Abstract
The molecular interactions between the bifidobacterial cell and its natural environment, namely, the gastrointestinal tract of its host, are particularly important in understanding the presumed positive effects of Bifidobacterium on the health status of the host. In this study an export-specific reporter system, designed for use in gram-positive organisms and based on the use of the staphylococcal nuclease (Nuc) as a reporter, was employed to identify exported proteins in Bifidobacterium breve UCC2003. A B. breve genomic library of translational fusions to the Nuc-encoding gene devoid of its own export signal was established in the shuttle vector pFUN (I. Poquet, S. D. Ehrlich, and A. Gruss, J. Bacteriol. 180:1904-1912, 1998) and screened for bifidobacterial export signals. Sequence analysis of the fusion proteins obtained that displayed a nuclease-producing phenotype in both Lactococcus lactis and B. breve predicted the presence of a classical signal peptide and/or single or multiple transmembrane domains, thus indicating that some of the export signals in B. breve are comparable to those used in L. lactis. Cell fractionation studies, zymograms, nuclease assays, and Western blotting were employed to confirm the function of the predicted signals and to determine the location and activity of the exported fusion proteins in B. breve and/or L. lactis.
Bifidobacteria are of increasing commercial interest due to their widespread use in the growing probiotics industry. Little is known about the molecular biology of bifidobacteria, despite the fact that they are among the most common bacteria in the human colon and have consistently had health-promoting properties attributed to them (for general reviews see references 11 and 15). Genetic characterization of bifidobacteria is essential in order to define their presumed beneficial activities as part of the intestinal microflora and to explore and potentially exploit any such beneficial properties. Exported proteins are of particular interest, as they may be critically important in the interaction between the microbes and their hosts (environment). Such exported proteins may be enzymes involved in the hydrolysis of biopolymers and may be synthesized as part of an adaptive response to changes in the environment, allowing the cells to benefit optimally from the available resources (12, 31). Furthermore, studies with Lactococcus lactis have previously implicated exported proteins in functions such as nutrient uptake, peptidoglycan assembly, environmental sensing, and protein folding (38). Characterization of secreted proteins in bifidobacteria is necessary to define their possible beneficial activities as part of the intestinal microflora and to explore and potentially exploit any such beneficial activities, while also allowing the production of heterologous exported proteins for use in biotechnology, in fermented food products, or in the digestive tracts of humans and animals.
The strategy of constructing random translational fusions between potential translocation signals and an export-specific reporter protein, designed to isolate genes encoding exported proteins, was first described for Escherichia coli, in which alkaline phosphatase (PhoA) was used as a reporter (18, 29). More recently, this strategy has been adopted for use in gram-positive bacteria (27, 37, 38) by using such reporters as staphylococcal nuclease (24, 38) and α-amylase from Bacillus (19). The reporter in such cases is translocation competent but is unable to direct its own export (due to removal of the signal peptide [SP]), while its activity depends on an extracytoplasmic location. Among a library of sequences N terminally fused to such a reporter, only those fusions having an appropriate export signal are directed by the Sec-dependent secretion machinery to be translocated. In most cases, a prerequisite for the release of the translocated protein from the membrane (and subsequent secretion into the medium) is removal of the SP by a signal peptidase (SPase) (48, 52). Notably, several integral membrane proteins retain their SPs and diffuse laterally from the translocase. Other proteins contain several membrane-spanning domains that are required for insertion into the cytoplasmic membrane.
At present, four major classes of amino-terminal SPs can be distinguished on the basis of the SPase recognition sequence. The first class is composed of classical SPs, which are present in preproteins that are cleaved by a type I SPase. A separate group of these SPs contains a so-called twin-arginine motif (RR motif), which may direct proteins into a distinct translocation pathway known as the twin-arginine translocation (Tat) pathway (for reviews see references 4, 53, and 55). The classical Sec-type SPs consist of an amino-terminal N domain containing at least one positively charged residue (7, 14), a central hydrophobic core (H region), and a C region with a consensus SPase recognition sequence, A-X-A at positions −3 to −1 relative to the SPase I cleavage site (39, 46, 54). The second major class of SPs is present in prelipoproteins, which are cleaved by the lipoprotein-specific (type II) SPase. Cleavage in this case occurs in front of a cysteine residue (39, 46, 54). The third major class is formed by SPs of prepilin-like proteins, in which the recognition sequence of the prepilin-specific SPase (unlike that of secretory proteins and lipoproteins) is localized between the N and H domains (28, 39). The fourth class of SPs is found in ribosomally synthesized bacteriocins and pheromones that are exported by dedicated ABC transporters (3, 36, 56). These SPs lack an H domain and are removed from the mature protein by a subunit of the ABC transporter.
Despite the assumed biotechnological importance of surface-located and extracellular proteins in Bifidobacterium, very few exported proteins have been identified in this genus to date. There is only one other report of an SP in Bifidobacterium, and this SP was identified in a probable extracellular β-galactosidase from Bifidobacterium bifidum (30). Approximately 200 proteins with probable Sec-type SPs were recently proposed based on a genomic sequence analysis of Bifidobacterium longum (41). In this study, the broad-host-range plasmid pFUN was utilized to identify exported proteins in Bifidobacterium breve by a strategy based on translational fusions with an export-specific reporter protein. The Staphylococcus aureus secreted nuclease (Nuc) devoid of its export signal (ΔSPNuc) was used as a reporter. Nuclease activity was shown to require an extracellular location in B. breve. ΔSPNuc translational fusions were constructed in which the export signal was provided by inserted B. breve chromosomal DNA. By using this strategy, seven previously unknown exported proteins were identified for B. breve UCC2003. From these results, combined with bioinformatics-based comparative analyses, it appears that protein translocation in several Bifidobacterium spp. occurs through a mechanism which is comparable to the mechanisms previously identified for a large number of gram-positive bacteria.
MATERIALS AND METHODS
Bacterial strains, media, and culture conditions.
