Abstract
Lactobacillus sakei 5, isolated from malted barley, produces three bacteriocins. Genetic and functional analysis of the purified bacteriocins showed that this strain produces a plasmid-encoded bacteriocin that is identical to sakacin P, as well as two novel, chromosomally encoded bacteriocins, which were designated sakacin T and sakacin X. The structural genes specifying sakacin T and sakacin X are part of the sakacin TX locus, which consists of two adjacent but divergently oriented gene clusters. The first gene cluster includes stxP, stxR, stxK, and stxT, which, based on functional and comparative sequence analysis, are believed to encode an inducing peptide and proteins involved in regulation and secretion of these bacteriocins. The second gene cluster includes the structural and immunity genes for sakacin T, a class IIb two-peptide bacteriocin composed of SakTα and SakTβ, and sakacin X, a class IIa bacteriocin. Interestingly, a so-called transport accessory protein was absent from the locus, and based on our results it appears that a dedicated accessory protein is not required for processing and transport of sakacin T and sakacin X.
Malted cereals contain high numbers of various microorganisms representing a wide range of bacteria, yeast, and fungal species (48). Microbial spoilage may result in a variety of technological impediments in the brewing and malting processes. These include damage to raw materials, filtration problems, and deleterious effects on both the fermentation process and the final product, such as turbidity of the beer and undesirable flavors and aromas (23). Any means by which the level of this undesirable microbial contamination can be controlled would be of interest to the brewing industry.
The last decade has seen a growing interest in the application of biopreservation through the use of microorganisms and/or their metabolites to prevent food spoilage and to extend the shelf life of foods (51). Lactic acid bacteria (LAB) are of particular interest as biopreservative organisms. The preserving effects of these organisms are partially due to the production of fermentation end products but may also be due to the formation of small, heat-stable inhibitory peptides, often referred to as bacteriocins (16, 18).
Bacteriocins are ribosomally synthesized, extracellularly released, bioactive peptides or peptide complexes that have a bactericidal or bacteriostatic effect on other (usually closely related) species. Bacteriocins of LAB have previously been arranged into three classes based on their composition, size, mode of action, mechanism of export, and inhibitory spectrum (16, 33, 45). The class I bacteriocins, the so-called lantibiotics, are posttranslationally modified. The class II bacteriocins, which are nonlantibiotic bacteriocins, are further divided into three subgroups. Class IIa bacteriocins are pediocin-like bacteriocins with strong antilisterial effects and a conserved N-terminal YGNGVXC consensus motif in the mature peptide, while class IIb bacteriocins consist of two peptides, both of which are required for full antimicrobial activity. Most class IIa and IIb bacteriocins are synthesized with a double-glycine type of leader sequence (26, 28). While all bacteriocins are formed with an N-terminal leader sequence, some small, heat-stable, and nonmodified bacteriocins are translated with sec-dependent leaders (36, 60). Due to their similarity to the class II bacteriocins these molecules have in the past been placed in a separate subgroup, class IIc (45). There are apparently hybrid bacteriocins which display characteristics of both class IIa and IIb subgroups(12, 13), which has resulted in an alternative class II classification scheme (16), in which subclass IIc is defined as other peptide bacteriocins which do not fulfill the criteria for class IIa or IIb. Class III was defined as large protein bacteriocins. A fourth class of bacteriocins was defined by Klaenhammer (33), and this class contains bacteriocins composed of undefined mixtures of proteins, lipids, and carbohydrates. However, experimental data suggest that the complex bacteriocinogenic activities of these molecules may be artifacts caused by interactions between constituents from the cells or growth medium and that the undefined bacteriocin activities are likely to be activities of regular peptide bacteriocins, and thus recognition of this separate class may not be valid. Kemperman et al. (32) have proposed a new class, class V, of bacteriocins that consist of ribosomally synthesized, nonmodified, head-to-tail-ligated, cyclic, antibacterial peptides, such as circularin A (32) and AS-48 (39).
Recent studies have revealed that there is considerable variation in the number of bacteriocins (11) produced by a particular strain (range, one to three bacteriocins), as well as considerable flexibility in the way in which bacteriocin loci can be organized (16). However, most genetically characterized class II bacteriocin gene clusters are composed of three gene modules, a module that includes the structural and immunity genes, a transport gene module, and a regulatory gene module. The bacteriocin structural gene specifies a prepeptide that is processed during secretion either by dedicated transport machinery or via the sec-dependent pathway. In the case of two-component bacteriocins, the two structural genes are located adjacent to each other. The structural gene for the bacteriocin is cotranscribed with the corresponding immunity gene located downstream, although there are exceptions to this genetic organization (21, 22). Most bacteriocins are produced as a precursor with an N-terminal double-glycine leader peptide sequence, which is removed upon externalization of the bacteriocin by dedicated secretion machinery, which consists of an ATP-binding cassette (ABC) transporter and a so-called accessory protein (16, 25, 26). The transport machinery normally tolerates some variation in the leader peptide sequence, which allows it to transport different precursor peptides (3, 15, 16). Accessory transport proteins are postulated to facilitate membrane translocation and/or help in processing of the leader peptide, although their specific role in the translocation process is not fully understood (18).
Another set of genes that is responsible for the control of the production of many, but not all, bacteriocins is the so-called three-component regulatory system (44), which consists of a secreted bacteriocin-like peptide pheromone, a histidine protein kinase, and a response regulator. This three-component regulatory system acts as a quorum-sensing device, coupling coordinated bacteriocin production by a strain to its cell density (16, 34).
In this paper, we describe functional characterization of a locus involved in the production of a novel class IIb bacteriocin, sakacin T, and a novel class IIa bacteriocin, sakacin X, produced by Lactobacillus sakei 5. Isolation of this strain from malt and purification of the bacteriocins that it produces have been described previously (47, 57).
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. Lactobacillus and Pediococcus strains were cultivated in MRS broth (Oxoid, Basingstoke, England) at 30°C for approximately 16 h prior to experimental use. To obtain nonproducing (Bac−) derivatives, the bacteriocin-producing (Bac+) strain L. sakei 5 was cultivated in wort, which was made by using a standard mashing procedure (Analytica-EBC 4.5.1; relative density, 1.061). Escherichia coli strains were grown at 37°C in Luria-Bertani broth (49) with vigorous agitation. Agar media were prepared by adding 1.5% (wt/vol) granulated agar (Difco) to liquid broth media; overlay agars were prepared by adding 0.7% (wt/vol) granulated agar to the liquid broth media. The antibiotics used in the selective media were added at the following concentrations: ampicillin, 100 μg ml−1; chloramphenicol, 10 μg ml−1; and erythromycin, 200 μg ml−1 (E. coli) or 10 μg ml−1 (Lactobacillus). All chemical reagents were obtained from Sigma, St. Louis, Mo.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Source and/or reference |
|---|---|---|
| Strains | ||
| L. sakei 5 | Producer of sakacin T, sakacin X, and sakacin P | Malt (47, 57) |
| L. sakei LMG2313 | Indicator strain sensitive to sakacin T, sakacin X, and sakacin P | LMGa |
| P. pentosaceus LMG2001 | Indicator strain sensitive to sakacin X only | LMG |
| L. sakei Lb790X | Plasmid-free derivative of L. sakei Lb790; Bac− | 6 |
| L. sakei Lb790(pSAK20) | L. sakei Lb790 containing pSAK20, host strain; Bac− Cmr | 5 |
| L. sakei 23K | Plasmid-free L. sakei | 8 |
| E. coli DH5α | φ80dlacZΔM15 recA1 endA1 gyrA96 thi-1 hsdR17(rK− mK+) supE44 relA1 deoR Δ(lacZYA-argF)U169 | Promega |
| E. coli EC101 | supE thi Δ(lacproAB) (F′ traD36 proAB lacIq ZΔM15) repA | 35 |
| Plasmids | ||
| pBluescript SK(+) | 3-kb E. coli cloning vector; αlacZ Apr | Stratagene |
| pTOPO | 3.9-kb pCR 2.1 TOPO cloning vector; Apr | Invitrogen |
| pNZ44 | E. coli-L. lactis vector containing P44 promoter; Cmr | 42 |
| pLPV111 | 4.2 kb; E. coli-L. plantarum/L. sakei shuttle vector; Emr | 6 |
| pSAK20 | 11.8 kb; contains the operon RIRorf4sapKRTE, necessary for transcriptional activation and processing and transport of sakacin A; Cmr | 6 |
| pLPT5 | pLPV111 sakIT | This study |
| pLPT6 | pLPV111 sakTβsakIT | This study |
| pLPT7 | pLPV111 sakTαsakTβsakIT | This study |
| pLPT8 | pLPV111 sakTαsakIT | This study |
| pLPX14 | pLPV111 sakX sakIX | This study |
| pLPXi23 | pLPV111 sakIX | This study |
| pZ235T | pNZ44 sakIT | This study |
| pZ233X | pNZ44 sakIX | This study |
| pSKΔTE | pSAK20 derivative with a 3.5-kb BamHI deletion; sapK+R+ΔsapTE | This study |
| pSK5T | pSKΔTE stxT | This study |
LMG, Laboratory of Microbial Gene Technology, Ås, Norway.
