Abstract
The Achères wastewater treatment plant, located just downstream of Paris, discharges its effluents into the lower Seine River. The effluents contain large numbers of heterotrophic bacteria, organic matter, and ammonium and are a source of nitrifying bacteria. As a result, degradation of organic matter by heterotrophic bacteria and subsequent oxygen depletion occur immediately downstream of the effluent outlet, whereas nitrifying bacteria apparently need to build up a significant biomass before ammonium oxidation significantly depletes the oxygen. We quantified the potential total nitrifying activity and the potential activities of the ammonia- and nitrite-oxidizing communities along the Seine River. In the summer, the maximum nitrifying activity occurs in the upper freshwater estuary, ∼200 km downstream of Achères. The quantities of nitrifying bacteria, based on amoA gene copy numbers, and of Nitrobacter organisms, based on 16S rRNA gene copy numbers, were correlated with the potential nitrifying activities. The species composition of ammonia-oxidizing bacteria was investigated at two sites: the Triel station just downstream from Achères (km 84) and the Seine freshwater estuary at the Duclair station (km 278). By means of PCR primers targeting the amoA gene, a gene library was created. Phylogenetic analysis revealed that the majority of the analyzed clones at both sites were affiliated with the genus Nitrosomonas. The Nitrosomonas oligotropha- and Nitrosomonas urea-related clones represented nearly 81% of the community of ammonia-oxidizing bacteria at Triel and 60% at Duclair. Two other ammonia-oxidizing clusters of the β subclass of the Proteobacteria, i.e., Nitrosomonas europaea- and Nitrosospira-like bacteria, were found in smaller numbers. The major change in the ammonia-oxidizing community between the two stations along the Seine River-upper estuary continuum was the replacement of the N. oligotropha- and N. urea-related bacteria by the Nitrosospira-affiliated bacteria. Although the diversities of the ammonia oxidizers appear to be similar for the two sites, only half of the restriction patterns are common to both sites, which could be explained by the differences in ammonium concentrations, which are much lower in the upper estuary than in the river at the effluent outlet. These results imply a significant immigration and/or selection of the ammonia-oxidizing bacterial population along the continuum of the Seine River from Paris to the estuary.
The Seine River downstream of Paris is greatly affected by the discharge of treated effluents from the Achères wastewater treatment plant (WWTP) (8.5 million inhabitant equivalent from Paris and its suburbs, subjected only to a secondary treatment). A typical feature of the ecological functioning of the lower Seine River is a high summer oxygen deficit immediately downstream of the Achères effluent outlet due to degradation of organic matter (18, 19, 48) and another depletion in the freshwater estuary due to nitrification of ammonium that is also contained in the effluent (41). Since 1989, studies of the Seine River have extended the knowledge of heterotrophic organic matter degradation (18, 48) and nitrification (8), as well as of the characteristics of wastewater effluents (49) and oxygen budgets (19). Moreover, nitrifying bacteria had previously been shown to originate from wastewater effluent that was not subjected to tertiary treatments (8). The plans for a tertiary-treatment facility (including nitrification) at the Achères WWTP in 2005, therefore, required further studies of the nitrification process and its associated bacteria in the continuum of the Seine River.
Nitrification, mainly due to autotrophic bacteria, is the oxidation of ammonia into nitrite and subsequently into nitrate. In freshwater environments, the first step is carried out by chemolithotrophic ammonia-oxidizing bacteria (AOB), which form a tight cluster within the β subclass of the Proteobacteria and contain members of the genera Nitrosomonas (as well as Nitrosococcus mobilis) and Nitrosospira (as well as Nitrosolobus and Nitrosovibrio) (23, 58, 60); AOB of the γ subclass of the Proteobacteria are only found in halophilic and marine environments. The second step of nitrification is carried out by chemolithotrophic nitrite-oxidizing bacteria (NOB) that belong to four phylogenetically distinct groups. One group, belonging to the α subclass of the Proteobacteria, is represented by the genus Nitrobacter. The two known species of the genus Nitrospira are members of a distinct phylum. Finally, the two marine species Nitrococcus mobilis and Nitrospina gracilis are assigned to the γ and the δ subclasses of the Proteobacteria, respectively. Several methods can be used to investigate nitrifying bacterial populations in situ; cultivation-dependent analysis often leads to significantly underestimated cell counts due to the presence of unculturable species, and the very low growth rate of the nitrifiers (62) makes it time-consuming. Measurements of potential nitrifying activities by the 14C method can be used to estimate biomass (7) but do not identify the bacterial populations involved in the two steps of the process. However, in the last 5 years, molecular techniques have proved to be well suited to studying these bacteria, both for quantifying the nitrifiers by competitive or real-time PCR and for assessing the composition of the nitrifying population by the construction of a specific clone library and subsequent sequencing. Primers targeting part of the amoA gene, which encodes ammonia monooxygenase, may serve to target the AOB by the use of PCR (42). PCR primers targeting the whole NOB functional group do not exist at present, but 16S ribosomal DNA (rDNA) primers are used to target a specific genus. The genus Nitrobacter, which can be targeted according to the method of Degrange and Bardin (12), was chosen as a representative of the nitrite-oxidizing population in the system studied here.
The study was carried out in summer, at low flow, from Paris to the coastal zone (Honfleur) along the aquatic continuum of the lower Seine River and its estuary, both of which are strongly impacted by the effluent inputs. In addition to determining the relative contribution of the nitrifying activity to the oxygen budget (19), the aims were (i) to quantify the bacterial populations involved in the two steps of the process in order to pursue quantitative modeling of the ecological functioning of the Seine River and estuary (4) and (ii) to identify the composition of the AOB community by molecular methods in order to investigate potential species selection or adaptation of the populations along the Seine River. The population composition was therefore studied at two sites: one immediately downstream of the effluent outlet of the Achères treatment plant and the other in the upstream part of the freshwater estuary, where nitrification generally reaches its maximum.
MATERIALS AND METHODS
Description of research area.