B. breve UCC2003 was routinely cultured in de Man-Rogosa-Sharpe medium (MRS) (9) (Oxoid Ltd., Basingstoke, Hampshire, England) supplemented with 0.2% (wt/vol) glucose. MRS was also supplemented with 0.05% (wt/vol) cysteine-HCl, and strains were grown at 37°C under anaerobic conditions maintained with the Anaerocult oxygen-depleting system (Merck, Darmstadt, Germany) in an anaerobic chamber. Escherichia coli DH5α (16) was grown in Luria-Bertani medium at 37°C with agitation (40). L. lactis subsp. cremoris MG1363 (13) and its derivatives were grown on M17 medium supplemented with 1% glucose at 30°C. Plasmids pFUN, pVE8009, and pVE8010 (38) and derivatives of these plasmids were maintained by antibiotic selection by using erythromycin (5 μg ml−1 for L. lactis) or ampicillin (100 μg ml−1 for E. coli). The E. coli vectors pBluescript (Stratagene Ltd., Cambridge, United Kingdom) and pCR2.1-TOPO (Invitrogen BV, Groningen, The Netherlands) were used for cloning purposes.
DNA manipulations.
Plasmid DNA was obtained from E. coli by using either an alkaline lysis method (5) or a QIAprep Spin plasmid miniprep kit (Qiagen GmbH, Hilden, Germany). L. lactis plasmid DNA was prepared by using either a modification of the alkaline lysis method as described previously (40) or a similarly modified QIAprep Spin plasmid miniprep kit procedure. The modification included addition of lysozyme (10 μg ml−1) after resuspension of the pelleted cells, followed by incubation at 37°C for 30 min. The normal manufacturer's protocol was used after this step. Large-scale production of total DNA from B. breve was performed as previously described (34). Restriction endonucleases, T4 DNA ligase, and calf intestinal alkaline phosphatase were purchased from Roche Diagnostics Ltd. (Lewes, East Sussex, United Kingdom) or New England Biolabs Ltd. (Hitchin, United Kingdom) and were used as recommended by the manufacturers. Electroporation of plasmid DNA into E. coli (40) and L. lactis (10) was performed essentially as previously described. Electrotransformation of B. breve with plasmid DNA was performed as follows. Mid-logarithmic-phase cells (optical density at 600 nm, 0.5 to 0.6) were chilled on ice for 20 min, and this was followed by centrifugation. The cell pellet was washed twice and resuspended in 0.5 M sucrose-1 mM citrate buffer (pH 5.8). The cells were incubated on ice for 10 min, and this was followed by electrotransformation with a Bio-Rad Gene Pulser II apparatus under the following conditions: 25 μF, 200 Ω, and 2.0 kV cm−1. Modified Rogosa medium (20) was added to the cells, and the mixture was incubated anaerobically at 37°C for 2.5 h prior to plating. PCRs were performed by using either the Taq PCR Master Mix (Qiagen) or the Expand Long Template PCR system (Roche Diagnostics GmbH, Mannheim, Germany) in accordance with the manufacturers' instructions. PCRs were performed with an Omnigene thermal cycler (Hybaid Ltd., Middlesex, United Kingdom).
Screening of genomic libraries and Nuc activity assays.
B. breve UCC2003 genomic libraries were constructed by cloning Sau3A fragments ranging in size from 0.5 to 1.8 kb into the unique dephosphorylated BamHI, BclI, or BglII sites of pFUN. Ligation mixtures were introduced into E. coli DH5α as the intermediate host by electrotransformation. Libraries were then established in L. lactis MG1363 by electroporation of plasmid DNA prepared from approximately 10,000 E. coli transformants. An insertion rate of approximately 70% was observed. Lactococcal transformants which were Nuc+ were selected for further study. A Nuc+ phenotype was detected by using a chromogenic toluidine blue-DNA-agar overlay (22, 23). Nuclease activity in the supernatants of cultures (see below) was determined spectrophotometrically, essentially as described previously (17). For all nuclease assay procedures, strains containing plasmids pVE8009 and pVE8010 (38) were used as positive and negative controls, respectively.
Sequence analysis and bioinformatics.
The pFUN derivatives of nuclease-positive clones that were obtained were characterized by PCR amplification of the pFUN insert from plasmid DNA preparations. Primers were designed for either side of the multiple cloning site; one primer corresponded to a sequence in the Rho-independent terminator (5′ GTTAGCTCACTCATTAGG 3′), and the other was complementary to the 5′ end of the truncated nuclease gene (5′ TGCACTTGCTTCAGGACC 3′). PCR products were purified by using the CONCERT rapid PCR purification system (GibcoBRL, Paisley, Scotland) and sequenced. Sequencing was performed by MWG-BIOTECH AG (Ebersberg, Germany). Sequence data assembly and analyses were performed by using the DNASTAR software (version 5.05; DNASTAR, Madison, Wis.). Database searches were performed by using nonredundant sequences at the National Center for Biotechnology Information web site (http://www.ncbi.nlm.nih.gov) and the tBlastN, tBlastX, and BlastP programs (1, 2). Sequence alignments were constructed by using the Clustal method of the MEGALIGN program of the DNASTAR software package. Functional domains in deduced proteins were identified by using the SMART database (42, 43; http://smart.embl-heidelberg.de). Classical SPs and their cleavage sites were predicted by using the Signal Peptide Prediction program with gram-positive data (32, 33; www.cbs.dtu.dk/services/SignalP). Likely transmembrane domains were determined by using the HMMTOP server (49; www.enzim.hu/hmmtop) and the DAS (Dense Alignment Surface) transmembrane prediction server (8; www.sbc.su.se/∼miklos/DAS/maindas.html).
Location studies, SDS-PAGE, Western blotting, zymograms, and quantitative nuclease assays.
The localization of all ΔSPNuc fusion proteins was examined in whole cells, culture supernatants, and membrane vesicles of mid-exponential-phase B. breve cells (optical density at 600 nm, 0.5 to 0.6). Whole cells and supernatant fractions were isolated essentially as described previously (38), with modifications for Bifidobacterium. Briefly, following harvesting and washing of the pellet, cells were resuspended in Tris-EDTA containing lysozyme (final concentration, 30 mg ml−1) and incubated at 37°C for 30 min. The cells were then lysed with sodium dodecyl sulfate (SDS) (final concentration, 4%) for 15 min on ice. Membrane vesicles were isolated essentially as described previously (35). The procedure was slightly modified for use with Bifidobacterium by using a variation of the lysozyme step (30 mg ml−1 for 30 min at 37°C). Equal volumes of loading buffer (60 mM Tris-HCl, 2% SDS, 10% glycerol, 0.01% bromophenol blue, 200 mM dithiothreitol) were added to cell pellet, supernatant, or membrane fractions, and 30 μl was loaded onto an SDS-polyacrylamide gel electrophoresis (PAGE) gel prior to electrophoresis.
