Abstract
The Veterans Affairs Hypertension Primary Care Longitudinal Cohort (VAHC) was initiated in 2003 as a pilot study designed to link the VA electronic medical record system with individual genetic data. Between June 2003 and December 2004, 1,527 hypertensive participants were recruited. Protected health information (PHI) was extracted from the regional VA data warehouse. Differences between the clinic and mail recruits suggested that clinic recruitment resulted in an over-sampling of African Americans. A review of medical records in a random sample of study participants confirmed that the data warehouse accurately captured most selected diagnoses. Genomic DNA was acquired non-invasively from buccal cells in mouthwash; ~ 96.5 per cent of samples contained DNA suitable for genotyping, with an average DNA yield of 5.02 ± 0.12 micrograms, enough for several thousand genotypes. The coupling of detailed medical databases with genetic information has the potential to facilitate the genetic study of hypertension and other complex diseases.
Keywords: electronic medical record, hypertension, primary care, privacy
Introduction
The electronic medical record (EMR) can be used to create very large-scale databases for genetic studies1–3 and `virtual cohorts' for the study of complex chronic diseases in a setting representative of clinical practice.4–8 To date, few publications have fully demonstrated the utility of EMRs as effective sources of phenotypic data for genetic studies.9, 10 Hypertension is an example of a complex trait, determined by the interplay of multiple genes, lifestyle and medical comorbid factors, that may be best studied in the primary care (or clinical cohort) arena rather than smaller experimental (clinical trial) settings. The most recent Joint National Committee guidelines for the management of hypertension include more aggressive blood pressure management below 130/80 mmHg for high-risk patients with diabetes or chronic renal disease.11 However, a substantial fraction of patients remain inadequately treated, even among managed care patients with access to antihypertensive medications,12, 13 and among high-risk patients with diabetes or chronic kidney disease.14 Ethnic differences in the response to blood pressure management and outcomes have been suggested,15 and these differences persist even with adequate access to healthcare.16, 17 African Americans, for example, have a disproportionate burden of hypertensive target organ damage,18 perhaps as a result of risk-associated genetic variance, lifestyle (environmental) factors, or differences in the prevalence of comorbid disease.
One of the key features of this study was to take advantage of the large-scale, comprehensive and integrated EMR in the Veteran's Administration Healthcare System known as VistA (Veterans Health Information Systems and Technology Architecture; http://www.va.gov/VISTA_ MONOGRAPH/) and to explore the predictive value of genetic markers for antihypertensive drug response and hypertension-related comorbid disease. The VA EMR can be accessed via a graphical user interface known as the Computerized Patient Record System (CPRS: http://www1.va.gov/cprsdemo/), an intuitive single interface for healthcare providers to review and update a patient's EMR. VASDHS is a part of the Veterans Integrated Service Network 22 (VISN-22), spanning Southern California and Southern Nevada. VISN-22 provides research support by abstracting electronic medical data from individual VA healthcare facilities and makes the data accessible to investigators for research purposes in a `data warehouse'.
In this study, we recruited patients from Veterans Affairs San Diego Healthcare System (VASDHS) in collaboration with physicians from General Internal Medicine, and were able to access protected health information (PHI) in bulk from the VISN-22 data warehouse. Here, we present a description of the VA Hypertension Primary Care Longitudinal Cohort (VAHC), as well as workable methods to surmount challenging issues concerning patient privacy, diagnosis coding validation, representative recruitment and DNA acquisition. Finally, we were able to find preliminary evidence for a novel predictor of angiotensin converting enzyme inhibitor efficacy, illustrating the power and utility of this cohort.
Methods
Privacy: Health Insurance Portability and Accountability Act (HIPPA) compliance and protected health information (PHI)
This study involved the integration of PHI collected from the personal EMR (CPRS) and a regional data warehouse (VISN-22) with genomic data on DNA extracted from patient buccal cells (Figure 1). In order to maintain compliance with federal HIPAA Privacy Rule (http://privacyruleandresearch.nih.gov/pdf/HIPAA_Booklet_4-14-2003.pdf),19 we worked interactively with the local Human Research Protection Program, as well as the Division of General Internal Medicine, to develop a protocol in which primary care providers (privileged to approach their own patients for research) were enlisted as co-investigators to assist in subject enrollment.