Bacteriocin activity and immunity assays.
Bacteriocin assays were performed by using either bacterial colonies, cell-free supernatant (CFS), or samples obtained at various stages of the bacteriocin purification procedure as described previously (57). Bacteriocin activity was quantified by critical dilution by using the direct, spot-on-lawn assay (40). The number of activity units per milliliter was determined by determining the inverse of the last dilution at which growth inhibition was still detectable. L. sakei LMG2313 was used as the indicator strain unless otherwise specified.
Bacteriocin purification and characterization of the purified peptides.
Bacteriocins were purified chromatographically and subjected to N-terminal amino acid sequencing and mass spectrometry as described previously (57).
General molecular cloning techniques.
Plasmid DNA was isolated from E. coli strains with a JetQuick plasmid miniprep spin kit (Genomed, Löhne, Germany) and from Lactobacillus strains by the method of Birnboim and Doly (9). E. coli and Lactobacillus strains were transformed by electroporation by using the methods of Sambrook et al. (49) and Aukrust and Blom (4), respectively. All electroporations were carried out with a Gene-Pulser apparatus (Bio-Rad). Restriction enzymes, T4 DNA ligase, and shrimp alkaline phosphatase were used as directed by the manufacturer (Roche Diagnostics, Mannheim, Germany). PCR amplification was performed by standard procedures by using the Expand High Fidelity or Long Template PCR system (Roche Diagnostics) and an Omnigene thermocycler (Hybaid, Ashford, United Kingdom). Primer synthesis and sequencing were performed by MWG Biotech (Ebersberg, Germany). DNA fragments were isolated and purified from agarose gels by using a QIAEX II agarose gel extraction kit (Qiagen, Crawley, United Kingdom). PCR products were purified by using the Concert PCR rapid purification system (Gibco/BRL). Southern blot hybridization onto Hybond-N+ nylon membranes (Amersham, Uppsala, Sweden) was performed by standard methods (49). An enhanced chemiluminesence kit (ECL; Amersham) was used to label the PCR-generated probes.
DNA sequencing.
Total DNA of L. sakei 5 was obtained by the alkaline lysis method of Anderson and McKay (2) and was used as a DNA template for PCRs and DNA sequencing. Total DNA of L. sakei 5 was digested with ClaI, EcoRI, EcoRV, HincII, and HindIII, and the DNA fragments obtained from each of the digests were ligated to dephosphorylated pBluescript II SK+ (Stratagene, La Jolla, Calif.) digested with ClaI, EcoRI, EcoRV, HincII, and HindIII, respectively. Each of the ligation reaction mixtures, representing a restriction fragment library of total DNA of L. sakei 5, was then used as a template to amplify DNA segments near the bacteriocin structural gene by using degenerate primers that were designed on the basis of the amino acid sequences of SakTβ and SakX (57). Two primers, T1D (5′-AARACNAAYTGGGGNTCNGTNGT-3′) and T2 (5′-ATNGCRTCYTGNCCNGCNCC-3′), were designed based on the SakTβ amino acid sequence, and two other primers, Bac-X1 (5′-GGNGGNAARTAYTAYGGNAAYGG-3′) and Bac-X2 (5′-TTCCANCCNGCNGCNCCNCC-3′), were designed based on the sakacin X N-terminal sequence. Specific PCR products were obtained from the ligation reaction templates when a primer designed from the vector sequence was used in combination with one of the degenerate primers. Purified PCR products were cloned into the vector pTOPO (Invitrogen). DNA sequencing was performed by MWG Biotech with the universal forward and reverse primers. Based on the sequence information obtained from these DNA fragments, new sequence-specific primers were synthesized, and the procedure described above was repeated until the complete sequence of a 17-kb chromosomal fragment of L. sakei 5 encompassing the bacteriocin locus was determined. The integrity of this sequence was verified by resequencing the same region with overlapping PCR products, which had been generated by using chromosomal DNA as the template. Assembly and analysis of the DNA sequences were performed by using the DNASTAR software package. Database searches were performed by using the BLAST program (1) with the latest release of the nonredundant databases of the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov).
Plasmid construction and analysis.
The DNA inserts in the pLPV111 derivatives pLPT5, pLPT6, and pLPT7 (Table 1) were generated by using the high-fidelity PCR system (Roche Diagnostics) and primers with introduced PstI and XbaI restriction sites which were ligated into the PstI and XbaI sites of pNZ44. The insert and the P44 promoter were excised from the plasmids by restriction with BglI and XhoI, which are compatible for ligation into the BamHI and XhoI sites of pLPV111. Plasmid pLPT5 contains the proposed immunity gene, sakIT, which is assumed to confer resistance to sakacin T. Plasmid pLPT6 contains only one of the proposed structural genes of sakacin T, sakT, upstream of the proposed immunity gene, while pLPT7 harbors both proposed structural genes for sakacin T activity upstream of the proposed immunity gene. The SOEingtechnique (29) was used to generate a PCR product encompassing the sakTα structural gene upstream of the sakIT gene, which was subsequently cloned into pLPV111 to generate pLPT8. A similar strategy was used for the genes thought to encode sakacin X (sakX) and its immunity protein (sakIX). Two PCR fragments with incorporated restriction sites were obtained; one of these fragments contained sakX and sakIX, and the other contained only sakIX. These two fragments were cloned into the BamHI and XhoI sites of pLPV111 to generate pLPX14 and pLPXi23, respectively. Two other constructs were made; these constructs, pZ235T and pZ233X, contained the putative sakacin T and sakacin X immunity genes, sakIT and sakIX, respectively. The latter genes were amplified with incorporated restriction sites by PCR and cloned into the PstI and XbaI sites of pNZ44.
Three derivatives of pSAK20, which contains the genes necessary for activation of transcription of the sakacin A structural gene, as well as the genes encoding the proteins needed for export and processing of presakacin A (6), were constructed. This plasmid has been successfully used for heterologous expression of other class II bacteriocins (5). All manipulations with pSAK20 were performed by using plasmids isolated from E. coli EC101. Restriction of pSAK20 with BamHI and subsequent self-ligation allowed construction of pSKΔTE, containing a 3.5-kb deletion of the sapT and sapE transport and accessory protein genes. A PCR fragment containing the stxT gene with flanking BamHI restriction sites was cloned into pSKΔTE to obtain plasmid pSK5T.
Induction factor synthesis and induction assays.