For immunoblot experiments, proteins separated by SDS-PAGE were blotted electrophoretically onto nitrocellulose filters (Schleicher and Schuell, Dassel, Germany) as described previously (25). Custom-made polyclonal anti-Nuc rabbit antibodies (raised against the peptide EFDKGQRTDKYGRG) were obtained from ProSci Inc. (Poway, Calif.) and used according to the instructions of the manufacturer. Immunodetection was performed with peroxidase-conjugated goat anti-rabbit immunoglobulins (Dako A/S, Glostrup,Denmark) and an enhanced chemiluminescence kit (Amersham Biosciences UK Limited, Little Chalfont, Buckinghamshire, United Kingdom) as recommended by the suppliers. Nuclease activity was evaluated on zymograms of SDS-PAGE gels after removal of SDS, as described previously (26). Samples of concentrated supernatant fractions of both B. breve and L. lactis were compared by using a spectrophotometric assay based on the release of acid-soluble oligonucleotides following nuclease digestion of DNA (17).
Nucleotide sequence accession numbers.
The sequences of the pFUN inserts encoding the polypeptides fused to ΔSPNuc have been deposited in the GenBank database under the following accession numbers: AY297716 (Sec1), AY297717 (Sec2), AY297718 (Sec3), AY297719 (Tmp1), AY297720 (Tmp2), AY297721 (Tmp3), and AY297722 (Tmp4).
RESULTS
Comparison of Sec machinery in B. breve, B. longum, and other gram-positive bacteria.
Database searches, alignment, and comparative analyses were performed for the known components of the Sec machinery of gram-positive bacteria by using the preliminary genome sequence of B. breve UCC2003 (S. Leahy, J. Moreno-Munoz, G. F. Fitzgerald, D. Higgins, and D. van Sinderen, unpublished data) and genome sequences of B. longum NCC2705 and DJO10A (Table 1). Components of the secretion machinery found in B. breve UCC2003 include the signal recognition particle proteins Ffh and FtsY, the chaperones GroEL, GroES, GrpE, DnaJ, DnaK, and Tf (trigger factor), and the SecA, SecE, SecG, SecY, and YajC components of the major translocation pathway (47). No SecDF homologue and no components of the Tat pathway were observed in any of the Bifidobacterium spp. Two YidC homologues and two PrsA-like peptidylprolyl isomerase homologues (involved in protein folding) were also identified. Two SPases I, an SPase II (for cleavage of lipoprotein SPs and coupling to membrane lipids), and three sortase homologues were also identified.
TABLE 1.
Homologues of export machinery identified in B. breve UCC2003
| Protein | % Amino acid identity in B. longum NCC2705 | % Amino acid identity in B. longum DJO10A |
|---|---|---|
| SecA | 88 | NDa |
| SecE | 93 | 92 |
| SecG | 50 | 51 |
| SecY | 94 | 91 |
| SPase I-1 | 61 | 61 |
| SPase I-2 | 55 | 72 |
| SPase II | 66 | 66 |
| DnaJ | 77 | 77 |
| DnaK | 95 | 95 |
| Tf | 92 | 96 |
| GroEL | 81 | 81 |
| GroES | 97 | 97 |
| GrpE | 67 | 67 |
| Ffh | 84 | 84 |
| FtsY | 73 | 73 |
| YajC | 75 | 75 |
| YidC 1 | 84 | 84 |
| YidC 2 | 95 | 95 |
| PrsA 1 | 97 | 97 |
| PrsA 2 | 96 | 97 |
| Sortase 1 | 78 | 78 |
| Sortase 2 | 78 | 88 |
| Sortase 3 | 92 | 90 |
ND, not detected (for any sequence matches that were not in the first 100 database hits).
Identification of B. breve exported proteins in L. lactis.
Libraries of B. breve UCC2003 genomic DNA were established in pFUN by using the unique BamHI, BclI, or BglII sites, which allowed construction of translational fusions in any of the three possible reading frames. A Nuc+ phenotype was observed in approximately 0.2% of the transformants obtained, which were subjected to DNA sequence analysis. Plasmids pVE8009 (positive control) and pVE8010 (negative control) are pFUN derivatives (38) containing translational fusions between usp45 (50) and Δnuc (nuclease gene lacking the export signal). For both of these plasmids, fusion expression is driven by the usp45 promoter and its original translational start site, followed by the intact Usp45 SP sequence in the case of pVE8009 or a deleted and therefore inactive version of this sequence in the case of pVE8010.
All vectors harboring ΔSPNuc fusions and displaying a Nuc+ phenotype were introduced into B. breve UCC2003 for location studies and activity assays. Successful transformation of the appropriate plasmid DNA into B. breve was confirmed by PCR and sequence analysis. All clones displaying a Nuc+ phenotype in L. lactis exhibited a similar Nuc+ phenotype in B. breve. Ten nonredundant fusions were identified among the clones sequenced. All seven fusions which displayed a strong Nuc+ phenotype contained a putative export signal (represented by a predicted SP sequence or putative transmembrane domains) (see below). Three fusions which had a delayed Nuc+ phenotype did not contain any recognizable export signal and probably corresponded to cytoplasmic proteins (data not shown). The phenotypes of these fusions may have been due to cell lysis or leakage, as described previously (38), and these proteins are not discussed further here.
Sec proteins.