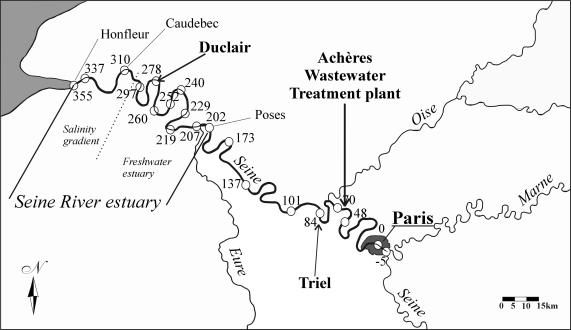
The lower Seine River extends from Paris (km 0) to the weir at Poses (km 202), which prevents tidal water from moving upstream and consequently forms the upstream limit between the river and the estuary (Fig. 1). Paris is at km 0, and the other stations are designated by their distances from Paris. The fluvial sector of the estuary extends up to Caudebec (km 310), which is the limit of saline intrusion. From Caudebec to Honfleur (km 356), at the mouth of the estuary, there is a continuous salinity gradient influenced mainly by tides and river flow.
FIG. 1.
Representation of the Seine River from Paris to the estuary. The numbered circles represent sampling points for process work. The two sites in boldface type are where the molecular characterization was done.
The lower Seine River and estuary receive all the inputs from a drainage basin characterized by great industrial activity, intensive agriculture, and a high population density, including, in particular, the city of Paris (Fig. 1). The domestic effluent of the 8.5 million inhabitants of Paris and its suburbs is treated by activated sludge in the WWTP of Achères, 70 km downstream from Paris. The lower Seine River is therefore enriched in organic matter and nutrients, especially ammonium, which is known to be completely converted into nitrate in the upstream estuary.
Sample collection and analytical methods.
Water samples were taken along the lower Seine River and estuary, from upstream of Paris to Honfleur (the mouth of the estuary), in September 1997, September 1998, and July 1999. The three longitudinal profiles included samples collected upstream and just downstream of the treatment plant outlet (at the stations of Maisons-Laffitte and Conflans-Ste Honorine, at km 48 and 70, respectively) (Fig. 1). At each station, a 10-liter water sample was collected for all laboratory analyses of water quality variables (especially suspended matter and nitrogen) and was stored at 4°C during transfer to the laboratory. DNA was extracted from the same samples but only from those collected in September 1998; an aliquot of 500 ml was immediately poured into sterile polypropylene centrifugation bottles (Nalgene) and adjusted to 0.1% sodium dodecyl sulfate and 1 mM EDTA. The samples were then stored at −20°C until the nucleic acids were extracted. Four hundred fifty milliliters of each water sample was used for the measurement of potential nitrifying activity and ammonia and nitrite oxidation rates. Ten milliliters was fixed with formaldehyde (final concentration, 2%) to determine the total bacterial abundance by DAPI (4′,6′-diamidino-2-phenylindole) staining and epifluorescence microscopy (35).
Measurement of potential nitrifying activity.
The potential nitrifying activities of all the water samples brought to the laboratory were measured according to the method described by Brion and Billen (7). The measurements were made under optimum conditions for oxygen (7.5 mg · liter−1) and ammonium concentration (2 mM NH4Cl) and at a constant temperature of 20°C. The potential nitrifying activities were determined by the differences between the amounts of H14CO3− incorporated after 20 to 24 h of incubation in the samples with and without added specific nitrification inhibitors (7, 51). N-Serve (2-chloro-6-trichloromethyl pyridine [also called Nitrapyrin]; 5 mg · ml−1) and sodium chlorate (NaClO3; 10 mM) were used in order to specifically inhibit ammonia and nitrite oxidation, respectively (1, 26, 56). Carbon incorporation is proportional to ammonia oxidation, with 0.11 mol of carbon incorporated per mol of NH4+ oxidized into nitrate (7). Under these optimal conditions, the nitrifying activity is roughly proportional to the nitrifying biomass (1, 7).
In order to separately estimate the ammonia and nitrite oxidation rates, we measured nitrite concentration changes in subsamples containing either N-Serve or NaClO3, which inhibit the oxidation of ammonia and nitrite, respectively. The samples were incubated in the dark, at room temperature, under optimum conditions for oxygen and with the addition of ammonium (NH4Cl; 2 mM) and nitrite (KNO2; 0.36 mM). The nitrite concentrations were measured at the beginning and end of the 24-h incubation period, during which the oxidation rates were shown to be linear. The ammonia or nitrite oxidation rates were calculated from the increase (ammonia oxidation) or decrease (nitrite oxidation) of nitrite concentrations during the incubation (3).
Measurements of water quality variables.
Ammonium and nitrite were measured by spectrophotometry in water filtered through a glass fiber membrane (GF/F; Whatman) according to the method of Slavyck and McIsaac (50). Nitrate was assessed spectrophotometrically in filtered water after Cd reduction to nitrite (40).Suspended matter was weighed on GF/F filters dried at 450°C.
DNA extraction and purification.
DNA was extracted from the Seine River water samples (only samples from September 1998) according to the protocol developed by Petit et al. (33). This method allows extraction of nucleic acids from particle-attached and free-living bacteria in the water. The DNA was further purified through an Elutip-d column (Schleicher & Schuell) according to the recommendations of the manufacturer. The purified DNA was then precipitated with ethanol, resuspended in Tris-EDTA buffer, and stored at −20°C until it was used.
Detection and quantification of ammonia oxidizers and Nitrobacter species by PCR and cPCR.
PCR amplification of a 491-bp fragment of the amoA gene was carried out by using the amoA-1F and amoA-2R primer set specific for ammonia oxidizers belonging to the β subclass of Proteobacteria (42). Nitrobacter, one of the most studied genera for which the tools to target the population were available, was chosen as a representative of the NOB for a quantitative study (2). Nitrobacter cells were detected by amplification of a 397-bp fragment of the Nitrobacter 16S rDNA gene with the FGPS 872 and FGPS 1269′ primers (12). All PCR amplifications were carried out in a total volume of 50 μl in 0.2-ml tubes by means of a DNA thermocycler (GeneAmp 2400 PCR system; Perkin-Elmer Cetus). The reaction mixtures were prepared in 1× buffer [75 mM Tris, pH 9.0, 20 mM (NH4)2SO4, 0.01% Tween 20], 1.5 mM MgCl2, 100 ng of DNA, and 0.5 U of Taq DNA polymerase (RedGoldtar; Eurogentec). The thermal profiles included an initial denaturing step consisting of 94°C for 45 s followed by 35 cycles of denaturation at 94°C for 45 s, annealing at 55°C with the amoA primers and at 50°C with the FGPS primers, and elongation at 72°C for 120 s. The cycle was ended by a final elongation step at 72°C for 10 min. Aliquots (10 μl) of the amplification products were analyzed by gel electrophoresis on 2% (wt/vol) agarose gels (Boehringer). Estimates of ammonia oxidizers (amoA gene copy numbers) and Nitrobacter cell numbers were made by competitive PCR (cPCR) as described by Stephen et al. (55) and Berthe et al. (2), respectively. For cPCR of AOB, we constructed a competitor as described by Stephen et al. (55). One of our clones (Duc27) was used as a DNA template. Amplification products were analyzed by agarose gel electrophoresis, and DNA band intensities were estimated with imaging and analysis software (Bio-Rad).