The proposed induction factor for sakacin T and sakacin X production (designated IP-TX) and the sakacin P-inducing peptide (IP-673) (10) were synthesized at the Facility for Molecular Biology at the University of Newcastle Upon Tyne (Newcastle Upon Tyne, United Kingdom) and were purified to >97% homogeneity by reverse-phase high-performance liquid chromatography with an acetonitrile-H2O gradient. The molecular weights of the high-performance liquid chromatography-purified peptides were verified by laser desorption mass spectrometry. To assay biological activity, 1-mg ml−1 stock solutions of both IP-TX and IP-673 were prepared in sterile distilled water. To generate a Bac− derivative of L. sakei 5, an overnight culture of this strain was inoculated (1%) into wort and subcultured up to five times at 37°C until the culture medium became turbid. Individual colonies, as well as CFS, were assayed for loss of inhibitory activity against L. sakei LMG2313 (which is sensitive to all three bacteriocins produced by L. sakei 5) and Pediococcus pentosaceus LMG2001 (which is sensitive only to sakacin X). The L. sakei LMG2313 derivatives pZ235T and pZ233X, which contained the putative sakacin T and sakacin X immunity genes sakIT and sakIX, respectively, were also used as indicator cultures. The resulting completely Bac− culture was inoculated (1%) into wort containing the inducing peptide IP-TX (this study) or IP-673 (10) at a concentration of 400 ng ml−1 and incubated at 37°C for 16 h. The cultures were then tested for bacteriocin production.
Nucleotide sequence accession number.
The sequence determined in this study has been deposited in the GenBank database under accession number AY206863.
RESULTS
Sequence analysis of the sakacin TX locus.
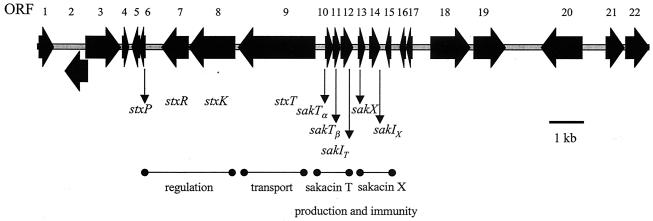
L. sakei 5 produces three bacteriocins, sakacin P, sakacin T, and sakacin X (previously referred to as sakacin 5P, sakacin 5T, and sakacin 5X), which were purified and subjected to N-terminal amino acid sequencing previously (57). Degenerate primers were designed on the basis of the amino acid sequences obtained for one component of sakacin T, SakTβ (see below), and sakacin X. Two PCR products that were approximately 130 bp long were generated, and these products were shown to contain coding regions that were consistent with the derived amino acid sequences (data not shown). Additional rounds of anchored PCR were performed until the complete nucleotide sequence of a 17,243-bp region, referred to as the sakacin TX locus, was determined by several rounds of ligation-anchored PCR. Sequence analysis revealed the presence of 14 complete putative open reading frames (ORFs) and eight incomplete ORFs arranged in divergent operons (Fig. 1) (the ORFs were designated ORFs 1 to 22, but some of them were renamed if functions could be assigned to the encoded products). The ORFs were identified based on the criterion that an ORF consists of least 25 codons preceded by a potential ribosome binding site at an appropriate distance from one of the commonly used initiation codons (24). A nine-gene cluster in the middle of the locus seems to contain all the genes needed for sakacin T and sakacin X production, regulation, and transport (see below).
FIG. 1.
Schematic representation of the sakacin TX locus sequenced to date, obtained by using a combination of ligation-anchored PCR and the primer walking strategy. A total of 22 potential ORFs are shown. The arrows indicate ORFs and the proposed directions of transcription. ORFs with deduced functions in production and secretion of active sakacin T and sakacin X are indicated by stx or sak. Sections of the stx locus assumed to be dedicated to production, immunity, regulation, and transport are indicated.
Analysis of the protein products encoded by the sakacin TX locus.
The protein products encoded by ORFs 6, 10, 11, 13, and 17′ all resemble class II bacteriocin precursors containing an N-terminal extension or leader sequence. The leader sequences of these ORFs conform closely to the consensus leader sequences proposed by Nes et al. (45). Two important features of these leaders are the presence of two glycine residues in the C terminus at positions −2 and −1 relative to the processing site and the remarkable degree of similarity in the hydropathic profiles. Defined distances separate conserved hydrophobic and hydrophilic residues between the conserved residues (Table 2).
TABLE 2.
Prepeptides of the sakacin TX locus containing double-glycine leader sequences
| Protein | Amino acid sequence of prepeptidea | Prepeptide size (amino acids) | Mature peptide size (amino acids) |
|---|---|---|---|
| ORF 6 (IP-TX) | MTNRKTLPKEELKKIKGG TPGGFDIISGGPHVAQDVLNAIKDFFK | 45 | 27 |
| ORF 10 (SakTα) | MKNVQSLSKEELVLVVGG YTAKQCLQAIGSWGIAGTGAGAAGGPAGAFVGAHVGVIAGSAVCIGGFLGQ | 69 | 51 |
| ORF 11 (SakTβ) | MKTANIKLLTNQEMIEIFGG KTNWGSVVGSCVAGGLVGALGGTPISIGAGCLVGAGQDWISQK | 63 | 43 |
| ORF 13 (SakX) | MEAIKKLDLQAMKGIVGG KYYGNGLSCNKSGCSVDWSKAISIIGNNAVANLTTGGAAGWKS | 61 | 43 |
| ORF 17′ | MRKFQKLNEQEMKRLMGGb SSKDCLKDIGKGIGAGTVAGAAGGGCLTGAIGSIWDQW | 56 | 38 |
| Consensusc | -XO-OOX--X-GG |
X, hydrophobic residue; O, hydrophilic residue. The arrow indicates the conserved cleavage site.
Frameshift occurs at this site.
Suggested consensus sequence (45).
Based on the amino acid sequencing results (57), the gene encoding sakacin X activity corresponds to ORF 13 and thus was designated sakX. The N-terminal sequence obtained for sakacin T corresponds to ORF 11, and this gene was designated sakTβ. The product of sakTβ was found to constitute part of a two-component bacteriocin (see below). The second part of this bacteriocin was shown to be encoded by ORF 10, which is located immediately upstream of sakTβ and was therefore designated sakTα. The products of ORF 12 and ORF 14, which were designated sakIT and sakIX, confer immunity to the antimicrobial activities of sakacin T and sakacin X, respectively (see below), and these genes are located immediately downstream of the corresponding bacteriocin structural genes.
The proteins encoded by ORF 6 (designated stxP), ORF 7 (stxR), and ORF 8 (stxK) exhibited significant sequence similarity to a small, cationic pheromone with a double-glycine leader and to response regulator and histidine protein kinase proteins, respectively. These three associated genes therefore seem to represent a so-called three-component regulatory system, similar to those involved in the control of several class II bacteriocins (34, 44).
The deduced protein product of ORF 9, designated stxT, exhibits highly significant similarity to ABC transporter proteins. It has been well established that secretion of bacteriocin peptides possessing a double-glycine leader is mediated by a dedicated membrane-associated translocator belonging to the HlyB ABC transporter superfamily (20, 25, 45). In addition, a so-called accessory protein has been implicated to play an essential, but still unidentified, role in this transport process. The genes encoding the dedicated bacteriocin ABC-type transporter and the accessory protein are usually located adjacent to each other as part of a bacteriocin production gene cluster. However, no complete homologue of an accessory-encoding gene was identified in the sakacin TX locus.
It has been found previously that many LAB contain remnants of bacteriocin gene clusters on both the chromosome and resident plasmids (16, 30). It is therefore not surprising that the sakacin TX locus harbors genes whose deduced proteins are related to bacteriocin production and immunity but which do not seem to be involved in the production of or immunity to sakacin T and sakacin X. ORF 16 displays 63% identity to brcI, the immunity gene for brochocin C. ORF 17, which is located immediately upstream of ORF 16, does not strictly conform to the ORF definition described above in that it is not preceded by a ribosome binding site. A putative leader peptide-encoding sequence designated ORF 17′ precedes ORF 17, but at the DNA sequence specifying the double-glycine residues an apparent frameshift has taken place. The 5′ part of ORF 17 is homologous brcA, while the 3′ part exhibits homology to brcB; brcA and brcB are the two genes encoding the two peptides of brochocin C (41). Furthermore, ORF 5 and ORF 15 appear to represent truncated bacteriocin-related genes, since they encode proteins that are between 45 and 50% identical to the first 45 amino acids (in the case of ORF 15) and 74 amino acids (in the case of ORF 5) to BrcD, the transport accessory protein for brochocin C. The protein products of the remaining ORFs identified in the sakacin TX locus exhibit significant similarity to various transposases and hypothetical proteins with unknown functions produced by different species of LAB (Table 3).