Three B. breve polypeptides isolated as active ΔSPNuc fusions, designated Sec (for putative secreted protein), contained a predicted N-terminal classical SP (i.e., an SP composed of an N-terminal positively charged region, a central hydrophobic core, and a C-terminal cleavage region) (Table 2). Sequence analysis with the sequenced genome of B. breve UCC2003 (Leahy et al., unpublished) revealed the full protein-encoding open reading frame in each case (Table 3). Sec1 is predicted to contain a classical SP with a single transmembrane region. The portion of this protein fused to the nuclease comprises the first 77 amino acids of a putative 602-amino-acid protein. This protein is significantly similar to the permease component of an ABC-type transport system, and clear homologues of the gene are found in B. longum NCC2705 (accession no. NP_695398) and DJO10A (ZP_00121339) (Table 3). Homologues of the gene are also found in Enterobacter (NP_815308) and Lactobacillus (NP_786841) (data not shown), both of which are present in the gastrointestinal tract, indicating that the gene may be involved in a function specific to bacterial inhabitants of the gastrointestinal tract.
TABLE 2.
Putative B. breve UCC2003 export signals (fused to ΔSPNuc) identified in this study
| ΔSPNuc fusions | Size (amino acids)a | Putative export signal sequenceb |
|---|---|---|
| Sec polypeptides | ||
| Sec1 | 77 | MAQRNTFREDEELEESINLHDIARVGKYLKPYISRIVRILAVVVSMSCIVVSVPYLTKIMIDDA↓IPNKDLGKLAMLA |
| Sec2 | 81 | MEHMKMFRRLSSVVVIVLLMPLILVAMPVPAAQA↓DQLPNPDWVAL-36 |
| Sec3 | 95 | 25-DSVNEAHRTLANMREQIAADDDRILQLQAQLQEERNKKSQGNTFASLGANAQQMLASA↓EQTSSELLERAK |
| Tmp polypeptides | ||
| Tmp1 | 66 | MTLMAGRERRSMMTGAQASHCGSVSAISLGLPVSTAIPEAKGILPKALFVGKAPISGKLKQRFVNE |
| Tmp2 | 226 | METSSIELWHGSSHVIKHPEYGMGKPNNDYGRGFYCTRSIELAKEWACAGLDDGFANRYTLTTDGLTFLDLSQPPYTILNWLALL VENRRFQPTTAVAAQ-126 |
| Tmp3 | 63 | MLTAPACGSSCGSRLAGQEDAVGVGSLAAGTQRVQQFVEPESLIQSGKRLLGGPHLQIILMIK |
| Tmp4 | 592 | 155-YAVSGGLHGVGISVVNALSTHVDIEVRRQGFHWTQTYVDQHPVAPLKQGEPMAEDESTGTSVTFWADPKIFETTIYDFETL RSRFQQMAFLNKGLKLSLTDERVTDQAGDEVAGDAEGEPGEKHQTVTYQYLNGIKDYVDYLVKVRKATPVEEDVISFEAEDLK LGISAELAMQWTTAYSEAVHTFANTISTTEGGTHEEGFRAALTSLVNRYARDKAILKDKDENLSGDDVREGLTAVISVKLTNPQF EGQTKTKLGNSEAKTFVQRVMTDKLGDWFDAHPAEAKNIIQKAIEASRARLAAKKARENTRRKSIFESAGMPDKLKDCQSSNPEE CELFIVEGDSAGGSAIQGRNPITQAILPLRGKILNTERASLDRMMKSDTIESLITAVGGGYGEDFDISKVRYHKVIIMADADVDGAH IATLNLTLFFRYMRPM |
Number of amino acids fused to ΔSPNuc.
For Sec and Tmp polypeptides, classical SPs and transmembrane domains, respectively, are shown. For the export signal sequences shown, charged amino acids are indicated by italics, stretches of hydrophobic amino acids predicted to form a transmembrane domain are underlined, SP cleavage regions (amino acids −3 to 1) are indicated by boldface type, and predicted cleavage sites are indicated by arrows. The numbers to the right of the export signal indicate the length of the polypeptide that is joined at that end in the fusion. The start sites correspond to the most-N-terminal methionine (M), valine (V), or leucine (L) following a consensus ribosome-binding site.
TABLE 3.
Homologues of B. breve UCC2003 polypeptides identified as active ΔSPNuc fusions
| ΔSPNuc fusion | Putative function | % Amino acid identity in B. longum DJO10A | % Amino acid identity in B. longum NCC2705 |
|---|---|---|---|
| Sec polypeptides | |||
| Sec1 | ABC transport system, permease component | 87 | 87 |
| Sec2 | Putative serine protease, acid phosphatase domain | 59 | 59 |
| Sec3 | Possible adhesion protein | 85 | 85 |
| Tmp polypeptides | |||
| Tmp1 | Hypothetical protein | 95 | 94 |
| Tmp2 | Hypothetical protein | 85 | 86 |
| Tmp3 | Hypothetical protein | NPa | 82 |
| Tmp4 | DNA gyrase, B subunit | 94 | 94 |
NP, not present.
Sec2 contains a classical SP and a single transmembrane domain. The cloned portion of this protein consists of the first 81 amino acids of a putative 482-amino-acid protein. The complete protein appears to be a homologue of a hypothetical secreted protein from B. longum NCC2705 (accession no. NP_695256) and is also encoded by the genomes of B. longum DJO10A (ZP_00121565) and Streptomyces coelicolor (NP_630514). Sec2 also displays significant similarity to serine proteases from Agrobacterium tumefaciens (NP_356094) and Corynebacterium glutamicum (NP_600418). A probable acid phosphatase domain (COG0671) was identified in Sec2, which indicates involvement in lipid metabolism. This domain was also identified in the A. tumefaciens and C. glutamicum homologues.
Sec3 is predicted to contain a classical Sec-type SP. The fused portion of this protein is homologous to the N-terminal regions of approximately 530-amino-acid hypothetical proteins from B. longum NCC2705 (accession no. NP_695526) and DJO10A (ZP_00121034). Sec3 also displays significant similarity to a hypothetical protein from Thermobifida fusca (ZP_00057200) and a putative M protein from Streptomyces (NP_824034).
Tmp proteins.
Four B. breve UCC2003 polypeptides (Table 2) containing at least one predicted transmembrane domain were identified as active ΔSPNuc fusions and were designated Tmp (for putative transmembrane protein). Sequence analysis of B. breve revealed the complete protein-encoding open reading frame in each case (Table 3). The fused portion of Tmp1 corresponds to the N-terminal region of a hypothetical protein and is predicted to have a C-out topology, which is expected for transmembrane proteins (Nuc activity of a fusion indicates that the ΔSPNuc domain is exported). Proteins similar to Tmp1 have been found only in B. longum NCC2705 (accession no. NP_696268) and DJO10A (ZP_00120937) (Table 3), although no function has been assigned to these proteins.