Construction and analysis of an amoA gene fragment library.
Two clone libraries were constructed for two sites along the Seine River continuum, Triel (km 84) and Duclair (km 278). The amplified amoA PCR fragments were excised from the agarose gel, purified with an agarose gel extraction kit (Sephaglass; Pharmacia), and eluted in Tris-EDTA buffer. The 491-bp amoA DNA fragments from the Duclair sample were cloned in a pPCR-Script Amp SK(+) vector (PCR-Script Amp cloning kit; Stratagene) according to the manufacturer's recommendations. The 491-bp amoA DNA fragments from the Triel sample were cloned in a pCR2.1 vector (TAcloning kit; Invitrogen), also according to the manufacturer's recommendations. Fifty clones from each library were randomly selected for further analysis. The cloned inserts were reamplified with amoA primers and then digested with the MspI enzyme. Restriction patterns were analyzed after gel electrophoresis on 2% agarose gels. Clones representative of each restriction pattern were chosen for sequence analysis. Selected plasmids were then subjected to double-strand service sequencing (Eurogentec). Phylogenetic analyses were conducted by aligning our amoA sequences with the amoA sequences from the GenBank database using Clustal W version 1.7 (59). Phylogenetic algorithms and trees (DNA-DIST, NEIGHBOR, and SEQBOOT) were operated through the PHYLIP version 3.5 package written by J. Felsenstein (16).
Nucleotide sequence accession numbers.
The sequences determined in this study were deposited in the GenBank database under the following accession numbers: Duc1 (AF367461), Duc2 (AF367462), Duc14 (AF367464), Duc17 (AY249149), Duc26 (AF367465), Duc27 (AF367463), Duc31 (AY249150), Duc32 (AF367466), Duc34 (AY249151), Duc47 (AY249152), T15 (AY249153), T21 (AY249154), T22 (AY249155), T27 (AY249156), T38 (AY249157), T42 (AY249158), T45 (AY249159), T47 (AY249160), and T48 (AY249161).
RESULTS
Nitrifying activities in the lower Seine River and estuary.
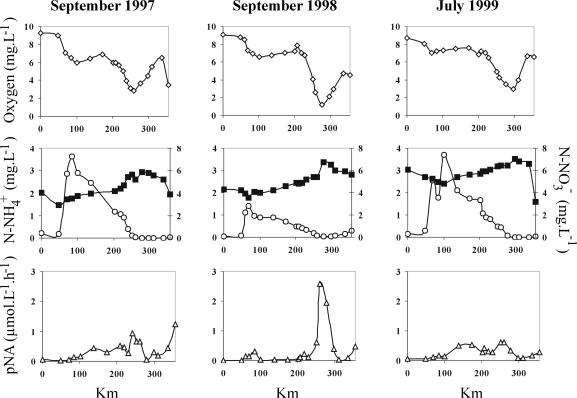
The samples collected in September 1997, September 1998, and July 1999 corresponded to summer low flows of 290, 230, and 240 m3 s−1, respectively. As mentioned above, the two oxygen-depleted sectors were monitored (Fig. 2). The first oxygen depletion (∼6 to 7 mg of O2 · liter−1) was observed immediately downstream from the outlet of the Achères WWTP and was linked to the decomposition of organic matter by heterotrophic bacteria (18, 19). The second occurred in the Seine estuary (with oxygen concentrations as low as 1.2 mg · liter−1) and was associated with the maximum of potential nitrifying activity (from 0.6 to 2.6 μmol N-oxidized · liter−1 · h−1). Despite a regular decrease in ammonium along the studied sector due (i) to dilution by the Oise River at km 100 and (ii) to a progressive nitrification increase, the ammonium supplied by the effluent from the Achères plant was completely oxidized in the estuarine sector. As a consequence, NO3− concentrations increased, typically from 4 mg of N · liter−1 downstream of Achères to 7 mg of N · liter−1 in the Seine estuary; therefore, low levels of denitrification occurred.
FIG. 2.
Variations in oxygen, ammonium (○), and nitrate (▪) concentrations and potential nitrifying activity (pNA) along a longitudinal profile of the Seine River (from Paris, km 0, to Honfleur, km 356) during September 1997, September 1998, and July 1999.
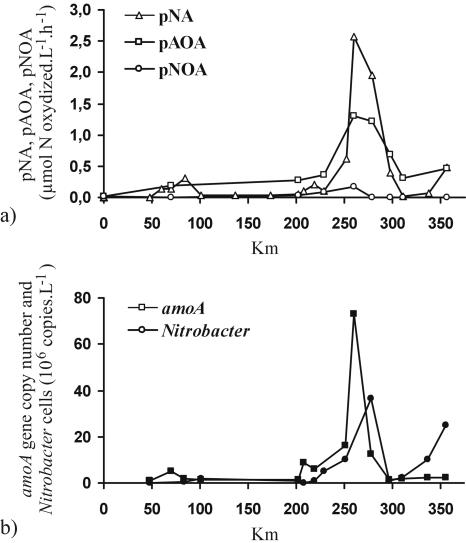
Comparison of potential ammonia and nitrite oxidation rates with the total potential nitrifying activity (by the 14C method) showed strong correlation (Fig. 3a). For the two steps of nitrification, the potential ammonia oxidation rate was ∼5 to 10 times higher than the nitrite oxidation rate, which can be explained by the much higher specific growth yield (the biomass formed per unit of substrate oxidized) of the former than of the latter (7).