TABLE 3.
Similarities of the proteins encoded by the identified ORFs of the sakacin TX gene cluster to their homologuesa
| ORF (gene) | Size of product (amino acids) | Homologue
|
||
|---|---|---|---|---|
| Identity | Homology | Reference or accession no. | ||
| ORF 1 | 148 | Transposase | 100% identity to TraISLp1 of Lactobacillus plantarum | AF459445b |
| Conserved domain Tra8 of IS30 family involved in DNA replication, recombination, and repair | COG2826b | |||
| ORF 2 | 220 | Tyrosyl-tRNA synthetase | 69% identity to TyrS-1 of Enterococcus faecalis | 14 |
| Conserved domain TyrS involved in translation, ribosomal structure, and biogenesis | COG0162b | |||
| ORF 3 | 332 | Transposase | 40% identity to BH3986 encoding transposase (24) of Bacillus halodurans | 52 |
| Conserved domain transposase_11 necessary for efficient DNA transposition | pfam01609b | |||
| ORF 4 | 34 | Transposase (fragment) | 100% identity to IS1163 of L. sakei L45 | 50 |
| ORF 5 | 88 | Transport accessory protein (fragment) | 45% identity to brcD of Brochothrix campestris | 41 |
| ORF 6 (stxP) | 45 | Inducing peptide | 38% identity to carnobacteriocin A precursor of Carnobacterium piscicola LV17A | 60 |
| ORF 7 (stxR) | 252 | Response regulator | 36% identity to entR of Enterococcus faecium | 46 |
| Conserved domain LytT, response regulator involved in transcription and signal transduction | COG3279b | |||
| ORF 8 (stxK) | 437 | Histidine kinase | 28% identity to entK of Enterococcus faecium | 46 |
| Conserved domain of signal transduction protein | COG2972b | |||
| ORF 9 (stxT) | 723 | ABC transporter | 53% identity to entT of Enterococcus faecium | 46 |
| Conserved domain SunT, ABC-type bacteriocin exporters containing an N-terminal double-glycine peptidase domain | COG2274b | |||
| ORF 10 (sakTα) | 69 | Bacteriocin | 57% identity to brcA of Brochothrix campestris | 41 |
| ORF 11 (sakTβ) | 63 | Bacteriocin | 52% identity to brcB of Brochothrix campestris | 41 |
| ORF 12 (sakIT) | 112 | Bacteriocin immunity protein | No homologue | |
| ORF 13 (sakX) | 61 | Bacteriocin | 71% identity to pisA of Carnobacterium piscicola JG126 | 31 |
| ORF 14 (sakIX) | 97 | Bacteriocin immunity protein | 59% identity to pisI of Carnobacterium piscicola JG126 | 31 |
| ORF 15 | 45 | Transport accessory protein (fragment) | 50% identity to brcD of Brochothrix campestris | 41 |
| ORF 16 | 52 | Bacteriocin immunity protein | 61% identity to brcI of Brochothrix campestris | 41 |
| ORF 17 | 56 | Bacteriocin | 69% identity to brcA of Brochothrix campestris | 41 |
| 47% identity to brcB of Brochothrix campestris | ||||
| ORF 18 | 370 | Transposase | 68% identity to Efae1527 of Enterococcus faecium | NZAAAK01000222b |
| Conserved domain transposase and inactivated derivatives | COG3547b | |||
| ORF 19 | 301 | Transposase | 45% identity to TNP-encoding IS1070 of Leuconostoc lactis | 58 |
| Conserved domain Tra8 of IS30 family involved in DNA replication, recombination, and repair | COG2826b | |||
| ORF 20 | 354 | Transposon | 25% identity to SMU207 of Streptococcus mutans UA159 | NC004350b |
| Conserved domain Rep_trans, replication initiation factor | pfam02486b | |||
| ORF 21 | 165 | Transcriptional regulator | 34% identity to SAG1991 of Streptococcus agalactiae | 54 |
| Conserved domain HTH_XRE, helix-turn-helix xenobiotic response element family of transcriptional regulators | cd00093b | |||
| ORF 22 | 219 | Hypothetical protein | 65% identity to Efae1295 of Enterococcus faecium | NZAAAK01000203b |
| Conserved domain XerD, site-specific recombinase | COG4974b | |||
Only the best matches are shown.
Accession number of sequence deposited directly in the National Center for Biotechnology Information database.
Immunity genes.
The assumption that sakIT and sakIX encode immunity proteins was based on the fact that in many bacteriocin-encoding gene clusters the immunity gene is located immediately downstream of the structural gene(s) and the fact that such immunity-conferring proteins are hydrophobic (45). Expression clone pZ235T, containing only sakIT, was constructed to confirm the role of this gene in immunity to sakacin T. This plasmid was introduced into L. sakei LMG2313, which is sensitive to sakacin T, and this strain was subsequently used as an indicator in the deferred antagonism assay. L. sakei LMG2313 harboring pZ235T was shown to be immune to sakacin T produced by clone pLPT7 (see below), demonstrating that the insert on the plasmid, sakIT, is capable of conferring immunity to a previously sensitive strain.
A similar construct, pZ233X containing sakIX, was made in the same manner. L. sakei LMG2313 is sensitive to sakacin X, and so this strain was used as an indicator. L. sakei LMG2313 containing pZ233X was shown to be immune to sakacin X produced by a colony of the pLPX14 clone, although it was not completely immune to purified sakacin X at concentrations greater than 800 activity units ml−1. Thus, the insert on pZ233X is capable of conferring sakacin X immunity to a previously sensitive strain.
To investigate whether the immunity genes in the sakacin TX locus conveyed cross-immunity to sakacin T and sakacin X, the L. sakei LMG2313 derivatives harboring pZ235T and pZ233X were used as indicators. The construct that included the sakacin T immunity gene did not provide immunity to sakacin X, and likewise, sakIX did not confer immunity to sakacin T to the indicator strain harboring it. Plasmid pNZ44 without an insert was used as a control in both the sakacin T and sakacin X immunity experiments and did not confer immunity.
Bacteriocin structural genes.