The portion of Tmp2 fused to ΔSPNuc contains a single transmembrane domain and has a C-out topology. Tmp2 is similar to hypothetical proteins from B. longum NCC2705 (accession no. NP_695850) and DJO10A (ZP_00121150) and to a lesser extent to a protein from Clostridium (NP_782940) and a hypothetical protein from Lactobacillus (ZP_00047154). No conserved domains were detected, and consequently no putative function could be assigned to this protein.
Tmp3 contains a single transmembrane domain. B. longum NCC2705 contains a homologue of this protein (accession no. NP_695675); however, Tmp3 appears to be unique to these two bacteria and does not exhibit similarity to any other protein identified to date (Table 2). It is notable that no homologue of Tmp3 was identified in B. longum DJO10A; however, it is possible that this putative protein has not been identified yet, as the available genome sequence is incomplete. No conserved domains were detected; thus, it is not possible to attribute a (putative) function to this protein.
Tmp4 contains two transmembrane domains. This protein exhibits highly significant similarity to DNA gyrase B subunits from B. longum NCC2705 (accession no. NP_695821) and DJO10A (ZP_00121174). Homologues of Tmp4 are found in a wide variety of bacterial species and are known to be involved in DNA replication and repair.
Locations and activities of fusion proteins.
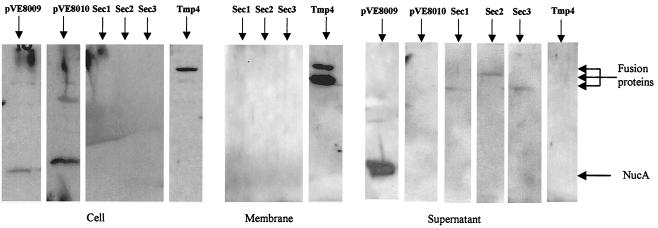
The locations of all exported ΔSPNuc fusions were examined (see Materials and Methods), and the results are shown in Fig. 1. The mature NucA form was found to be mainly associated with the pellet fraction in the case of pVE8010 and was almost exclusively located in the supernatant fraction in the case of pVE8009 (Fig. 1). Sec1-ΔSPNuc, Sec2-ΔSPNuc, and Sec3-ΔSPNuc were found almost exclusively in the medium, thus confirming that these fusion proteins are secreted (Fig. 1). Full-size Tmp4-ΔSPNuc was located mainly in the membrane vesicle fraction (Fig. 1). Similar results were obtained for Tmp1-ΔSPNuc, Tmp2-ΔSPNuc, and Tmp3-ΔSPNuc (data not shown). Zymograms performed for SDS-PAGE gels by using concentrated supernatant samples of B. breve UCC2003 derivatives harboring Sec1-ΔSPNuc, Sec2-ΔSPNuc, and Sec3-ΔSPNuc demonstrated the enzymatic activity of the full-length fusion forms (data not shown).
FIG. 1.
Locations of Usp-ΔSPNuc (pVE8009), ΔSPUsp-ΔSPNuc (pVE8010), Sec1-ΔSPNuc (Sec1), Sec2-ΔSPNuc (Sec2), Sec3-ΔSPNuc (Sec3), and Tmp4-ΔSPNuc (Tmp4) fusion proteins in B. breve UCC2003. The identities of fusion proteins are indicated above the lanes. Cell, membrane, and supernatant fractions from mid-exponential-phase cultures of the strains were analyzed by SDS-PAGE and Western blotting by using polyclonal Nuc antibodies. Putative forms of each fusion are indicated. Results similar to those presented for Tmp4-ΔSPNuc were obtained for Tmp1-ΔSPNuc, Tmp2-ΔSPNuc, and Tmp3-ΔSPNuc. The mature NucA band indicated is a faint band, but its location and activity were confirmed by zymograms.
Quantitative nuclease assay with supernatant fractions of L. lactis and B. breve.
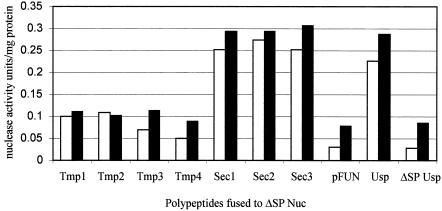
Quantitative nuclease assays (Fig. 2) revealed that Sec clones exhibited the highest activity in both L. lactis and B. breve. This indicates that the SPs (identified in L. lactis) act in comparable ways in the secretion process in these two bacteria.
FIG. 2.
Nuclease activities of the polypeptides identified, as determined by the method of Heins et al. (17). pFUN harboring no insert was used as a negative control. Usp corresponds to plasmid pVE8009, and ΔSP Usp corresponds to plasmid pVE8010. Open bars, fusion proteins in L. lactis MG1363; solid bars, fusion proteins in B. breve UCC2003.
DISCUSSION
In this work we aimed to study protein export in B. breve. As the SPs identified in this study were shown to function in both L. lactis and B. breve, it was expected that the machinery by which Bifidobacterium species export proteins would be comparable to that of other gram-positive bacteria. Homology studies indicated that all expected components of the gram-positive Sec machinery (with the exception of a SecDF homologue) are present in the genomes of B. breve UCC2003 and B. longum NCC2705 and DJO10A (see above). The apparent lack of a SecDF homologue has also been reported recently for a Lactobacillus species (21). SecDF is required for efficient secretion of proteins; however, it is not essential for translocation in B. subtilis, and its precise function has yet to be established (6). Also present are homologues of signal recognition particle proteins and general chaperones, as well as a number of predicted SPases and sortases. Two YidC homologues were found that may also play a role in the secretion pathway, as it has been shown that in E. coli YidC associates with the Sec translocase (44). No components of a Tat pathway were identified in the genomes of the three Bifidobacterium spp. investigated. As it is known that the Tat pathway is involved in exporting folded proteins, this indicates that Bifidobacterium predominantly exports proteins in an unfolded conformation. The Tat pathway also appears to be missing from the genome of Lactobacillus mentioned above (21). Thus, the results of this study indicate that the secretory machinery of Bifidobacterium is comparable to that of other gram-positive bacteria.