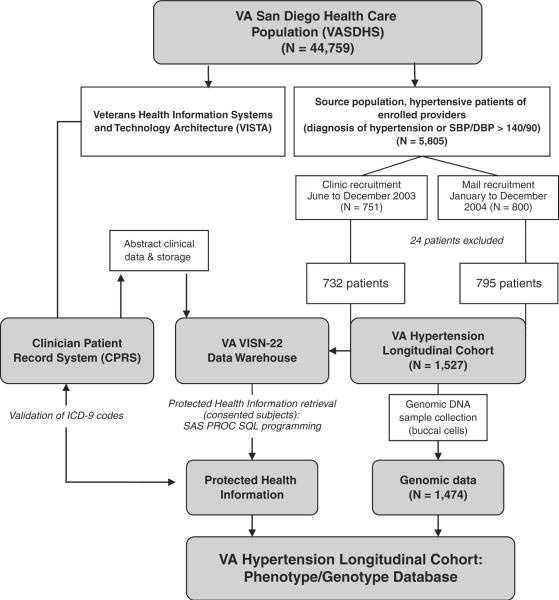
Figure 1.
Patient recruitment and development of database. This study incorporated 1,527 hypertensive patients. Phenotype information from regional data warehouse (VISN-22) was collected using SAS PROC SQL interface, and validated against the EMR (using CPRS). Buccal DNA was obtained and genotype data was merged with the phenotype data
Patient recruitment
Patients with a diagnosis of hypertension (ICD-9 codes 401.1 and 401.9), or a blood pressure over 140/90 mmHg confirmed by chart review, were eligible to participate. Between June and December 2003, participants were recruited directly (face-to-face) in clinic by study personnel. Signed consent was obtained and buccal/mouthwash samples were collected on site. From January to December 2004, those not recruited in clinic were invited to participate by mail; a second mailer was sent six to eight weeks later if there was no response to the first invitation. The mailer included a consent form with a contact number to answer questions or review details and materials to collect a mouthwash DNA sample.
EMR data collection
Each subject completed a consent form containing a line item asking the participant to authorize a review of medical records for research purposes. The consent form listed the sources of medical data review (progress notes, billing information, pathology reports, etc.), and had line item consents for drug, alcohol, mental health, and HIV/AIDS diagnosis and treatment. Additional precautions were used to insure patient privacy: the analytic data sets had all 18 HIPAA personal identifiers removed (name, geographic ID, specific dates related to patient, telephone number, fax number, email address, social security number, medical record number, health plan number, account number, certificate or license number, device identifiers and serial numbers, URLs, internet protocol (IP) addresses, biometric identifiers, full face photographs and comparable images, and any other unique identifier), and dates were de-identified by adding a random number (i.e. ± 1 to 7 days). Each subject was then sequentially assigned a de-identifying unique subject ID. All data were encrypted and stored on institutional servers behind firewalls; access was password controlled and limited to study investigators.
With the guidance of the VA Information Resource Center (ViReC) unit in Chicago (http://www.virec.research.va.gov/Support/Training-NewUsersToolkit/IntroToVAData.htm), we utilized the VA VISN-22 (http://www.desertpacific.va.gov/) Data Warehouse to access medical information stored in VISN-22 digital data files dating back to October 1999, retrieved using a SAS PROC SQL interface to a Microsoft SQL-Server database. This process is extensively described for VISN-20 (http://www.visn20.med.va.gov/V20/DataWarehouse/Documents/OverHistory.asp). Queries are submitted to the server to retrieve medical files on consented subjects based on VA patient identification numbers. This database was used to collate the following parameters: demographics, vital signs, drug treatments, selected diagnoses by billing (ICD-9) codes, drug allergies, laboratory measurements of interest (such as serum creatinine, cholesterol profile), and anthropo-metric measures such as blood pressure, heart rate, and body mass index (Figure 1). Since most measures were repeated multiple times, baseline measures at time of enrollment are reported in this article. While this initial study focused on blood pressure, this database was also developed to study other end-points of interest in this patient population – comorbid illnesses such as coronary artery disease and type 2 diabetes, commonly found in the same study subject.
We linked blood pressure (vital signs) profiles with antihypertensive medication use. Medication use time intervals for major classes of antihypertension prescriptions were identified using each subject's electronic medical record. These classes include beta-adrenergic receptor blockers, angiotensin converting enzyme and receptor blockers, thiazide and loop diuretics, calcium channel blockers, central alpha agonists, peripheral vasodilators and nitrates. Since the VA uses a limited drug formulary, most drug classes are represented by only one or two drugs. Therefore, analysis by drug class was deemed appropriate. Information on subclasses and drug dose was also collected and cataloged for future study.
Study population and data validation
In order to explore whether the study sample cohort differed from the source hypertensive patient population, a de-identified database of hypertensive patients who did not enroll was also developed.20 We also compared differences between clinic and mail recruits. ANOVA or chi-square tests were used for comparisons; all p-values are two sided, and standard deviations are presented unless specified.