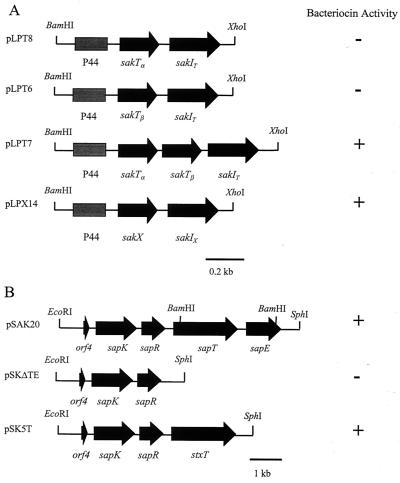
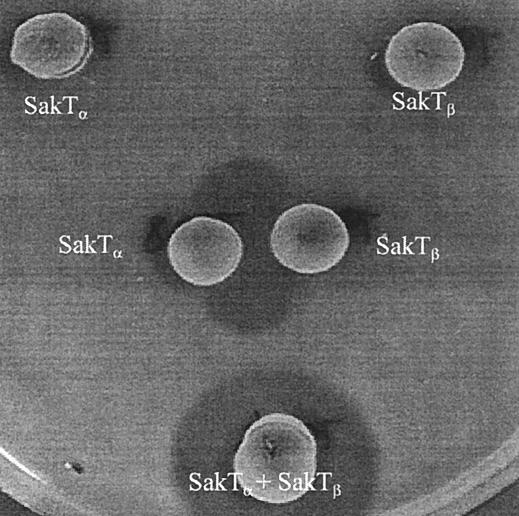
A system for heterologous expression of class II bacteriocins has been constructed previously, and this system is based on introducing two plasmids into a bacteriocin-negative L. sakei strain (6). The first plasmid (pSAK20) contains the genes necessary for export and processing of several bacteriocin precursors, while the second plasmid (a pLPV111 derivative) supplies the structural and immunity genes for the bacteriocins (5). To investigate whether both sakTα and sakTβ are essential for sakacin T production, sakTα and sakTβ were cloned individually and in combination upstream of an intact sakIT immunity gene, under control of the constitutive lactococcal promoter P44, in pLPV111 in order to produce plasmids pLPT6, pLPT7, and pLPT8 (see Materials and Methods) (Table 1). These three plasmids were introduced into L. sakei Lb790X or a derivative of this strain harboring plasmid pSAK20. Only L. sakei Lb790X transformants containing both pLPT7 and pSAK20 were capable of producing antimicrobial activity. L. sakei Lb790X clones containing either pLPT6 or pLPT8 along with pSAK20 were assayed for inhibitory activity by using the deferred antagonism assay with the indicator L. sakei LMG2313. No zones of inhibition were observed surrounding the colonies of either the pLPT6 or pLPT8 transformants (Fig. 2). However, when colonies of each transformant type were grown adjacent to each other, a clear zone of inhibition was observed between the colonies (Fig. 3). These results clearly show that sakacin T requires two peptides for full activity and that this bacteriocin therefore is a member of the class IIb bacteriocins.
FIG. 2.
(A) PCR-derived inserts in pLPV111 derivatives with bacteriocin structural and immunity genes. The plasmid designations are indicated on the left, and the phenotypes when the plasmids are introduced (in conjunction with pSAK20) into L. sakei Lb790 are indicated on the right. (B) Inserts in pSAK20, containing the genes necessary for processing and transport of the bacteriocins used, and pSAK20 derivatives used in this study. The phenotypes when these constructs were introduced into L. sakei Lb790 or L. sakei 23K harboring the pLPV111 derivatives with bacteriocin structural and immunity genes are indicated.
FIG. 3.
Bacteriocin activity and complementation of the SakTα and SakTβ peptides. L. sakei Lb790(pSAK20) containing pLPT8 (SakTα) and L. sakei Lb790(pSAK20) containing pLPT6 (SakTβ) were grown and overlaid with the sensitive indicator strain L. sakei LMG2313. Cultures were grown adjacent to each other and as a mixed culture (SakTα + SakTβ↓).
To establish heterologous expression of sakacin X, sakX was cloned upstream of the sakIX immunity gene under control of the P44 promoter in the vector pLPV111. The resulting construct, designated pLPX14, was introduced into L. sakei strain Lb790 containing pSAK20 as described above. The resulting strain was shown to exhibit bacteriocin activity (Fig. 2), which was purified as described previously (57) (see Materials and Methods). The constructs pLPT5 and pLPXi23, which contained the immunity genes for sakacin T and sakacin X, respectively, without the structural genes under investigation, did not exhibit any inhibitory activity when they were transformed into L. sakei Lb790(pSAK20).
Sakacin P genes are plasmid encoded, while the sakacin TX locus is chromosomally encoded.
In addition to sakacin X and sakacin T, a bacteriocin identical to sakacin P was purified from L. sakei 5 (57). Several genes in the assumed sakacin P locus were PCR amplified and sequenced, which showed that the gene organization and sequence of this locus are identical to the gene organization and sequence described previously (10, 30, 55; results not shown). Total or plasmid DNA preparations isolated from L. sakei 5 were digested with a range of restriction enzymes and used in Southern hybridization experiments performed with the PCR products corresponding to the structural genes of sakacin P, sakacin X, and sakacin T. Hybridization signals were observed with a number of distinct bands of various sizes obtained from total DNA, whereas no hybridization signals were obtained when plasmid DNA was probed with the PCR-amplified sakacin T and sakacin X fragments. However, positive signals were detected when the plasmid DNA was probed with the PCR fragment corresponding to the DNA region encoding sakacin P (data not shown). These results show that the sakacin T and sakacin X structural genes are chromosomally encoded, whereas the sakacin P structural gene is plasmid encoded.
Bacteriocin production is inducible.
Spontaneous loss of bacteriocin production was not observed when standard cultures of L. sakei 5 were diluted in MRS medium, as observed previously for many class II bacteriocin-producing bacteria in which bacteriocin production is regulated by a so-called three-component regulatory system (44). However, loss of production was observed when L. sakei 5 was grown in wort at temperatures above 35°C. Different indicators were used to determine the identities of the bacteriocins which were produced during isolation and induction of the Bac− strains. Sakacin X inhibits P. pentosaceus LMG2001, and it was shown by using this indicator that isolates of L. sakei 5 could be obtained which did not exhibit sakacin X production but which still produced sakacin P. Total bacteriocin production was abolished when L. sakei 5 was grown in wort at an elevated temperature, 37°C. Once L. sakei 5 had lost the ability to produce bacteriocin (Bac−), subcultures of it continued to be Bac− when they were grown in wort. Addition of sterile CFS derived from a Bac+ culture of L. sakei 5 induced stable bacteriocin production in wort at 37°C, whereas CFS derived from a Bac− culture did not. To determine whether the product of stxP was the induction factor for sakacin T and sakacin X production, chemically synthesized mature peptide IP-TX was used to induce bacteriocin production in a Bac− culture of L. sakei 5 grown in wort. Addition of IP-TX at a concentration of 400 ng ml−1 restored sakacin T and sakacin X production. To demonstrate that sakacin T production and sakacin X production were switched on simultaneously upon addition of IP-TX, the L. sakei LMG2313 derivatives harboring pZ235T and pZ233X with immunity gene inserts (see above) were used as indicators. Both indicator strains were inhibited when IP-TX was added to the wort. To investigate whether the sakacin P-inducing peptide (10) could induce sakacin X production, chemically synthesized mature peptide IP-673 was used to induce bacteriocin production in a Bac− culture of L. sakei 5. Sakacin P was produced, which inhibited L. sakei LMG2313 but not P. pentosaceus LMG2001, which indicated that sakacin X was not produced. When inoculated into wort without IP-TX, the Bac− culture failed to produce bacteriocin and served as a negative control. In all cases, whether bacteriocin production was switched on or off following growth in wort, subsequent subculturing in MRS broth resulted in bacteriocin-producing cultures.
Transport of sakacin X and sakacin T does not appear to require an accessory protein.
To establish the role of the putative transport protein StxT and the lack of a gene encoding an accessory protein in the native L. sakei 5 bacteriocin locus, two derivatives of pSAK20 were constructed. Deletion of the sapT and sapE transport and accessory protein genes from pSAK20 resulted in construction of pSKΔTE. The stxT gene, encoding the putative native transport protein, was cloned into pSKΔTE to obtain plasmid pSK5T. Each transport construct (pSAK20, pSKΔTE, or pSK5T) was transformed into host strain L. sakei Lb790X. Then one of the structural gene constructs, pLPT7 (sakacin T) or pLPX14 (sakacin X), was introduced into the resulting transformants. L. sakei Lb790X clones containing either pLPT7 or pLPX14 in conjunction with one of the transport constructs (pSAK20, pSKΔTE, or pSK5T) were assayed for inhibitory activity against the indicator strain L. sakei LMG2313. The results of this series of experiments for both sakacin T and sakacin X were the same and are discussed collectively below. Bacteriocin production was observed for the pSAK20 and pSK5T transformants. No zones of inhibition were detected surrounding the colonies of the pSKΔTE transformants. However, it is known that while the host strain L. sakei Lb790X does not produce bacteriocin, it does contain homologues of the spp gene cluster (30, 43), and this could have affected the results obtained with the transport gene constructs by gene complementation. The bacteriocin tests were therefore repeated with a different host strain, L. sakei 23K, which does not appear to contain spp gene homologues (L. Axelsson, personal communication). Bacteriocin production by the pSAK20 and pSK5T transformants was observed. However, no inhibitory activity was observed for pSKΔTE (Fig. 2).
