti, and M. Gobbetti. 2000. Purification and characterization of novel antifungal compounds from the sourdough Lactobacillus plantarum strain 21B. Appl. Environ. Microbiol. 66:4084-4090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee, J. S., C. O. Chun, H. J. Kim, Y. J. Joo, H. J. Lee, C. S. Park, J. S. Ahn, Y. H. Park, and T. I. Mheen. 1996. Analysis of cellular fatty acid methyl esters (FAMEs) for the identification of Leuconostoc strains isolated from kimchi. J. Microbiol. 34:225-228. [Google Scholar]
- 14.Lindgren, S. E., and W. J. Dobrogosz. 1990. Antagonistic activities of lactic-acid bacteria in food and feed fermentations. FEMS Microbiol. Rev. 87:149-164. [DOI] [PubMed] [Google Scholar]
- 15.Magnusson, J., and J. Schnürer. 2001. Lactobacillus coryniformis subsp. coryniformis strain Si3 produces a broad-spectrum proteinaceous antifungal compound. Appl. Environ. Microbiol. 67:1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Magnusson, J., K. Ström, S. Roos, J. Sjögren, and J. Schnürer. 2003. Broad and complex antifungal activity among environmental isolates of lactic acid bacteria. FEMS Microbiol. Lett. 219:129-135. [DOI] [PubMed] [Google Scholar]
- 17.Matsumoto, G. I., K. Watanuki, and T. Torii. 1988. Hydroxy acids in Antarctic lake sediments and their geochemical significance. Org. Geochem. 13:785-790. [Google Scholar]
- 18.Niku-Paavola, M. L., A. Laitila, T. Mattila-Sandholm, and A. Haikara. 1999. New types of antimicrobial compounds produced by Lactobacillus plantarum. J. Appl. Microbiol. 86:29-35. [DOI] [PubMed] [Google Scholar]
- 19.Parks, O. W. 1977. Isolation and characterization of nonesterified 3-hydroxy acids in milk. J. Dairy Sci. 60:718-720. [Google Scholar]
- 20.Pitt, J. J., and A. D. Hocking. 1999. Fungi and food spoilage, 2nd ed. Aspen Publications, New York, N.Y.
- 21.Rizzo, A. F., H. Korkeala, and I. Mononen. 1987. Gas chromatography analysis of cellular fatty acids and neutral monosaccharides in the identification of Lactobacilli. Appl. Environ. Microbiol. 53:2883-2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schildknecht, H., and K. Koob. 1971. Myrmicacin, the first insect herbicide. Angew. Chem. Int. Ed. 10:124-125. [DOI] [PubMed] [Google Scholar]
- 23.Steinbüchel, A., and H. V. Valentin. 1995. Diversity of bacterial polyhydroxyalkanoic acids. FEMS Microbiol. Lett. 128:219-228. [Google Scholar]
- 24.Stiles, M. E. 1996. Biopreservation by lactic acid bacteria. Antonie Leeuwenhoek 70:331-345. [DOI] [PubMed] [Google Scholar]
- 25.Ström, K., J. Sjögren, A. Broberg, and J. Schnürer. 2002. Lactobacillus plantarum MiLAB 393 produces the antifungal cyclic dipeptides cyclo(l-Phe-l-Pro) and cyclo(l-Phe-trans-4-OH-l-Pro) and 3-phenyllactic acid. Appl. Environ. Microbiol. 68:4322-4327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van Dyk, M. S., J. L. F. Kock, and A. Botha. 1994. Hydroxy long chain fatty-acids in fungi. World J. Microbiol. Biotechnol. 10:495-504. [DOI] [PubMed] [Google Scholar]
- 27.Wanikawa, A., H. Shoji, K. Hosoi, and K. Nakagawa. 2002. Stereospecificity of 10-hydroxystearic acid and formation of 10-ketostearic acid by lactic acid bacteria. J. Am. Soc. Brew. Chem. 60:14-20. [Google Scholar]
- 28.Yamada, Y., H. Uemura, H. Nakaya, K. Sakata, T. Takatori, M. Nagao, H. Iwase, and K. Iwadate. 1996. Production of hydroxy fatty acid (10-hydroxy-12(Z)-octadecenoic acid) by Lactobacillus plantarum from linoleic acid and its cardiac effects to guinea pig papillary muscles. Biochem. Biophys. Res. Commun. 226:391-395. [DOI] [PubMed] [Google Scholar]