A subset of N = 105 randomly selected study participants (N = 50 clinic and N = 55 mail recruits) was used to validate several hypertension-related ICD-9 (billing codes) diagnoses extracted from the VISN-22 Data Warehouse. The medical problem list was first reviewed; discrepancies between the problem list and electronic ICD-9 diagnoses were resolved by reviewing clinic notes. Concordance (kappa or agreement correlation scores) between the electronic extraction and medical records (problem list and/or text notes) were calculated for each diagnosis. Because veterans occasionally receive healthcare outside the VA system, an additional random sample of n = 150 was reviewed to determine whether the use of non-VA medications was adequately captured in the database. This was done by first reviewing the `non-VA medication' field, a separate CPRS entry used by VA providers to document non-VA medication use. Clinic notes were then reviewed to check for medications prescribed by outside medical providers.
Genomics: DNA acquisition, genotyping, and pharmacogenetic association
Buccal cell genomic DNA was obtained from a mouthwash sample which contains a bacterostatic agent (cetylpyridinium chloride) as well as ethanol; this method has been shown to yield good quality DNA.21 Volunteers submitted the sample at the time of recruitment in the clinic or were given instructions on mailing a sample in a clean plastic screw-cap tube placed in a pre-addressed, crushproof/padded, franked envelope. The mouthwash DNA was extracted from buccal cells using PureGene extraction columns (Gentra Biosystems, MN) and quantified by PicoGreen fluorescence (specific for double-stranded DNA), followed by a diagnostic PCR for amplifiability. DNA samples were stored in a −70 °C freezer with controlled access for future analyses. We then identified 110 Caucasian individuals with sequential blood pressure readings off and then on monotherapy with an angiotensin concerting enzyme inhibitor. The plasminogen/plasmin system (especially via tissue plasminogen activator) represents an alternative pathway for production of angiotensin.22 The PAI-1 gene was typed for a simple sequence repeat (G4/G5) in the proximal promoter region, rs1799768, a variant that confers functional change on promoter strength.23
Results
Privacy, HIPAA, and the EMR: cohort enrollment
Nineteen providers agreed to participate in this study, resulting in a source population of 5805 with a diagnosis of hypertension from administrative records in VASHDHS. Between June and December 2003, 751 patients were directly recruited in primary care clinics at their scheduled appointments. Those who were not recruited in clinic were sent mailed invitations (5054); of these, an additional 800 patients consented to participate. Of the 1551 recruits, 24 subjects were excluded; one withdrew from study, four consented twice, and 19 did not regularly use VASDHS services or were not veterans (including seven who were not found in VASDHS medical records and two employees). The final cohort consisted of 1,527 VASDHS (N = 732 clinic and N = 795 mail) patients, approximately 26 per cent (1,527/5805) of the source population.
Study population
The average age was 64.3 ± 12.7 years. Approximately half the participants had VA service-connected medical diagnoses (49%); the majority was male (95.7%), self-reported Caucasian (68%), and married (77%). Study participants had 133,530 months of follow-up and an average of 87.4 ± 22.9 months of follow-up per person. Digital data extraction (of patient encounters from 1999 to the present) yielded substantial quantities of longitudinal data, including 346,628 outpatient encounters (average ~ 226/subject), 48,504 sets of vital signs (blood pressures, ~ 39.5/subject), 115,891 prescriptions (~ 76/subject), 119,766 prescription refills (~ 78/subject), 403,328 procedure code entries (~ 263/subject), and 931,625 laboratory determinations (~ 608/subject; data not shown).
Blood pressure was generally well controlled: the average mean arterial pressure or MAP was 98 ± 13 mmHg (normal < 105–107 mmHg). Average estimates for renal function, LDL and HDL cholesterol and hemoglobin A1C levels were also within normal ranges (Table 1).
Table 1.