FIG. 3.
(a) Potential nitrifying activity (pNA) compared with potential ammonia-oxidizing activity (pAOA) and potential nitrite-oxidizing activity (pNOA). (b) amoA gene copy numbers and Nitrobacter 16S rDNA gene copy numbers determined by cPCR (samples from September 1998).
Detection and quantification of ammonia oxidizers and Nitrobacter cells.
The approaches used for the bacterial populations involved in the two steps of the nitrification process were conceptually different. We used the specific primer set published by Rotthauwe et al. (42) to target a stretch of the amoA gene, which encodes the active site of ammonia monooxygenase, a key enzyme in AOB, whereas a primer set targeting only Nitrobacter 16S rDNA (12) was used for the detection of a fraction of the nitrite-oxidizing community. As Nitrobacter cells have one copy of the 16S rRNA operon (31), the number of 16S rDNA copies determined by cPCR represents a true evaluation of the abundance of Nitrobacter cells, whereas the abundance of the AOB can be determined only by a factor of 1 to 3, as the cells contain from one to three copies of the amoA gene (28, 32).
Following DNA extraction, application of the bacterial 16S rDNA primers pA and pHr (14) generated a fragment of 1.5 kbp for all samples examined, suggesting that no inhibitory substances (such as humic acids) were present in the DNA samples. Application of the amoA primer set generated a 491-bp DNA fragment for nearly all water samples collected along the longitudinal profile, except for those from the Maisons-Laffitte station upstream of the Achères plant. For NOB, amplification of a 397-bp region of the Nitrobacter 16S rDNA gene was achieved in all water samples, from Paris to Honfleur, including the station upstream of the WWTP. Quantification of Nitrobacter abundances and the numbers of amoA gene copies (by cPCR) along the longitudinal profile led to a pattern fairly similar to the one for the potential nitrifying activity (Fig. 3). The number of amoA gene copies increased ∼75-fold (from 9.7 × 105 to 7.3 × 107 amoA copies liter−1), and the Nitrobacter abundance increased 370-fold (from 9.8 × 104 to 3.6 × 107 Nitrobacter cells liter−1) between Maisons Laffitte (km 48, upstream of the Achères effluent output) and La Bouille (km 260)-Duclair (km 278), the sector where the potential nitrifying activity was at its maximum. We noticed an increase in the number of amoA gene copies immediately downstream from the Achères WWTP that was not observed for Nitrobacter. In the lower estuary, a sharp decrease in both nitrifying populations was observed at Caudebec (km 300), the limit of the saline intrusion; Nitrobacter abundance then increased again in the turbidity maximum and salinity gradient.
The concentration of the total bacterial community (by epifluorescence microscopy) showed a rough increase along the continuum, from 8.3 × 109 bacteria liter−1 at the station upstream of Achères (Maisons Laffitte) to 3 × 1010 bacteria liter−1 in the estuary. Considering the whole longitudinal profile, the β-AOB and Nitrobacter represented, on average, 0.11 and 0.03%, respectively, of the total bacterial community, with maximum values at La Bouille and Duclair (0.70% for β-AOB; 0.17% for Nitrobacter).
Characterization of the communities of AOB at two sites on the Seine River.
Evaluations of the natural diversity of amoA genes, as obtained by PCR amplification, cloning, and sequence analysis, were conducted at two key stations on the river continuum: at Triel, strongly influenced by the effluent of the Achères WWTP, and at Duclair, in the estuary (Fig. 1), where the highest value for potential nitrifying activity and the lowest concentration of ammonium were measured. The amoA PCR products retrieved from these water samples were used for the generation of two amoA gene libraries. A restriction enzyme analysis (restriction fragment length polymorphism) was performed on amoA fragments from 50 clones from each library.
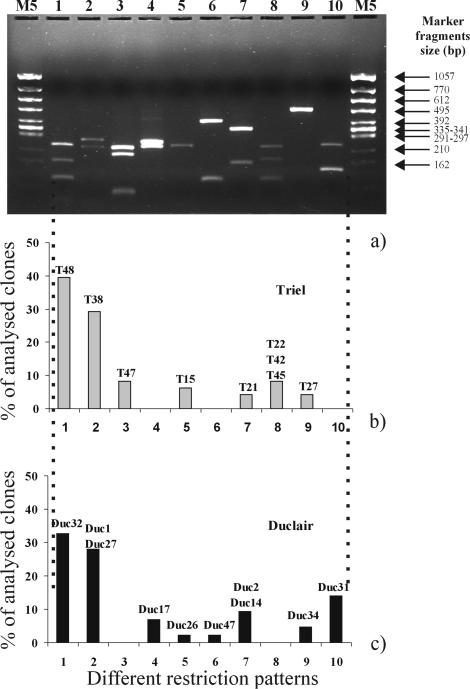
Digestion with MspI revealed 10 different restriction patterns (Fig. 4) among the 100 analyzed clones. The analyzed Triel (T) clones were clustered in seven different patterns, while the analyzed Duclair (Duc) clones were clustered in eight different patterns. The distribution of the analyzed clones in the different patterns is indicated in Fig. 4. The two major groups were the same at both sites (restriction groups 1 and 2) and represented 60 and 70% of the Duclair and Triel clones, respectively. Two groups were unique to the Triel station (restriction groups 3 and 8), and three were unique to Duclair (restriction groups 4, 6, and 10).
FIG. 4.
(a) Gel with 10 different MspI restriction patterns of analyzed amoA clones from Triel and Duclair; the size marker (M5; Eurogentec) was a HincII digest of phage φX174 DNA. (b and c) Proportions of analyzed clones in each restriction pattern for the two clone libraries of Triel and Duclair. The names of the representative clones that were sequenced and used for phylogenetic analyses are indicated above the bars (samples from September 1998).
In order to identify the dominant ammonia oxidizers, one clone representative of all pattern groups was chosen for sequence analysis, but for some groups two were selected, i.e., a total of 19 sequences were analyzed (Fig. 4). The similarity among all amoA sequences (from the two samples from the Seine River) ranged from 71 to 99%, according to the results from the Edtaln program. BLASTN searches of the GenBank database confirmed that all of the sequenced clones corresponded to amoA-like sequences and showed that some clones were very close to sequences available in the databases (Table 1). All of the analyzed clones showed strong homology (from 87 to 99%) with uncultured environmental clones. The majority were retrieved from various nitrifying WWTPs (29), except T27 and T47, which seemed closely related to clones retrieved from freshwater habitats.