DISCUSSION
In this paper we describe genetic characterization of a bacteriocin cluster of L. sakei 5, which was originally isolated from malt. The genetic analysis focused on a 17-kb chromosomal fragment, which was shown to contain the genetic information responsible for production of sakacin X and the two-component bacteriocin sakacin T. This strain also produces sakacin P (57), which was found to be plasmid encoded by Southern blot experiments. The ability to produce several different bacteriocins is expected to endow the producer with a wider inhibitory capacity and therefore an enhanced ability to compete with other bacteria in the same environment. Alternatively, these bacteriocins may act synergistically with each other or with other antimicrobial peptides in a particular environment, as was shown recently (37).
In many cases, bacteriocins produced by an LAB inhibit species that are closely related to the producing strain (38). This is particularly relevant in the brewing industry, as LAB account for a large percentage of spoilage bacteria in the brewing environment (7, 27). In a previous study, sakacin X was shown to inhibit a range of beer spoilage LAB (57), and this was the driving force for characterizing the genetic information of this and other bacteriocins produced by L. sakei 5. This strain was shown to produce a single peptide bacteriocin, sakacin X, as well as sakacin T, which requires the complementary activity of two small, unmodified hydrophobic peptides. This finding identifies sakacin T as a member of the class IIb bacteriocins. Both peptides are required for full activity as neither component exhibited bacteriocin activity on its own when it was produced in a heterologous host. However, one component of sakacin T, SakTβ, exhibits a low but detectable level of activity at high concentrations, as observed during purification of this peptide (57).
The organization of the (putative) genes required for sakacin T and sakacin X production and regulation is similar to that of other class II bacteriocin gene clusters (45). The adjacent genetic elements sakTα, sakTβ, and sakIT were identified as the structural and immunity genes for sakacin T. The neighboring genes sakX and sakIX were demonstrated to be the structural and immunity genes, respectively, for sakacin X.
Sakacin T and sakacin X seem to be controlled by a three-component regulatory system consisting of IP-TX, StxK, and StxR (10, 44). In a situation similar to that observed for ABP-118 production by Lactobacillus salivarius UCC118 (19), L. sakei 5 does not lose the Bac+ phenotype upon extreme dilution of the culture, a strategy used successfully for obtaining Bac− derivatives of other bacteriocin systems (10). Obtaining a Bac− derivative of L. sakei 5 was further complicated by the ability of the organism to produce multiple bacteriocins and by the presence of more than one regulatory system controlling the production of sakacin P on the one hand and sakacin T and sakacin X on the other hand. Production of the latter two bacteriocins was specifically induced by addition of chemically synthesized IP-TX to the medium, while supplementation of wort with synthetic peptide IP-673 exclusively induced sakacin P production.
It is not unusual for LAB to produce more than one bacteriocin and to contain a diverse range of bacteriocin-related genes that are dispersed over the chromosome and plasmids (15, 16, 30, 43). For example, the sakacin P gene cluster ends with an incomplete gene, orfX, that is likely to encode a bacteriocin (10, 17). ORF 3 of the abp-118 locus (19) encodes a peptide that is almost identical to presalivaricin B, a bacteriocin produced by L. salivarius M6 (53). Four genes in the sakacin TX locus exhibit homology to genes required for brochocin C production. A leader peptide sequence precedes ORF 17, but at the sequence specifying the predicted double-glycine motif an apparent frameshift seems to have taken place. This suggests that these genes are the remains of a gene cluster encoding a brochocin C-like bacteriocin. The presence of what seems to be a putative intact immunity gene, ORF 16, might provide L. sakei 5 protection against a brochocin C-type bacteriocin produced by competing strains in the same environment.
The sequenced chromosomal fragment flanking the bacteriocin gene cluster in L. sakei 5 contains several genes whose products have not been characterized and have unknown functions. The protein products of these genes display significant similarity to hypothetical proteins found in other LAB, which resemble transposases or IS elements. The presence of these proteins, the finding that sakacin P production is plasmid encoded rather than chromosomally encoded (10, 43), and the rearrangement of the brc-type genes are interesting and indicate that bacteriocin gene clusters are highly mobile and subject to strong evolutionary pressures.
There is a notable absence of an accessory transport gene in the sakacin TX locus. In the present study, we obtained evidence which suggests that an accessory protein is not required for processing or transport of sakacin T and sakacin X, although we cannot completely rule out the possibility that this function is provided by the host strains used in this study. It may be that there is some form of complementation by an unknown chromosomal gene fulfilling the role of an accessory protein. While the specific role of accessory transport proteins in bacteriocin processing is still not fully understood (18), it has been demonstrated for several bacteriocins that such proteins are needed for production (6, 56, 59). Sakacin T and sakacin X, while different from previously described bacteriocins, nevertheless exhibit homology to other bacteriocins and seem to share many characteristics with other standard class II bacteriocins, and so the absence of an accessory transport gene in the locus is unexpected. This adds to the genetic complexity of bacteriocin production in LAB mentioned above.
The antimicrobial properties of the multiple bacteriocins produced by malt isolate L. sakei 5 may be used to enhance the microbiological stability of the brewing process and its product, beer. In addition to the sakacin T and sakacin X bacteriocin structural genes, L. sakei 5 appears to contain genetic information encoding additional bacteriocin-related peptides, and while some proteins may be inactive, the presence of at least four immunity proteins, which provide immunity to sakacin P, sakacin T, sakacin X, and the brochocin C-like bacteriocin, should give this bacterium a competitive advantage in its environment. Since L. sakei 5 was isolated from malt and since it also inhibits LAB that cause problems for the brewing industry, this bacteriocin-producing strain might be a suitable candidate for use in the industry.
Acknowledgments
This work was supported by the Department of Agriculture and Food (contracts 97/R&D/C/131 and 00/R&D/C/53), by Heineken Ireland Ltd. through a Heineken Ireland fellowship to A. Vaughan, and by Enterprise Ireland through the International Collaboration Programme (RE:IC/1999/065).