Baseline demographic and biomedical parameters
| VAHC cohort | VAHC source population | P | VAHC clinic | VAHC mail | P | |
|---|---|---|---|---|---|---|
| N | 1,527 | 4278 | 732 | 795 | ||
| Demographics | ||||||
| Ageyears (mean±SD) | 64.3±12.7 | 65.2±13.1 | 0.04 | 60.8±12.4 | 67.5±12.1 | <0.001 |
| Sex male (%) | 1461 (95.7%) | 4145 (97%) | 696 (95.3%) | 763 (96%) | ||
| Service connected (%) | 750 (49%) | 2178 (51%) | 381 (52%) | 369 (46%) | ||
| Married (%) | 714 (77%) | 2190 (51%) | 301 (41%) | 413 (52%) | ||
| White, not Hispanic (%) | 493 (69%) | 1721 (69%) | 439 (60%) | 595 (74.8%) | ||
| Black, not Hispanic (%) | 125 (16%) | 221 (8%) | 161 (22%) | 64 (8.1%) | ||
| Biomedical (mean±SD) | ||||||
| MAP mmHg | 97.8±13.2 | 97.7±13.3 | 0.80 | 99.7±13.7 | 96.0±12.5 | <0.001 |
| Serum creatinine (mg/dl) | 1.20±0.76 | 1.15±0.49 | 0.004 | 1.10±0.68 | 1.56±7.41 | 0.14 |
| HDL cholesterol (mg/dl) | 41.1 ± 11.9 | 41.1 ± 12.0 | 0.92 | 44.8±13.5 | 46.3±15.1 | 0.20 |
| LDL cholesterol (mg/dl) | 111.4±33.9 | 117.6±37.7 | <0.001 | 112.5±37.9 | 112.6±33.3 | 0.10 |
| Hemoglobin AIC (%) | 6.4±1.2 | 6.4±1.3 | 0.4 | 6.5±1.5 | 6.5±1.4 | 0.91 |
Comparison to source population
There were clinically minor biomedical differences between VAHC study subjects and source population (Table 1). Compared to the source population, study participants were slightly younger (64.3 versus 65.2 years, P = 0.04), had higher creatinine (1.20 versus 1.15 mg/dl; P = 0.004) and had lower LDL cholesterol (111.4 versus 117.6; P < 0.0001). Self-identified ethnicity differed in the sample versus the source population (P = 0.013); inspection of the categories revealed that this difference was principally attributable to over-ascertainment of African American subjects (at 16% versus 8%). Comparison of baseline and biochemical parameters of the VAHC by recruitment status showed that the clinic population was younger compared to the mail recruits (60.8 versus 67.5 years, P < 0.001). They were more likely to be service connected (52% versus 46%) and African American (22% versus 8.1%), and less likely to be married (41% versus 51%). The clinic recruits also had higher baseline MAPs (99.7 versus 96.0 mmHg, P < 0.001).
Antihypertensive medication use
While most were prescribed two or more antihypertensive medications at some portion of this period, 1218 participants had time periods of mono therapy during the follow-up period, resulting in a total of 24,972 months (average of 20.5 months per patient) of follow-up on mono therapy. Of those on mono therapy, 552 participants were on an angiotensin converting enzyme inhibitor, 374 were on a beta-adrenergic receptor blocker, and 363 were on a diuretic, with an average of 15.7, 13.4, and 10.5 months of follow-up, respectively (Table 2). Of those on dual therapy, 850 were on an angiotensin converting enzyme inhibitor and diuretic, 776 on a diuretic and beta-adrenergic receptor blocker, and 740 on an angiotensin converting enzyme inhibitor and beta-adrenergic receptor blocker, with an average of 39.3, 35.7, and 37.3 months of follow-up, respectively (Table 2).
Table 2.
Antihypertensive medication use
| Description | Total follow-up (months) | Mean follow-up per subject (months±SD) | Unique subjects |
|---|---|---|---|
| Total (all) | 133,530 | 87.4 (22.6) | 1,527 |
| Number of antihypertensive medications | |||
| One | 24,972 | 20.5 (21.3) | 1,218 |
| Two | 31,580 | 24.8 (20.1) | 1,271 |
| Three | 26,886 | 25.3 (19.2) | 1,064 |
| Four | 15,680 | 22.1 (17.9) | 711 |
| Five | 6,370 | 17 (15.7) | 375 |
| Six | 1,335 | 12 (13) | 111 |
| Seven | 159 | 8.8 (6.3) | 18 |
| Monotherapy (most common occurrences) | |||
| Angiotensin converting enzyme inhibitor | 8,673 | 15.7 (19.8) | 552 |
| Beta-adrenergic receptor blocker | 5,017 | 13.4 (18.3) | 374 |
| Diuretic | 3,820 | 10.5 (14.3) | 363 |
| Calcium channel blocker | 3,242 | 11.5 (15.9) | 281 |
| Angiotensin receptor blocker | 852 | 11.5 (14.6) | 74 |
| Dual therapy (most common occurrences) | |||
| Angiotensin converting enzyme inhibitor and diuretic | 33,389 | 39.3 (28.5) | 850 |
| Angiotensin converting enzyme inhibitor and beta-adrenergic receptor blocker | 27,608 | 37.3 (27.8) | 740 |
| Diuretic and beta-adrenergic receptor blocker | 27,686 | 35.7 (26.7) | 776 |
| Diuretic and calcium channel blocker | 23,022 | 36.7 (27.1) | 627 |
| Angiotensin converting enzyme inhibitor and calcium channel blocker | 22,006 | 35.4 (26.7) | 621 |
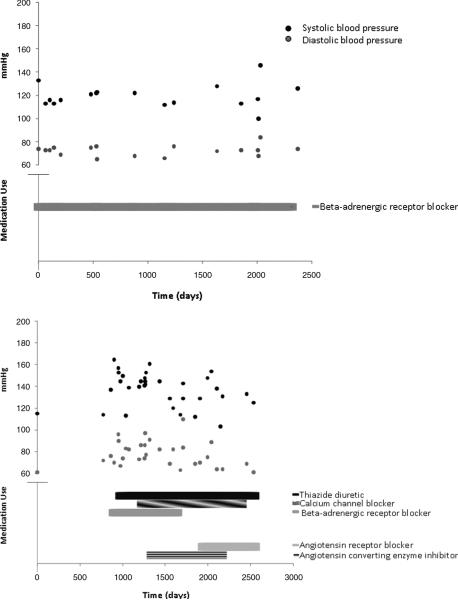
We then coupled the time periods of antihypertensive medication use to blood pressure readings from outpatient visits (hospital and emergency room blood pressures were excluded). While some patients appeared to be effectively managed by a single antihypertensive medication (Figure 2, first panel), there were others who had elevated blood pressure readings despite the use of several medications (Figure 2, second panel).