TABLE 1.
Description of closest relatives of Duclair and Triel clones found in GenBank database
| Clone | Closest relative sequence | % of similarity | Environment | Reference |
|---|---|---|---|---|
| Duc1 | Uncultured AOB | 87 | Biofilm | 21 |
| B2-3 (AF293067) | ||||
| B2-1 (AF293065) | ||||
| Duc2 | Uncultured AOB | 98 | WWTP | 38 |
| BF1-2 (AF272483) | ||||
| SBBR1-5 (AF272426) | ||||
| Duc14 | Uncultured AOB | 98 | WWTP | 38 |
| BF1-2 (AF272483) | ||||
| SBBR1-5 (AF272426) | ||||
| Duc17 | Uncultured AOB | 97 | WWTP | 38 |
| amoa17 (AF272507) | ||||
| Duc26 | Uncultured AOB | 97 | WWTP | 38 |
| amoa17 (AF272507) | ||||
| Duc27 | Uncultured AOB | 99 | WWTP | 38 |
| PoII-5 (AF272474) | ||||
| Duc31 | Uncultured AOB | 97 | WWTP | 38 |
| amoa17 (AF272507) | ||||
| Duc32 | Uncultured AOB | 99 | WWTP | 13 |
| M-20 (AF420299) | ||||
| Duc34 | Uncultured AOB | 98 | Activated sludge | 38 |
| GLII-9 (AF272472) | ||||
| Duc47 | N. nitrosa (AF272404) | 97 | Activated sludge | 38 |
| T15 | Uncultured AOB | 96 | WWTP | 38 |
| amoa17 (AF272507) | ||||
| T21 | Uncultured AOB | 97 | WWTP | 38 |
| IA-32 (AF272442) | ||||
| T22 | Nitrosomonas sp. strain Nm47 (AY123830) | 88 | WWTP | 38a |
| T27 | Uncultured AOB | 97 | Freshwater | 52 |
| pMpC.1 (AJ388579) | ||||
| T38 | Uncultured AOB | 97 | WWTP | 38 |
| PoII-8 (AF272475) | ||||
| T42 | Nitrosomonas sp. strain Nm47 (AY123830) | 88 | WWTP | 38a |
| T45 | Nitrosomonas sp. strain Nm47 (AY123830) | 87 | WWTP | 38a |
| T47 | Uncultured AOB | 97 | Freshwater | 52 |
| pDtC.1 (AJ388575) | ||||
| T48 | Uncultured AOB | 98 | WWTP | 13 |
| M-20 (AF420299) |
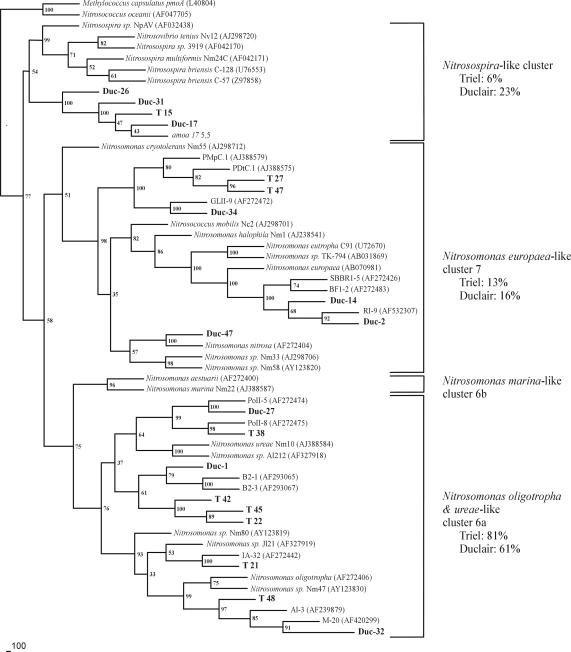
A phylogenetic tree was obtained from the analysis of a stretch of 417 bp (within the 491-bp amoA PCR product cloned) (Fig. 5). The majority of these amoA fragments retrieved from the two sites on the Seine River belonged to three clusters, with a majority belonging to the genus Nitrosomonas. The analyzed sequences did not form a monophyletic cluster but were affiliated with different Nitrosomonas species. The Duc1, Duc27, Duc32, T21, T22, T38, T42, T45, and T48 clones belonged to the cluster represented by the Nitrosomonas oligotropha- and Nitrosomonas urea-like sequences. These clones represented 60 and 81%, respectively, of the total analyzed clones from the Duclair and Triel stations (Fig. 5). In the Nitrosomonas europaea-like cluster, two clones from Duclair, Duc2 and Duc14 (belonging to the same MspI restriction group), formed a common branch with Nitrosomonas europaea, Duc47 was closely related to Nitrosomonas nitrosa, and Duc34 formed another branch. The last four sequenced clones made up 16% of the total analyzed clones from Duclair. For the Triel station, the proportion of the N. europaea-like cluster was roughly the same: clones T27 and T47 represented 13% of the total analyzed clones (Fig. 5) and were in the same separate branch as Duc34. Finally, the third cluster, Nitrosospira-like bacteria, comprised three sequenced clones from Duclair (Duc17, Duc26, and Duc31) but only one from Triel (T15), making up 23 and 6%, respectively, of the total analyzed clones. These Nitrosospira-like sequences formed a separate branch within the Nitrosospira cluster (Fig. 5).
FIG. 5.
Fitch-Margoliash tree of β subgroup Proteobacteria amoA sequences. The clones obtained in this work (in boldface type) are positioned in relation to cultured or environmental ammonia oxidizers (references are cited in Table 1). The tree was rooted with the pmoA sequence of Methylococcus capsulatus and the amoA sequence of Nitrosococcus oceanus, an AOB of the γ subclass of the Proteobacteria. The tree scale is in the lower left corner.