Wort used in this study was kindly provided by Declan Goode of the Department of Food Science, Technology and Nutrition, National University of Ireland, Cork. We are grateful to Lars Axelsson, MATFORSK, Norwegian Food Research Institute, Ås, Norway, for providing results before publication and for providing L. sakei Lb790 and plasmids pLPV111 and pSAK20. We thank Monique Zagorec, Unité FLEC, INRA, Domaine di Vilvert, Jouy-en-Josas, France, for providing L. sakei 23K.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, D. G., and L. L. McKay. 1983. Simple and rapid methods for isolating large plasmid DNA from lactic streptococci. Appl. Environ. Microbiol. 46:549-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderssen, E. L., D. B. Diep, I. F. Nes, V. G. H. Eijsink, and J. Nissen-Meyer. 1998. Antagonistic activity of Lactobacillus plantarum C11: two new two-peptide bacteriocins, plantaricins EF and JK, and the induction factor plantaricin A. Appl. Environ. Microbiol. 64:2269-2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aukrust, T., and H. Blom. 1992. Transformation of Lactobacillus strains used in meat and vegetable fermentations. Food Res. Int. 25:253-261. [Google Scholar]
- 5.Axelsson, L., T. Katla, M. Bjswsl]ornslett, V. G. H. Eijsink, and A. Holck. 1998. A system for heterologous expression of bacteriocins in Lactobacillus sake. FEMS Microbiol. Lett. 168:137-143. [DOI] [PubMed] [Google Scholar]
- 6.Axelsson, L., and A. Holck. 1995. The genes involved in production of and immunity to sakacin A, a bacteriocin from Lactobacillus sake Lb706. J. Bacteriol. 177:2125-2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Back, W. 1994. Secondary contamination in the filling area. Brauwelt Int. 4:326-328. [Google Scholar]
- 8.Berthier, F., M. Zagorec, M. C. Champomier-Vergès, S. D. Erlich, and F. Morel-Deville. 1996. Efficient transformation of Lactobacillus sake by electroporation. Microbiology 142:1273-1279. [DOI] [PubMed] [Google Scholar]
- 9.Birnboim, H. C., and J. Doly. 1979. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 7:1513-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brurberg, M. B., I. F. Nes, and V. G. H. Eijsink. 1997. Pheromone-induced production of antimicrobial peptides in Lactobacillus. Mol. Microbiol. 26:347-360. [DOI] [PubMed] [Google Scholar]
- 11.Casaus, P., T. Nilsen, L. M. Cintas, I. F. Nes, P. E. Hernández, and H. Holo. 1997. Enterocin B, a new bacteriocin from Enterococcus faecium T136 which can act synergistically with enterocin A. Microbiology 143:2287-2294. [DOI] [PubMed] [Google Scholar]
- 12.Cintas, L. M., P. Casaus, C. Heranz, L. S. Håvarstein, H. Holo, P. E. Hernández, and I. F. Nes. 2000. Biochemical and genetic evidence that Enterococcus faecium L50 produces enterocins L50A and L50B, the sec-dependent enterocin P, and a novel bacteriocin secreted without an N-terminal extension termed enterocin Q. J. Bacteriol. 182:6806-6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cintas, L. M., P. Casaus, L. S. Håvarstein, P. E. Hernández, and I. F. Nes. 1997. Biochemical and genetic characterization of enterocin P, a novel sec-dependent bacteriocin from Enterococcus faecium P13 with a broad antimicrobial spectrum. Appl. Environ. Microbiol. 63:4321-4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Connil, N., Y. Le Breton, X. Dousset, Y. Auffray, A. Rince, and H. Prevost. 2002. Identification of the Enterococcus faecalis tyrosine decarboxylase operon involved in tyramine production. Appl. Environ. Microbiol. 68:3537-35544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Diep, D. B., L. S. Håvarstein, and I. F. Nes. 1996. Characterization of the locus responsible for the bacteriocin production in Lactobacillus plantarum C11. J. Bacteriol. 178:4472-4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eijsink, V. G. H., L. Axelsson, D. B. Diep, L. S. Håvarstein, H. Holo, and I. F. Nes. 2002. Production of class II bacteriocins by lactic acid bacteria; an example of biological warfare and communication. Antonie Leeuwenhoek 81:639-654. [DOI] [PubMed] [Google Scholar]
- 17.Eijsink, V. G. H., M. Skeie, P. H. Middelhoven, M. B. Brurberg, and I. F. Nes. 1998. Comparative studies of class IIa bacteriocins of lactic acid bacteria. Appl. Environ. Microbiol. 64:3275-3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ennahar, S., T. Sashihara, K. Sonomoto, and A. Ishizaki. 2000. Class IIa bacteriocins: biosynthesis, structure and activity. FEMS Microbiol. Rev. 24:85-106. [DOI] [PubMed] [Google Scholar]
- 19.Flynn, S., D. van Sinderen, G. M. Thornton, H. Holo, I. F. Nes, and J. K. Collins. 2002. Characterization of the genetic locus responsible for the production of ABP-118, a novel bacteriocin produced by the probiotic bacterium Lactobacillus salivarius subsp. salivarius UCC118. Microbiology 148:973-984. [DOI] [PubMed] [Google Scholar]
- 20.Franke, C. M., J. Tiemersma, G. Venema, and J. Kok. 1999. Membrane topology of the lactococcal bacteriocin ATP-binding cassette transporter protein LcnC. J. Biol. Chem. 274:8484-8490. [DOI] [PubMed] [Google Scholar]
- 21.Franz, C. M. A. P., M. J. van Belkum, R. W. Worobo, J. C. Vederas, and M. E. Stiles. 2000. Characterization of the genetic locus responsible for production and immunity of carnobacteriocin A: the immunity gene confers cross-protection to enterocin B. Microbiology 146:621-631. [DOI] [PubMed] [Google Scholar]
- 22.Franz, C. M. A. P., R. W. Worobo, L. E. N. Quadri, U. Schillinger, W. H. Holzapfel, J. C. Vederas, and M. E. Stiles. 1999. Atypical genetic locus associated with constitutive production of enterocin B by Enterococcus faecium BFE900. Appl. Environ. Microbiol. 65:2170-2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haikara, A., and A. Laitila. 1995. Influence of lactic acid starter culture on the quality of malt and beer, p. 249-256. In Proceedings of the 25th Congress of European Brewing Convention, Brussels. IRL Press, Oxford, United Kingdom.
- 24.Harley, C. B., and R. P. Reynolds. 1987. Analysis of E. coli promoter sequences. Nucleic Acid Res. 15:2343-2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Håvarstein, L. S., D. B. Diep, and I. F. Nes. 1995. A family of bacteriocin ABC transporters carry out proteolytic processing of their substrates concomitant with export. Mol. Microbiol. 16:229-240. [DOI] [PubMed] [Google Scholar]
- 26. Håvarstein, L. S., H. Holo, and I. F. Nes. 1994. The leader peptide of colicin V shares consensus sequences with leader peptides that are common amongst peptide bacteriocins produced by Gram-positive bacteria. Microbiology 140:2383-2389. [DOI] [PubMed] [Google Scholar]
- 27.Hollerová, I., and P. Kubizniaková. 2001. Monitoring Gram positive bacterial contamination in Czech breweries. J. Inst. Brew. 107:355-358. [Google Scholar]
- 28.Holo, H., Ø. Nilssen, and I. F. Nes. 1991. Lactococcin A, a new bacteriocin from Lactococcus lactis subsp. cremoris: isolation and characterization of the protein and its gene. J. Bacteriol. 173:3879-3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Horton, R. M., Z. L. Cai, S. N. Ho, and L. R. Pease. 1990. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. BioTechniques 8:528-535. [PubMed] [Google Scholar]
- 30.Hühne, K., A. Holck, L. Axelsson, and L. Kroeckel. 1996. Analysis of the sakacin P gene cluster from Lactobacillus sake Lb674 and its expression in sakacin-negative L. sake strains. Microbiology 142:1437-1448. [DOI] [PubMed] [Google Scholar]
- 31.Jack, R. W., J. Wan, J. Gordon, K. Harmark, B. E. Davidson, A. J. Hillier, R. E. Wettenhall, M. W. Hickey, and M. J. Coventry. 1996. Characterization of the chemical and antimicrobial properties of piscicolin 126, a bacteriocin produced by Carnobacterium piscicola JG126. Appl. Environ. Microbiol. 62:2897-2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kemperman, R., A. Kuipers, H. Karsens, A. Nauta, O. Kuipers, and J. Kok. 2003. Identification and characterization of two novel clostridial bacteriocins, cicularin A and closticin 574. Appl. Environ. Microbiol. 69:1589-1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klaenhammer, T. R. 1993. Genetics of bacteriocins produced by lactic acid bacteria. FEMS Microbiol. Rev. 12:39-85. [DOI] [PubMed] [Google Scholar]
- 34.Kleerebezem, M., L. E. Quadri, O. P. Kuipers, and W. M. de Vos. 1997. Quorum sensing by peptide pheromones and two-component signal-transduction systems in Gram-positive bacteria. Mol. Microbiol. 24:895-904. [DOI] [PubMed] [Google Scholar]
- 35.Law, J., G. Buist, A. Haandrikman, J. Kok, G. Venema, and K. Leenhouts. 1995. A system to generate chromosomal mutations in Lactococcus lactis, which allows fast analysis of targeted genes. J. Bacteriol. 177:7011-7018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leer, R. L., J. M. B. M. van der Vossen, M. van Giezen, J. M. van Noort, and P. H. Pouwels. 1995. Genetic analysis of acidocin B, a novel bacteriocin produced by Lactobacillus acidophilus. Microbiology 141:1629-1635. [DOI] [PubMed] [Google Scholar]
- 37.Lüders, T., G. A. Birkemo, G. Fimland, J. Nissen-Meyer, and I. F. Nes. 2003. Strong synergy between a eukaryotic antimicrobial peptide and bacteriocins from lactic acid bacteria. Appl. Environ. Microbiol. 69:1797-1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marciset, O., M. C. Jeronimus-Stratingh, B. Mollet, and B. Poolman. 1997. Thermophilin 13, a nontypical antilisterial poration complex bacteriocin, that functions without a receptor. J. Biol. Chem. 272:14277-14284. [DOI] [PubMed] [Google Scholar]
- 39.Marinez-Bueno, M., M. Maquedam A. Galvez, B. Samyn, J. van Beeumen, J. Coyette, and E. Valdivia. 1994. Determination of the gene sequence and the molecular structure of the enterococcal peptide antibiotic AS-48. J. Bacteriol. 176:6334-6339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mayr-Harding, A., A. J. Hedges, and R. C. W. Buckley. 1972. Methods for studying bacteriocins. Academic Press, New York, N.Y.