Figure 2.
Longitudinal blood pressure profiles and antihypertensive medication use. The first panel illustrates long-term blood pressure control with a single antihypertensive medication. The second panel illustrates a more complex blood pressure pattern with multiple antihypertensive medications and changes
Validation of warehouse digital data extraction (VISN-22) against individual EMR (CPRS) notes
Medical records were reviewed in a random sample of n = 105 participants to validate the concordance between hypertension-related diagnoses extracted from the VISN-22 data warehouse and patient EMR text notes (in CPRS). There was excellent concordance for coronary artery disease (ICD-9 414.00–414.07, 414.8–414.9), cerebrovascular disease (430, 431, 433.×1, 434.×1), congestive heart failure (428.0–428.9), atrial fibrillation (427.31), and sleep apnea (780.51, 780.53, 780.57): kappa scores ranged from 0.84 for congestive heart failure to 1.0 for atrial fibrillation. Patients with chronic kidney disease (serum creatinine > 1.5 mg/dl on two occasions) were identified using several ICD-9 diagnostic codes including chronic kidney disease (585.0–585.9), glomerulonephritis (582.0–582.9), nephritis (583.0–583.9), disorders resulting from impaired renal function (588.0–588.9), hypertensive renal disease (403.0–404.93), renal failure (586), and renal sclerosis (587). There was poor concordance for the 38 patients with a single diagnosis of diabetes (type unspecified, ICD-9 250); a review of medical records for these 38 participants identified 21 patients with type 2 diabetes and 17 patients with glucose intolerance (data not shown).
Based on review of an additional random sample of n = 150 participants, 11 (7.3%) received medications outside the VASDHS. Of these, eight were identified in the `non-VA medications' electronic field, while three were identified only in the text notes. Thus, the `non-VA medications' field is a high-fidelity (only 3/150, or 2% false negative rate) estimator of whether non-VASDHS-prescribed medications might contribute to the final BP values; free-form text notes were a backup source of information.
DNA and novel association of ACE inhibitor response with a functional PAI-1 (plasminogen activator inhibitor) promoter polymorphism
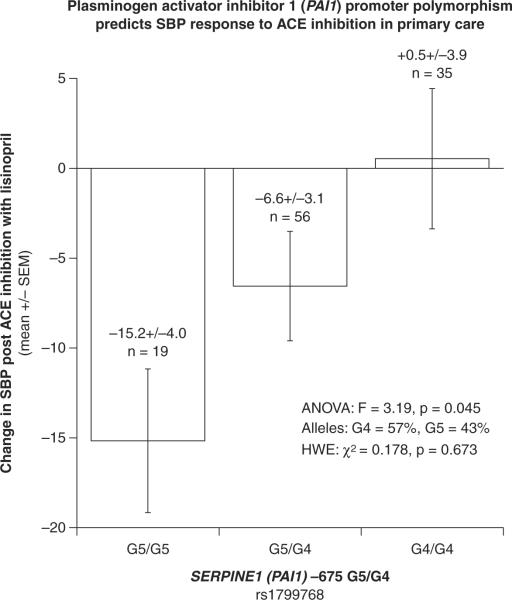
A total of 1474 (~96.5%) buccal cell samples yielded good quality (PCR-amplifiable) DNA. The average DNA yield was 5.02 ± 0.12 micrograms, by PicoGreen fluorescence (specific for double-stranded DNA). There was a significant association at the PAI-1 promoter polymorphism and blood pressure response to an angiotensin converting enzyme inhibitor (p = 0.045). PAI-1 G5/G5 homozygotes experienced a substantial decrease (−15.2 ± 4.0 mmHg), G4/G5 heterozygotes an intermediate decrease (−6.6 ± 3.1 mmHg), while G4/G4 homozygotes experienced essentially no change (+0.5 ± 3.9 mmHg) in systolic blood pressure, suggesting an additive effect of this PAI-1 variant on the BP response trait (Figure 3).