DISCUSSION
The treated effluent discharged into the Seine River sharply increased the ammonium concentration in the river water. After dilution by the Oise River at km 110, the concentration slowly decreases down to the estuary, presumably oxidized by nitrifying bacteria. As indicated by the potential nitrifying activity, the transformation of ammonium into nitrate was highest in the freshwater section of the estuary (Fig. 2), by which point the population of nitrifying bacteria had built up a large biomass. This generally occurred in summer, at a high temperature, when the water residence time was sufficiently long (8, 19).
Summer oxygen depletion in the fluvial part of the Seine estuary is a constant feature of the ecological functioning of the system (Fig. 2); the oxygen budget established for the whole section investigated here showed that nitrification was a major controlling process, probably typical of large river systems strongly impacted by domestic and industrial effluents (19).
Even after treatment, domestic effluents are sources not only of chemical compounds, but also of microorganisms that can be active in the river, and they may play a fundamental role in the ecological functioning of the system (18, 22). Heterotrophic bacteria deplete the oxygen immediately downstream from the input of the effluent (17, 18), whereas the nitrifying bacteria deplete it far downstream in the upper estuary. However, whereas heterotrophic bacteria are massively supplied by the effluent (18), the input of nitrifying bacteria is much weaker (8), which is corroborated by the molecular approaches originally used in the Seine River study. The DNAs of ammonia and nitrite oxidizers were detected even in wastewater at the heads of effluent collectors (20). The presence of nitrifying bacteria in the domestic wastewater suggests that the biofilm covering the walls of the collectors favors the growth of nitrifying bacteria. It has been reported that Nitrosomonas strains can survive in low oxygen concentrations, and they have been observed to constitute about one-fourth of the microbial biomass in an anoxic trickling filter biofilm (44).
Despite the fact that allochthonous heterotrophic bacteria supplied by the effluent were active in the river immediately downstream of the effluent input, they have been shown to disappear rapidly (17, 18). In contrast to observations of the heterotrophs, the Achères effluent was previously seen as seeding the system with nitrifying bacteria, and the allochthonous populations were thought to grow along the aquatic continuum, depending on the residence time of the system, and to build up a large biomass in the estuary (8). The molecular approach has led to a change in this view.
Indeed, the observed decrease in the numbers of amoA gene copies immediately after the WWTP input could be explained by the fact that the nitrifying bacteria supplied by the WWTP disappeared from the river water because they were less competitive than the autochthonous bacteria. The total number of amoA gene copies remained low until Poses (km 202), the entrance of the freshwater estuary, where conditions seemed to allow an AOB population, possibly different from the one from Achères, to build up a large biomass. The number of amoA gene copies increased sharply concomitantly with the potential ammonia-oxidizing activity (pAOA) (Fig. 3b), with a peak at the Bouille-Duclair stations and a sudden decrease at the saline intrusion limit. In the salinity gradient, the pAOA increased again, but not the number of β-amoA gene copies, which suggests a shift from the freshwater AOB community to another that is better adapted to the saline environment and that was not targeted in this study. In the saline estuarine sector, halophilic populations, probably belonging to the γ subclass of Proteobacteria, might be found.
As for the NOB, the lack of any increase downstream from the Achères WWTP might be due to the limitation of our research to the genus Nitrobacter (see Materials and Methods). According to the literature, the NOB of the WWTP belong mainly to the genus Nitrospira (13, 27), but they were not investigated here. Note that although the potential nitrite-oxidizing activity was very weak, the performance of the molecular biology tools still allowed us to detect and quantify the presence of Nitrobacter as a representative of this functional group of NOB. The genus Nitrobacter is detected in various environments (10, 43, 45). Our results clearly show that Nitrobacter cells were able to grow in the Seine River, their abundance increasing noticeably together with the potential nitrifying activity, reaching a maximum at the Duclair station in the estuary. According to the specific-activity values for various species of Nitrobacter (from 5 to 42 fmol of NO3− produced cell−1 h−1, according to Prosser [37]) and to the estimate of Nitrobacter abundance, assuming one operon per cell (12), rough calculations show that Nitrobacter might be responsible for the whole nitrite oxidation step in the Seine River. However, the presence of other NOB cannot be excluded, especially in the area directly impacted by the wastewater effluent. As several articles have recently reported a dominance of Nitrospira spp. in freshwater systems, activated sludge, and nitrifying fluidized bed reactors (24, 25, 27, 46, 61), further investigations into the NOB of the lower Seine River will be needed. On the basis of their different Km values, which are lower for Nitrospira than for Nitrobacter (15, 45), low nitrite concentrations would allow a Nitrospira population to dominate the nitrite-oxidizing communities in many environments.
The molecular approach based on restriction fragment length polymorphism that was used to analyze the diversity of the ammonia-oxidizing community led to the identification of 10 different restriction patterns for 100 analyzed clones; this is a relatively high number compared to the four groups obtained from the analysis of amoA sequences retrieved from heavy-metal-contaminated soils (55) and continuous culture enrichments (5). This result may indicate a greater diversity of ammonia oxidizers in waters polluted by domestic effluent, such as the lower Seine River. The different lineages in which the sequenced clones were found corresponded to the different restriction pattern groups, except for the clones Duc2, Duc14, and T21, which belonged to the same restriction group but were found in two different phylogenetic clusters. However, all the restriction patterns have been confirmed by the positions of the MspI enzyme sites found on the nucleic acid sequences.
The majority of the clones (from Triel and Duclair) have sequences closely related to those of bacteria commonly detected in WWTPs (Table 1) (38). However, this result could be biased, as WWTPs, together with soils, are mostly studied from the point of view of AOB diversity; thus, the majority of sequences available in databases are taken from these two types of environments. Due to the scarcity of studies of nitrifying populations in freshwater systems, we cannot conclude that all the clones originate from the WWTP effluent. As a consequence, the nitrifying community at Triel would represent a mixture of allochthonous bacteria from the Achères WWTP and autochthonous bacteria from the river upstream, whereas the population at Duclair would be the result of growth, between Paris and the estuary, of bacteria that have become adapted to their environment.
The proportions of the analyzed clones must be considered only as indicative, because the phylogenetic study has qualitative rather than quantitative value. We know that the various AOB species can have from one to three amoA genes with the same sequence resulting from a duplication phenomenon and not from interspecies transfer (32). Nitrosomonas strains usually carry two gene copies, whereas most Nitrosospira strains carry three (28, 32). Therefore, more than one clone might have been isolated from the same bacterium, so the proportions of the different bacterial groups cannot be extrapolated to the environmental populations.