This is one of the first reports of identification of exported proteins in the genus Bifidobacterium. In fact, there has been only one other report of an SP in Bifidobacterium, which was identified in a probable extracellular β-galactosidase from B. bifidum (30). The nuclease gene, which was first isolated from S. aureus, has previously been used to identify exported proteins in a number of gram-positive hosts, including Lactococcus (38) and Corynebacterium (26). It was demonstrated in this study that nuc (in the vector pFUN [38]) is a reliable reporter for export studies in Bifidobacterium.
Three Sec-dependent SPs were identified in this study, and they have SP cleavage regions comparable to those previously identified for other gram-positive bacteria (39, 46, 54). The SPs appear to be longer in Bifidobacterium than in other gram-positive bacteria, comprising between 35 and 84 amino acids (this study), compared with an average length of 15 to 30 amino acids (51). This may be due to differences in the cell wall structure of Bifidobacterium; however, a preliminary investigation of the average SP length in B. longum NCC2705 yielded a length of 44 amino acids (unpublished data). The fusion proteins are also consistently longer than the proteins described in similar studies (in which other reporter systems were used). This result is consistent with results previously obtained with the pFUN vector (38). The −3 to −1 cleavage regions for the three SPs (Table 1) also appear to be similar to the consensus A-X-A motif. Also noteworthy are the two helix-breaking proline residues located at positions −5 and −7 in Sec2. It has previously been reported that 50% of SPs in Bacillus subtilis contain a helix-breaking residue (proline or glycine) at a position between positions −7 and −4 relative to the predicted processing site for SPase I (47).
Sec1 is a probable permease component of an ABC transport system, a function that is consistent with its membrane location; however, the fact that this protein is subject to secretion is inconsistent with its assumed permease function. It is possible that fortuitous cloning of this fragment of the permease resulted in secretion of the nuclease as this moiety functions as an artificial SP. Alternatively, the complete protein may harbor a C-terminal anchor motif, which would target the permease for retention in the cell wall or membrane. As only the N-terminal region of this (putative) protein was cloned in this study, it is possible that the permease component was erroneously released from the membrane and secreted into the medium. Sec2 harbors an acid phosphatase domain and also displays homology to a serine protease. Sec3 has similarity with a cell surface-associated M protein and may be involved in adhesion of the bifidobacterial strain to host epithelial cells. Four of the SPs identified are putative transmembrane proteins harboring one or more transmembrane domains. Tmp3 harbors two predicted stretches of hydrophobic acids at either end of the fusion, which may facilitate insertion of the protein into the membrane in a hairpin-like manner. The position of a putative DNA gyrase (Tmp4) at the cell surface is both unexpected and inconsistent with the function of this protein. It is more likely that the transmembrane domains (fused to ΔSPNuc) caused improper insertion of the nuclease into the cell membrane, thus translocating the nuclease to the cell surface.
Tmp1 and Tmp3 appear to be unique to the three Bifidobacterium strains examined, as no homologues in other bacterial species were identified. Tmp3 was notably absent from B. longum DJO10A, although this may have been because the entire genome sequence was not available. Homologues of Tmp2 have been found in Clostridium and Lactobacillus, in addition to the two B. longum strains investigated. No functions could be assigned to these three putative exported proteins due to a lack of conserved domains or homology with proteins having known functions.
Western blotting with anti-Nuc polyclonal antibodies confirmed that the three Sec fusion proteins were exported to the culture supernatant, and the activities of the full-length mature forms were demonstrated by using zymograms. The nuclease activities of the secreted proteins were demonstrated to be substantially higher (in the supernatant fractions of both L. lactis and B. breve) than the nuclease activities of the other clones and were actually higher than the activity of the positive control (see above). Each of the four transmembrane proteins was shown by Western blotting to be mainly associated with the membrane fraction. Taken together, the results described above indicate that the polypeptides that are signals for protein export operate at comparable levels in L. lactis and B. breve.
There are a number of limitations when the screening strategy is employed in L. lactis. Screening for exported proteins of bifidobacterial origin in Lactococcus may result in a bias in the SPs identified, due in part to differences in both the G+C content and promoter motifs. Thus, there are likely to be (as-yet-unidentified) SPs in B. breve which do not target the nuclease for export in a heterologous host (namely, L. lactis). Although it would be preferable to screen for exported proteins directly in B. breve, this is not possible at present due to low electrotransformation frequencies (102 to 103 transformants per μg of DNA for this strain [unpublished results]). There is also a paucity of tools available for genetic manipulation of bifidobacterial species. It is noteworthy, for the purposes of future molecular cloning, that the pAMβ1 origin (45) of pFUN, which replicates in various gram-positive bacteria, is also active in Bifidobacterium. We found that the low copy number of this replicon in B. breve is roughly comparable to the copy number in L. lactis (data not shown).
It can be concluded from this study that proteins exported by Bifidobacterium contain either an SP similar to that of other gram-positive bacteria or a number of transmembrane region(s). Also, a number of these signals are heterologous, in that they function in both Lactococcus and Bifidobacterium. It is likely that a number of these exported proteins are important in the interactions of B. breve with its host environment. Future work should focus on functional analysis of the identified exported proteins and should result in a greater understanding of Bifidobacterium protein export. In addition, optimization of such mechanisms may greatly enhance the probiotic properties of members of this genus.
Acknowledgments
We are very grateful to Isabelle Poquet for providing plasmids pFUN, pVE8009, and pVE8010 and for helpful discussions during this study.