Figure 3.
Novel pharmacogenetic association. Participants with the PAI-1 G5 variant appeared to have a better systolic blood pressure response to treatment with an angiotensin converting enzyme inhibitor (p = 0.045)
Discussion
Overview
The VAHC was developed to explore the role of genetic variation in blood pressure, drug response, and hypertensive comorbid illness in a primary care setting with access to detailed electronic medical records. In this initial study, we developed the operational protocols used to extract patient data from the large VA data warehouse (VISN-22) and validated the accuracy of this information against the EMR (using CPRS text notes), developing a detailed phenotypic database. Participants were recruited in formal collaboration with VASDHS primary care doctors, enrolling 1,527 participants, approximately 26 per cent of the source VASDHS hypertension source population. While the majority (69%) were self-identified Caucasians, we over-sampled African Americans (N = 225, 16% of the cohort). Using a simple protocol, we were also able to prepare adequate buccal cell DNA from mouthwash samples for genotyping in 96.5 per cent of the study participants.
Patient privacy and the EMR
The use of the EMR to create virtual, anonymous cohorts should facilitate clinical research, especially for the study of multi-factorial complex disease requiring large study populations with data on medical comorbid illness, medication use, and patient demographics. However, patient privacy and compliance with US Federal HIPPA regulations need to be carefully considered. Despite these challenges to clinical investigators, we developed a practical, ethical, and successful approach to obtaining both EMR and genomic information from a large number of human subjects. Because of HIPPA limitations, only patients of physicians who participated as study co-investigators were recruited by mail, potentially limiting our source population to patients on their respective panels. Informed consent was obtained to enable study investigators to review the EMR and collect DNA samples for genomic analysis.
Population sampling
Our strategy yielded a final sample that was generally representative of the source population in age; however, we over-sampled particular minority populations (i.e. African Americans) and women. Hypertension takes an inordinate toll of target end organ disease on African Americans,15–17 and the benefits of hypertension treatment in women are incompletely understood;24 therefore, such over-sampling of particular population subgroups is desirable and seemed to be facilitated by direct patient contact. There appeared to be some differences as a function of recruitment method (clinic versus mail). The clinic subjects were younger, had higher MAPs, and had higher LDL cholesterol levels, suggesting a more complicated patient group who perhaps required more frequent follow-up. As shown in other research, the VA VISN databases appear to accurately capture diagnostic coding25, 26 and most hypertension-related diagnoses were accurately captured using the large VISN-22 administrative database in this study. Finally, the `non-VA medications' field in CPRS appears to be a high-fidelity estimator of whether non-VASDHS medications were prescribed, suggesting that misclassification of antihypertensive treatment should not be a significant source of error in these analyses.
DNA acquisition and pharmacogenetic association with blood pressure enabled by the longitudinal EMR
We were able to obtain high-quality DNA from participants non-invasively with a simple mouth-wash protocol. This method worked well even at a distance (by mail), with ~ 96.5 per cent of buccal cell samples generating DNA at an average yield of ~ 5 micrograms/person, sufficient for > 1000 SNP genotypes. We used the EMR to uncover a novel association between ACE inhibitor and a novel variant in the PAI-1 locus. Observational cohorts with detailed phenotypic information may prove to be valuable resources to uncover novel complex genetic disease determinants. However, such genomic predictions will require replication in independent groups before widespread acceptance.