Among the three identified clusters of β-AOB, the proportions of analyzed clones present in the N. europaea-like cluster were about the same for the two sites (13 and 16%, respectively, at the Triel and Duclair stations), whereas the 6a cluster (N. oligotropha-N. ureae) and the Nitrosospira-like cluster show noticeable differences between the two stations (81 and 60% for the former and 6 and 23% for the latter at Triel and Duclair, respectively).
The N. ureae-N. oligotropha-like cluster, to which the majority of the sequenced clones were closely affiliated, has also been found to be the predominant group of ammonia oxidizers in the estuary of the Elbe River (53) and in the freshwater part of the Schelde estuary (11). Bacteria showing affinity with the Nitrosomonas cluster 6a (closely affiliated with the cultured species N. urea and N. oligotropha) have previously been found to be the dominant AOB in soil, freshwater, and freshwater sediment (34, 52, 54) due to input of AOB from untreated wastewater (13, 38, 42). This cluster, well adapted to low ammonium concentrations, can grow in various environments (5, 6). Moreover, isolates of this lineage (e.g., N. oligotropha, N. ureae, and Nitrosomonas sp. strains JL21 and AL212) are inhibited at high ammonium concentrations (exceeding 10 mM) (5, 9, 29, 39, 53, 57). Usually, these bacteria have a high affinity for ammonium, with Km values ranging from 0.075 to 0.03 mM NH4+ (53). The conditions in the Seine River water, therefore, appear to favor their growth, i.e., to have high ammonium concentrations compared to other freshwater systems (0.128 mM N-NH4+ downstream from the WWTP, 0.09 mM N-NH4+ at Triel, and 3.8 μM N-NH4+ at Duclair) but lower concentrations than those (10 mM or more) found in reactors, cultures, or WWTPs.
The N. europaea-like clones (cluster 7) were present at both sites on the Seine River, but in small proportions. Due to its low substrate affinities (high Km values, ranging from 0.4 to 7 mM NH4+) (30, 37, 53), this cluster needs high ammonium concentrations for its growth (6, 9, 36). The concentrations found in the Seine River would therefore not favor it. A small proportion (6%) of Nitrosospira-like bacteria were found at the Triel station, while at Duclair they represented 23% of the analyzed clones. Like the Nitrosomonas cluster 6a, Nitrosospira bacteria, based on their low Km values (40 μM NH4+) (39, 47), would be more competitive in an environment with a low ammonium concentration, for example, in the downstream part of the river, where most of the ammonium has been oxidized.
In agreement with Bollmann et al. (6), we suggest that N. europaea and other members of Nitrosomonas cluster 7 could be classified as r strategists in accordance with their need for a substrate-rich environment (ammonium in this case) and their ability to colonize various environments. On the other hand, Nitrosomonas cluster 6a and Nitrosospira-like bacteria would both represent K strategists, as defined by their high affinity with the resources required for their growth and their demand for a selective environment.
This study combined a functional approach (i.e., nitrifying activities) with molecular tools to analyze the nitrifying bacterial population by quantifying the functional groups of both AOB and NOB and by determining the phylogenetic composition of ammonia oxidizers at two key points in the Seine River. The quantitative approach to estimating the nitrifying populations clearly fit the spatial distribution obtained from the potential activity and, furthermore, allowed us to estimate the numbers of amoA gene copies and Nitrobacter cells in a range from 1 · 105 to 7 · 107 cells liter−1 compared to the 109 cells liter−1 commonly found for whole populations of bacteria. These results are fundamental to quantitative ecological studies (see Billen et al. [4] and references therein). Moreover, the studies of community composition tend to modify our previous view of nitrifier dynamics in this sector that is strongly impacted by domestic effluent; nitrifying bacteria originating from the effluents and disappearing in the river would not represent seeding of the Seine River. However, more studies are needed to fully understand the role of allochthonous versus autochthonous nitrifying bacteria.
Acknowledgments
This work was undertaken within the framework of two French programs: PIREN-Seine (CNRS) and Seine-Aval (Région Haute-Normandie).
We are indebted to André Ficht from the SNS (Service de Navigation de la Seine) for his kind help in the field. Many thanks are due to R. Laanbroek for constructive remarks on the manuscript.
REFERENCES
- 1.Belser, L. W., and E. L. Mays. 1982. Use of nitrifier activity measurements to estimate the efficiency of viable nitrifier count in soils and sediments. Appl. Environ. Microbiol. 43:945-948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berthe, T., J. Garnier, and F. Petit. 1999. Quantification of nitrifying bacteria of the genus Nitrobacter in an aquatic system (Seine estuary, France). C. R. Acad. Sci. 322:517-526. [Google Scholar]
- 3.Billen, G., J. Garnier, T. Berthe, P. Servais, N. Brion, A. Ficht, S. Even, and M. Poulin. 1999. L'oxygène: un témoin du fonctionnement microbiologique. IFREMER Edition, Plouzané, France.