This work was financially supported by Enterprise Ireland (grant BR/1998/202), by the Higher Education Authority Programme for Research in Third Level Institutions, and by the Science Foundation Ireland Centre for Science Engineering and Technology.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tools. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schafer, J. Hang, Z. Hang, W. Miller, and D. J. Lipman. 1997. Gapped Blast and PSI-Blast: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banerjee, S., and J. N. Hansen. 1988. Structure and expression of a gene encoding the precursor of subtilin, a small protein antibiotic. J. Biol. Chem. 263:9508-9514. [PubMed] [Google Scholar]
- 4.Berks, B. C., F. Sargent, and T. Palmer. 2000. The Tat protein export pathway. Mol. Microbiol. 5:260-274. [DOI] [PubMed] [Google Scholar]
- 5.Birnboim, H. C., and J. Doly. 1979. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 7:1513-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bolhuis, A., C. P. Broekhuizen, A. Sorokin, M. L. van Roosmalen, G. Venema, S. Bron, W. J. Quax, and J. M. van Dijl. 1998. SecDF of Bacillus subtilis, a molecular Siamese twin required for the efficient secretion of proteins. J. Biol. Chem. 273:21217-21224. [DOI] [PubMed] [Google Scholar]
- 7.Chen, M., and V. Nagarajan. 1994. Effect of alteration of charged residues at the N termini of signal peptides on protein export in Bacillus subtilis. J. Bacteriol. 176:5796-5801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cserzo, M., E. Wallin, I. Simon, G. von Heijne, and A. Elofsson. 1997. Prediction of transmembrane alpha-helices in procariotic membrane proteins: the dense alignment surface method. Protein Eng. 10:673-676. [DOI] [PubMed] [Google Scholar]
- 9.de Man, J., M. Rogosa, and M. E. Sharpe. 1960. A medium for the culture of lactobacilli. J. Appl. Bacteriol. 23:130-135. [Google Scholar]
- 10.de Vos, W. M., P. Vos, H. de Haards, and I. Boerrigter. 1989. Cloning and expression of the Lactococcus lactis subsp. cremoris SK11 gene encoding an extracellular serine protease. Gene 85:169-176. [DOI] [PubMed] [Google Scholar]
- 11.Dunne, C. 2001. Adaptation of bacteria to the intestinal niche: probiotics and gut disorder. Inflamm. Bowel Dis. 7:136-145. [DOI] [PubMed] [Google Scholar]
- 12.Ferrari, E., A. S. Jarnagin, and B. F. Schmidt. 1993. Commercial production of extracellular enzymes, p. 917-937. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and other gram-positive bacteria. American Society for Microbiology, Washington, D.C.
- 13.Gasson, M. J. 1983. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J. Bacteriol. 154:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gennity, J., J. Goldstein, and M. Inouye. 1990. Signal peptide mutants of Escherichia coli. J. Bioenerg. Biomembr. 22:233-269. [DOI] [PubMed] [Google Scholar]
- 15.Guarner, F., and J.-R. Malagelada. 2003. Gut flora in health and disease. Lancet 360:512-519. [DOI] [PubMed] [Google Scholar]
- 16.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 17.Heins, J., J. Suniano, H. Taniuchi, and C. Anfinsen. 1967. Characterization of a nuclease produced by Staphylococcus aureus. J. Biol. Chem.242:1016-1020. [PubMed]
- 18.Hoffman, C. S., and A. Wright. 1985. Fusions of secreted proteins to alkaline phosphatase: an approach for studying protein secretion. Proc. Natl. Acad. Sci. 82:5107-5111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hols, P., A. Baulard, D. Garmyn, B. Delplace, S. Hogan, and J. Delcour. 1992. Isolation and characterisation of genetic expression and secretion signals from Enterococcus faecalis through the use of broad-host-range α-amylase probe vectors. Gene 118:21-30. [DOI] [PubMed] [Google Scholar]
- 20.Iwata, M., and T. Morishita. 1989. The presence of plasmids in Bifidobacterium breve. Lett. Appl. Microbiol. 9:165-168. [Google Scholar]
- 21.Kleerebezem, M., J. Boekhorst, R. van Krankenberg, D. Molenaar, O. P. Kuipers, R. Leer, R. Tarchini, S. A. Peters, H. M. Sandbrink, M. W. E. J. Fiers, W. Stiekema, R. M. Klein Lankhorst, P. A. Bron, S. M. Hoffer, M. N. N. Groot, R. Kerkhoven, M. de Vries, B. Ursing, W. M. de Vos, and R. J. Siezen. 2003. Complete genome sequence of Lactobacillus plantarum WCFS1. Proc. Natl. Acad. Sci. 100:1990-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lachica, R. V. F., C. Genigeorgis, and P. D. Hoeprich. 1971. Metachromatic agar-diffusion methods for detecting staphylococcal nuclease activity. Appl. Microbiol. 21:585-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Le Loir, Y., A. Gruss, S. D. Ehrlich, and P. Langella. 1994. Direct screening of recombinants in gram-positive bacteria using the secreted staphylococcal nuclease as a reporter. J. Bacteriol. 176:5135-5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Le Loir, Y., S. Nouaille, J. Commissaire, L. Bretigny, A. Gruss, and P. Langella. 2001. Signal peptide and propeptide optimisation for heterologous protein secretion in Lactococcus lactis. Appl. Environ. Microbiol. 67:4119-4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liebl, W., and F. Götz. 1986. Studies on lipase directed export of Escherichia coli β-lactamase in Staphylococcus carnosus. Mol. Gen. Genet. 204:166-173. [DOI] [PubMed] [Google Scholar]
- 26.Liebl, W., A. J. Sinskey, and K. H. Schleifer. 1992. Expression, secretion, and processing of a staphylococcal nuclease by Corynebacterium glutamicum. J. Bacteriol. 174:1854-1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lim, E. M., J. Rauzier, J. Timm, G. Tooea, A. Murray, B. Gicquel, and D. Portnoi. 1995. Identification of Mycobacterium tuberculosis DNA sequences encoding exported proteins by using phoA gene fusions. J. Bacteriol. 177:59-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lory, S. 1994. Leader peptidase of type IV prepilins and related proteins, p. 17-29. In G. von Heijne (ed.), Signal peptidases. R. G. Landes Company, Austin, Tex.
- 29.Manoil, C., and J. Beckwith. 1985. TnphoA: a transposon probe for protein export signals. Proc. Natl. Acad. Sci. 82:8129-8133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Møller, P. L., F. Jørgensen, O. C. Hansen, S. M. Madsen, and P. Stougaard. 2001. Intra- and extracellular β-galactosidases from Bifidobacterium bifidum and B. infantis: molecular cloning, heterologous expression, and comparative characterization. Appl. Environ. Microbiol. 67:2276-2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Msadek, T., F. Kunst, and G. Rapoport. 1993. Two-component regulatory systems, p. 729-745. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and other gram-positive bacteria. American Society for Microbiology, Washington, D.C.