Study limitations
This non-interventional, observational study presents several limitations. Because of HIPPA regulations, our source population was limited to hypertensive patients of primary care physicians who participated in this study. Despite repeat mailings, the response rate to the postal invitations was also low (~ 16%); thus, future recruitment efforts will emphasize sending additional reminders in addition to direct (face-to-face) recruitment from the outpatient clinic. Even though participants were from a primary care setting, results may not be readily generalized to women (relatively small numbers in this source population) or other patient populations not present in sufficient numbers within this particular target population. Some loss to follow-up and/or co-managed care with non-VA providers is expected to be an issue in a longitudinal study such as this. Although antihypertensive medication adherence has been shown to be good in another VA based study,27 adherence in this cohort remains to be studied. Non-VA medication use, however, appears to be adequately documented in an EMR field. As it is not feasible to execute a detailed examination of each individual patient's medical record, we are limited to use of the structured data such as ICD codes, medication class, anthropometric parameters, and laboratory data. The advancement in the development and utilization of coding systems for text mining within EMRs will further facilitate the process of cohort generation and phenotype determination for genetic studies.28, 29
Conclusion
The VAHC was established to explore potential genetic determinants of hypertension, comorbid illness, and drug responses in an ethnically diverse primary care setting, with longitudinal follow-up and access to a detailed EMR. With careful attention to privacy/HIPAA concerns, we developed a successful strategy to capture ~ 26 per cent of the source hypertensive population using two recruitment methods (in clinic or by mail), and the enrolled sample effectively mirrored the primary care source population. Direct recruitment from the clinic allowed us to over-sample African Americans, an important consideration since African Americans have a higher burden of hypertension and associated end organ disease. Non-invasive sampling yielded abundant genomic DNA in a majority of subjects, providing the opportunity to demonstrate a novel genetic effect on the longitudinal response to antihypertensive drug treatment. The use of similar electronic medical databases with accurate characterization of comorbid diseases and longitudinal follow-up has the potential to enhance the study of genetic determinants for complex chronic disease traits.
Acknowledgements
We are grateful for the participation and support of the VASDHS primary care physicians from the Division of General Internal Medicine. We would also like to thank Brenda Thomas LVN (patient recruitment and consent), Guangfa Zhang PhD (database development), and Neurita Salva BS (patient recruitment, DNA handling and processing). The funding sources for this study are: National Institutes of Health (K23 RR020822, HL58120, EXPORT/CRCHD MD000220, GCRC RR000827), Department of Veterans Affairs.
References
- 1.Hu H, Brzeski H, Hutchins J, et al. Biomedical informatics: development of a comprehensive data warehouse for clinical and genomic breast cancer research. Pharmacogen. 2004;5:933–941. doi: 10.1517/14622416.5.7.933. [DOI] [PubMed] [Google Scholar]
- 2.Murphy S, Churchill S, Bry L, et al. Instrumenting the health care enterprise for discovery research in the genomic era. Genome Res. 2009;19:1675–1681. doi: 10.1101/gr.094615.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.An afternoon at UK Biobank. Lancet. 2009;373:1146. doi: 10.1016/S0140-6736(09)60664-4. [DOI] [PubMed] [Google Scholar]
- 4.Wilke RA, Berg RL, Peissig P, et al. Use of an electronic medical record for the identification of research subjects with diabetes mellitus. Clin Med Res. 2007;5:1–7. doi: 10.3121/cmr.2007.726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fultz SL, Skanderson M, Mole LA, et al. Development and verification of a `virtual' cohort using the National VA Health Information System. Med Care. 2006;44:S25–30. doi: 10.1097/01.mlr.0000223670.00890.74. [DOI] [PubMed] [Google Scholar]
- 6.Siegel D, Meier J, Maas C, Lopez J, Swislocki AL. The effect of body mass index on fasting blood glucose after initiation of thiazide therapy in hypertensive patients. Am J Hypertens. 2008;21:438–442. doi: 10.1038/ajh.2007.75. [DOI] [PubMed] [Google Scholar]
- 7.Kristianson KJ, Ljunggren H, Gustafsson LL. Data extraction from a semi-structured electronic medical record system for outpatients: a model to facilitate the access and use of data for quality control and research. Health Inform J. 2009;15:305–319. doi: 10.1177/1460458209345889. [DOI] [PubMed] [Google Scholar]