- 41.McCormick, J. K., A. Poon, M. Sailer, Y. Gao, K. L. Roy, L. M. McMullen, J. C. Vederas, M. E. Stiles, and M. J. van Belkum. 1998. Genetic characterization and heterologous expression of brochocin C, an antibotulinal, two-peptide bacteriocin produced by Brochothrix campestris ATCC 43754. Appl. Environ. Microbiol. 64:4757-4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McGrath, S., G. F. Fitzgerald, and D. van Sinderen. 2001. Improvement and optimization of two engineered phage resistance mechanisms in Lactococcus lactis. Appl. Environ. Microbiol. 67:608-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Møretrø, T. 2000. Optimization of production of the bacteriocin sakacin P by Lactobacillus sakei cultural conditions and genetics. Ph.D. thesis. Agricultural University of Norway, Ås, Norway.
- 44.Nes, I. F., and V. G. H. Eijsink. 1999. Regulation of group II peptide bacteriocin synthesis by quorum-sensing mechanisms, p. 175-192. In G. M. Dunny and S. C. Winans (ed.), Cell-cell signaling in bacteria. American Society for Microbiology, Washington, D.C.
- 45.Nes, I. F., D. B. Diep, L. S. Håvarstein, M. B. Brurberg, V. G. H. Eijsink, and H. Holo. 1996. Biosynthesis of bacteriocins in lactic acid bacteria. Antonie Leewenhoek 70:113-128. [DOI] [PubMed] [Google Scholar]
- 46.O'Keeffe, T., C. Hill, and R. P. Ross. 1999. Characterization and heterologous expression of the genes encoding enterocin A production, immunity, and regulation in Enterococcus faecalis DPC1146. Appl. Environ. Microbiol. 65:1506-1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.O'Mahony, A. T., F. O'Sullivan, Y. Walsh, A. Vaughan, M. Maher, G. F. Fitzgerald, and D. van Sinderen. 2000. Characterization of antimicrobial producing lactic acid bacteria from malted barley. J. Inst. Brew. 106:403-410. [Google Scholar]
- 48.O'Sullivan, T. F., Y. Walsh, A. O'Mahony, G. F. Fitzgerald, and D. van Sinderen. 1999. A comparative study of malthouse and brewhouse microflora. J. Inst. Brew. 105:55-61. [Google Scholar]
- 49.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 50.Skaugen, M. 1994. Transposition in Lactobacillus sake and its abolition of lactocin S production by insertion of IS1163, a new member of the IS3 family. Appl. Environ. Microbiol. 60:2818-2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stiles, M. E. 1996. Biopreservation by lactic acid bacteria. Antonie Leeuwenhoek 70:331-345. [DOI] [PubMed] [Google Scholar]
- 52.Takami, H., K. Nakasone, Y. Takaki, G. Maeno, R. Sasaki, N. Masui, F. Fuji, C. Hirama, Y. Nakamura, N. Ogasawara, S. Kuhara, and K. Horikoshi. 2000. Complete genome sequence of the alkaliphilic bacterium Bacillus halodurans and genomic sequence comparison with Bacillus subtilis. Nucleic Acids Res. 28:4317-4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.ten Brink, B., M. Minekus, J. M. van der Vossen, R. J. Leer, and J. H. Huis in't Veld. 1994. Antimicrobial activity of lactobacilli: preliminary characterization and optimisation of production of acidocin B, a novel bacteriocin produced by Lactobacillus acidophilus M46. J. Appl. Bacteriol. 77:140148. [DOI] [PubMed] [Google Scholar]
- 54.Tettelin, H., V. Masignani, M. J. Cieslewicz, J. A. Eisen, S. Peterson, M. R. Wessels, I. T. Paulsen, K. E. Nelson, I. Margarit, T. D. Read, L. C. Madoff, A. M. Wolf, M. J. Beanan, L. M. Brinkac, S. C. Daugherty, R. T. DeBoy, A. S. Durkin, J. F. Kolonay, R. Madupu, M. R. Lewis, D. Radune, N. B. Fedorova, D. Scanlan, H. Khouri, S. Mulligan, H. A. Carty, R. T. Cline, S. E. Van Aken, J. Gill, M. Scarselli, M. Mora, E. T. Iacobini, C. Brettoni, G. Galli, M. Mariani, F. Vegni, D. Maione, D. Rinaudo, R. Rappuoli, J. L. Telford, D. L. Kasper, G. Grandi, and C. M. Fraser. 2002. Complete genome sequence and comparative genomic analysis of an emerging human pathogen, serotype V Streptococcus agalactiae. Proc. Natl. Acad. Sci. 99:12391-12396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tichaczek, P. S., J. Nissen-Meyer, I. F. Nes, R. F. Vogel, and W. P. Hammes. 1992. Characterization of the bacteriocins curvacin A from Lactobacillus curvatus LTH1174 and sakacin P from Lactobacillus sake LTH673. Syst. Appl. Microbiol. 15:460-468. [Google Scholar]
- 56.Van Belkum, M. J., and M. E. Stiles. 1995. Molecular characterization of genes involved in the production of the bacteriocin leucocin A from Leuconostoc gelidum. Appl. Environ. Microbiol. 61:3573-3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vaughan, A., V. G. H. Eijsink, T. F. O'Sullivan, K. O'Hanlon, and D. van Sinderen. 2001. An analysis of bacteriocins produced by lactic acid bacteria isolated from malted barley. J. Appl. Microbiol. 91:131-138. [DOI] [PubMed] [Google Scholar]
- 58.Vaughan, E. E., and W. M. de Vos. 1995. Identification and characterization of the insertion element IS070 from Leuconostoc lactis NZ6009. Gene 155:95-100. [DOI] [PubMed] [Google Scholar]
- 59.Venema, K., J. Kok, J. D. Marugg, M. Y. Toonen, A. M. Ledeboer, G. Venema, and M. L. Chikindas. 1995. Functional analysis of the pediocin operon of Pediococcus acidilactici PAC1.0: PedB is the immunity protein and PedD is the precursor processing enzyme. Mol. Microbiol. 17:515-522. [DOI] [PubMed] [Google Scholar]
- 60.Worobo, R. W., T. Henkel, M. Sailer, K. L. Roy, J. C. Vederas, and M. E. Stiles. 1994. Characteristics and genetic determinant of a hydrophobic peptide bacteriocin, carnobacteriocin A, produced by Carnobacterium piscicola LV17A. Microbiology 140:517-526. [DOI] [PubMed] [Google Scholar]