- 32.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. A neural network method for identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Int. J. Neural Syst. 8:581-599. [DOI] [PubMed] [Google Scholar]
- 33.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 34.O'Riordan, K. 1998. Studies on antimicrobial activity and genetic diversity of Bifidobacterium species: molecular characterisation of a 5.75 kb plasmid and a chromosomally encoded recA gene homologue from Bifidobacterium breve. Ph.D. thesis. National University of Ireland, Cork, Ireland.
- 35.Otto, R., R. G. Lageveen, H. Veldkamp, and W. N. Konings. 1982. Lactate efflux-induced electrical potential in membrane vesicles of Streptomyces cremoris. J. Bacteriol. 149:733-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paik, S. H., A. Chakicherla, and J. N. Hansen. 1998. Identification and characterization of the structural and transporter genes for, and the chemical and biological properties of, sublancin 168, a novel lantibiotic produced by Bacillus subtilis 168. J. Biol. Chem. 273:23134-23142. [DOI] [PubMed] [Google Scholar]
- 37.Pearce, B. J., Y. B. Yin, and H. R. Masure. 1993. Genetic identification of exported proteins in Streptococcus pneumoniae. Mol. Microbiol. 9:1037-1050. [DOI] [PubMed] [Google Scholar]
- 38.Poquet, I., S. D. Ehrlich, and A. Gruss. 1998. An export-specific reporter designed for gram-positive bacteria: application to Lactococcus lactis. J. Bacteriol. 180:1904-1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pugsley, A. P. 1993. The complete general secretory pathway in gram-negative bacteria. Microbiol. Rev. 57:50-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 41.Schell, M. A., M. Karmirantzou, B. Snel, D. Vilanova, B. Berger, G. Pessi, M.-C. Zwahlen, F. Desiere, P. Bork, M. Delley, R. D. Pridmore, and F. Arigoni. 2002. The genome sequence of Bifidobacterium longum reflects its adaptation the human gastrointestinal tract. Proc. Natl. Acad. Sci. 99:14422-14427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schultz, J., R. R. Copley, T. Doerks, C. P. Ponting, and P. Bork. 2000. SMART: a web based tool for the study of genetically mobile domains. Nucleic Acids Res. 28:231-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schultz, J., F. Milpetz, P. Bork, and C. P. Ponting. 1998. SMART, a simple modular architecture research tool: identification of signalling domains. Proc. Natl. Acad. Sci. 95:5857-5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scotti, P. A., M. L. Urbanus, J. Brunner, J. W. de Gier, G. von Heijne, C. van der Does, A. J. Driessen, B. Oudega, and J. Luirink. 2000. YidC, the Escherichia coli homologue of mitochondrial Oxa1p, is a component of the Sec translocase. EMBO J. 19:542-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Simon, D., and A. Chopin. 1988. Construction of a vector plasmid family and its use for molecular cloning in Streptococcus lactis. Biochimie 70:559-566. [DOI] [PubMed] [Google Scholar]
- 46.Simonen, M., and I. Palva. 1993. Protein secretion in Bacillus species. Microbiol. Rev. 57:109-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tjalsma, H., A. Bolhuis, J. D. Jongbloed, S. Bron, and J. M. van Dijl. 2000. Signal peptide-dependent protein transport in Bacillus subtilis: a genome-based survey of the secretome. Microbiol. Mol. Biol. Rev. 64:515-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tjalsma, H., A. Bolhuis, M. L. van Roosmalen, T. Wiegert, W. Schumann, C. P. Broekhuizen, W. J. Quax, G. Venema, S. Bron, and J. M. van Dijl. 1998. Functional analysis of the secretory precursor processing machinery of Bacillus subtilis: identification of a eubacterial homolog of archaeal and eukaryotic signal peptidases. Genes Dev. 12:2318-2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tusnady, G. E., and I. Simon. 1998. Principles governing amino acid composition of integral membrane proteins: applications to topology prediction. J. Mol. Biol. 283:489-506. [DOI] [PubMed] [Google Scholar]
- 50.van Asseldonk, M., G. Rutten, M. Oteman, R. J. Siezen, W. M. de Vos, and G. Simons. 1990. Cloning of usp45, a gene encoding a secreted protein from Lactococcus lactis subsp. lactis MG1363. Gene 95:155-160. [DOI] [PubMed] [Google Scholar]
- 51.van Dijl, J. M. 1990. Protein export in Bacillus subtilis and Escherichia coli. Ph.D. thesis. Groningen University, Groningen, The Netherlands.
- 52.van Dijl, J. M., A. Bolhuis, H. Tjalsma, J. H. D. Jongbloed, A. de Jong, and S. Bron. 2001. Protein transport pathways in Bacillus subtilis: a genome-based road map, p. 337-355. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.). Bacillus subtilis and its closest relatives: from genes to cells. American Society for Microbiology, Washington, D.C.
- 53.van Dijl, J. M., P. G. Braun, C. Robinson, W. J. Quax, H. Antelmann, M. Hecker, J. Müller, H. Tjalsma, S. Bron, and J. D. H. Jongbloed. 2002. Functional genomic analysis of the Bacillus subtilis Tat pathway for protein secretion. J. Biotechnol. 98:243-254. [DOI] [PubMed] [Google Scholar]
- 54.von Heijne, G. 1988. Transcending the impenetrable: how proteins come to terms with membranes. Biochim. Biophys. Acta 947:307-333. [DOI] [PubMed] [Google Scholar]
- 55.Wu, L.-F., B. Ize, A. Chanal, Y. Quentin, and G. Fichant. 2000. Bacterial twin-arginine signal peptide-dependent protein translocation pathway: evolution and mechanism. J. Mol. Microbiol. Biotechnol. 2:170-189. [PubMed] [Google Scholar]
- 56.Yanouri, A., R. A. Daniel, J. Errington, and C. E. Buchanan. 1993. Cloning and sequencing of the cell division gene pbpB, which encodes penicillin-binding protein 2B in Bacillus subtilis. J. Bacteriol. 175:7604-7616. [DOI] [PMC free article] [PubMed] [Google Scholar]