- 8.West SL, Blake C, Zhiwen L, McKoy JN, Oertel MD, Carey TS. Reflections on the use of electronic health record data for clinical research. Health Inform J. 2009;15:108–121. doi: 10.1177/1460458209102972. [DOI] [PubMed] [Google Scholar]
- 9.Chen DP, Weber SC, Constantinou PS, Ferris TA, Lowe HJ, Butte AJ. Novel integration of hospital electronic medical records and gene expression measurements to identify genetic markers of maturation. Pac Symp Biocomput. 2008:243–254. [PMC free article] [PubMed] [Google Scholar]
- 10.Wood GC, Still CD, Chu X, et al. Association of chromosome 9p21 SNPs with cardiovascular phenotypes in morbid obesity using electronic health record data. Genomic Med. 2008;2:33–43. doi: 10.1007/s11568-008-9023-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chobanian AV, Bakris GL, Black HR, et al. The Seventh Report of the Joint National Committee on Prevention, Detection, Evaluation, and Treatment of High Blood Pressure: the JNC 7 report. J Am Med Assoc. 2003;289:2560–2572. doi: 10.1001/jama.289.19.2560. [DOI] [PubMed] [Google Scholar]
- 12.Alexander M, Tekawa I, Hunkeler E, et al. Evaluating hypertension control in a managed care setting. Arch Intern Med. 1999;159:2673–2677. doi: 10.1001/archinte.159.22.2673. [DOI] [PubMed] [Google Scholar]
- 13.Bramley TJ, Gerbino PP, Nightengale BS, Frech-Tamas F. Relationship of blood pressure control to adherence with antihypertensive monotherapy in 13 managed care organizations. J Manag Care Pharm. 2006;12:239–245. doi: 10.18553/jmcp.2006.12.3.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jami P, Smith P, Moningi S, et al. Compliance with Joint National Committee 7 guidelines in hypertension management in a teaching institution. Am J Med Qual. 2007;22:251–258. doi: 10.1177/1062860607303293. [DOI] [PubMed] [Google Scholar]
- 15.El-Gharbawy AH, Kotchen JM, Grim CE, et al. Predictors of target organ damage in hypertensive blacks and whites. Hypertension. 2001;38:761–766. doi: 10.1161/hy1001.092613. [DOI] [PubMed] [Google Scholar]
- 16.Hicks LS, Shaykevich S, Bates DW, Ayanian JZ. Determinants of racial/ethnic differences in blood pressure management among hypertensive patients. BMC Cardiovasc Disord. 2005;5:16. doi: 10.1186/1471-2261-5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rehman SU, Hutchison FN, Hendrix K, Okonofua EC, Egan BM. Ethnic differences in blood pressure control among men at Veterans Affairs clinics and other health care sites. Arch Intern Med. 2005;165:1041–1047. doi: 10.1001/archinte.165.9.1041. [DOI] [PubMed] [Google Scholar]
- 18.Kotchen TA, Piering AW, Cowley AW, et al. Glomerular hyperfiltration in hypertensive African Americans. Hypertension. 35:822–826. doi: 10.1161/01.hyp.35.3.822. [DOI] [PubMed] [Google Scholar]
- 19.Gunn PP, Fremont AM, Bottrell M, Shugarman LR, Galegher J, Bikson T. The Health Insurance Portability and Accountability Act Privacy Rule: a practical guide for researchers. Med Care. 2004;42:321–327. doi: 10.1097/01.mlr.0000119578.94846.f2. [DOI] [PubMed] [Google Scholar]
- 20.Watts PL, Hynes DM, Kopp A. Research implications of the privacy standards under the Health Insurance Portability and Accountability Act of 1996 (HIPPA) VIRec (Technical Bulletin) 2003:4. [Google Scholar]
- 21.Keltai M, Johnson JA, Kowey PR, Ried LD, Tueth M. INVEST substudies: design and patient characteristics. Clin Cardiol. 2001;24:V9–11. doi: 10.1002/clc.4960241704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Belova LA. Angiotensin II-generating enzymes. Biochemistry (Mosc) 2000;65:1337–1345. doi: 10.1023/a:1002848402911. [DOI] [PubMed] [Google Scholar]
- 23.Ding Z, Pan JQ. Distribution of PAI-1 promoter 4G/5G polymorphism in healthy Chinese and functional characterization using a luciferase reporter vector. Ann Hematol. 2005;84:183–187. doi: 10.1007/s00277-004-0926-z. [DOI] [PubMed] [Google Scholar]
- 24.Samad Z, Wang TY, Frazier CG, Shah SH, Dolor RJ, Newby LK. Closing the gap: treating hypertension in women. Cardiol Rev. 2008;16:305–313. doi: 10.1097/CRD.0b013e31817f9350. [DOI] [PubMed] [Google Scholar]
- 25.Borzecki AM, Wong AT, Hickey EC, Ash AS, Berlowitz DR. Identifying hypertension-related comorbidi-ties from administrative data: what's the optimal approach? Am J Med Qual. 2004;19:201–206. doi: 10.1177/106286060401900504. [DOI] [PubMed] [Google Scholar]
- 26.Szeto HC, Coleman RK, Gholami P, Hoffman BB, Goldstein MK. Accuracy of computerized outpatient diagnoses in a Veterans Affairs general medicine clinic. Am J Manag Care. 2002;8:37–43. [PubMed] [Google Scholar]
- 27.Siegel D, Lopez J, Meier J. Antihypertensive medication adherence in the Department of Veterans Affairs. Am J Med. 2007;120:26–32. doi: 10.1016/j.amjmed.2006.06.028. [DOI] [PubMed] [Google Scholar]
- 28.Rector AL, Brandt S. Why do it the hard way? The case for an expressive description logic for SNOMED. J Am Med Inform Assoc. 2008;15:744–751. doi: 10.1197/jamia.M2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rector AL, Qamar R, Marley T. Binding ontologies and coding systems to electronic health records and messages. Appl Ontol. 2009;4:51–69. [Google Scholar]