- 4.Billen, G., J. Garnier, A. Ficht, and C. Cun. 2001. Ecological modeling of the 50 last years of anthropogenic impact in the Seine estuary, Estuaries 24:977-993. [Google Scholar]
- 5.Bollmann, A., and H. J. Laanbroek. 2001. Continuous culture enrichments of ammonia-oxidizing bacteria at low ammonium concentrations. FEMS Microbiol. Ecol. 1279:1-11. [Google Scholar]
- 6.Bollmann, A., M.-J. Bär-Gilissen, and H. J. Laanbroek. 2002. Growth at low ammonium concentrations and starvation response as potential factors involved in niche differentiation among ammonia-oxidizing bacteria. Appl. Environ. Microbiol. 68:4751-4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brion, N., and G. Billen. 1998. Une réévaluation de la méthode d'incorporation de H14CO3− pour mesurer la nitrification autotrophe et son application pour estimer des biomasses de bactéries nitrifiantes. Rev. Sci. Eau 11:283-302. [Google Scholar]
- 8.Brion, N., and G. Billen. 2000. Wastewater as a source of nitrifying bacteria in river systems: the case of the river Seine downstream from Paris. Water Res. 12:3213-3221. [Google Scholar]
- 9.Burrell, P. C., C. M. Phalen, and T. A. Hovanec. 2001. Identification of bacteria responsible for ammonia oxidation in freshwater aquaria. Appl. Environ. Microbiol. 67:5791-5800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coskuner, G., and T. P. Curtis. 2002. In situ characterization of nitrifiers in an activated sludge plant: detection of Nitrobacter spp. J. Appl. Microbiol. 93:431-437. [DOI] [PubMed] [Google Scholar]
- 11.DeBie, M. J., A. G. Speksnijder, G. A. Kowalchuk, T. Schuurman, G. Zwart, J. R. Stephen, O. E. Diekmann, and H. J. Laanbroek. 2001. Shifts in the dominant populations of ammonia-oxidizing β-subclass Proteobacteria along the eutrophic Shelde estuary. Aquat. Microb. Ecol. 23:225-236. [Google Scholar]
- 12.Degrange, V., and R. Bardin. 1995. Detection and counting of Nitrobacter populations in soil by PCR. Appl. Environ. Microbiol. 61:2093-2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dionisi, H. M., A. C. Layton, G. Harms, I. R. Gregory, K. G. Robinson, and G. S. Sayler. 2002. Quantification of Nitrosomonas oligotropha-like ammonia-oxidizing bacteria and Nitrospira spp. from full-scale wastewater treatment plants by competitive PCR. Appl. Environ. Microbiol. 68:245-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Edwards, U., T. Rogall, H. Blocker, and E. C. Bottger. 1989. Isolation and direct complete nucleotide determination of entire genes. Characterisation of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 17:7843-7853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ehrich, S., D. Behrens, E. Lebedeva, W. Ludwig, and E. Bock. 1995. A new obligately chemolithoautotrophic, nitrite-oxidizing bacterium, Nitrospira moscoviensis sp. nov., and its phylogenetic relationship. Arch. Microbiol. 164:16-23. [DOI] [PubMed] [Google Scholar]
- 16.Felsenstein, J. 1993. PHYLIP: phylogeny inference package (version 3.5c). University of Washington, Seattle.
- 17.Garnier, J., P. Servais, and G. Billen. 1991. Bacterioplankton in the Seine River: impact of the Parisian urban effluents. Can. J. Microbiol. 38:56-64. [Google Scholar]
- 18.Garnier, J., G. Billen, and P. Servais. 1992. Physiological characteristics and ecological role of small and large sized bacteria in a polluted river (Seine river, France). Arch. Hydrobiol. Ergebn. Limnol. 37:83-94. [Google Scholar]
- 19.Garnier, J., P. Servais, G. Billen, M. Akopian, and N. Brion. 2001. The oxygen budget in the Seine estuary: balance between photosynthesis and degradation of organic matter. Estuaries 24:964-977. [Google Scholar]
- 20.Garnier, J., G. Billen, T. Berthe, A. Martinez, S. Pinault, M. Desruelle, L. Laroche, and A. Cébron. 2002. Nitrification de l'ammonium de Paris à l'estuaire. Rapport synthèse Piren-Seine, 1998-2001. CNRS, Paris, France.
- 21.Gieseke, A., U. Purkhold, M. Wagner, R. Amann, and A. Schramm. 2001. Community structure and activity dynamics of nitrifying bacteria in a phosphate-removing biofilm. Appl. Environ. Microbiol. 67:1351-1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goñi-Urriza, M., M. Capdepuy, C. Arpin, N. Raymond, P. Caumette, and C. Quentin. 2000. Impact of an urban effluent on antibiotic resistance of riverine Enterobacteriaceae and Aeromonas spp. Appl. Environ. Microbiol. 66:125-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Head, I. M., W. D. Hiorns, T. M. Embley, A. J. McCarthy, and J. R. Saunders. 1993. The phylogeny of autotrophic ammonia-oxidizing bacteria as determined by analysis of 16S ribosomal RNA gene sequences. J. Gen. Microbiol. 139:1147-1153. [DOI] [PubMed] [Google Scholar]
- 24.Hovanec, T. A., and E. F. DeLong. 1996. Comparative analysis of nitrifying bacteria associated with freshwater and marine aquaria. Appl. Environ. Microbiol. 62:2888-2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hovanec, T. A., L. T. Taylor, A. Blakis, and E. F. Delong. 1998. Nitrospira-like bacteria associated with nitrite oxidation in freshwater aquaria. Appl. Environ. Microbiol. 64:258-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hynes, R. K., and R. Knowles. 1983. Inhibition of chemoautotrophic nitrification by sodium chlorite: a reexamination. Appl. Environ. Microbiol. 45:1178-1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Juretschko, S., G. Timmermann, M. Schmid, K. H. Schleifer, A. Pommerening-Roser, H.-P. Koops, and M. Wagner. 1998. Combined molecular and conventional analyses of nitrifying bacterium diversity in activated sludge: Nitrosococcus mobilis and Nitrospira-like bacteria as dominant populations. Appl. Environ. Microbiol. 64:3042-3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klotz, M. G., and J. M. Norton. 1998. Multiple copies of ammonia monooxygenase (amo) operons have evolved under biased AT/GC mutational pressure in ammonia-oxidizing autotrophic bacteria. FEMS Microbiol. Lett. 168:303-311. [DOI] [PubMed] [Google Scholar]
- 29.Koops, H. P., U. C. Böttcher, A. Möller, A. Pommerening-Röser, and G. Stehr. 1991. Classification of eight new species of ammonia-oxidizing bacteria: Nitrosomonas communis sp. nov., Nitrosomonas ureae sp. nov., Nitrosomonas aestuarii sp. nov., Nitrosomonas marina sp. nov., Nitrosomonas nitrosa sp. nov., Nitrosomonas eutropha sp. nov., Nitrosomonas oligotropha sp. nov., Nitrosomonas halophila sp. nov. J. Gen. Microbiol. 137:1689-1699. [Google Scholar]
- 30.Laanbroek, H. J., and J. W. Woldendorp. 1994. Activity of chemolithotrophic nitrifying bacteria under stress in natural soils, p. 275-304. In J. Gwynfryn Jones (ed.), Advances in microbial ecology. Plenum Press, New York, N.Y.
- 31.Navarro, E., M. P. Fernandez, P. Normand, and R. Bardin. 1992. Genomic heterogeneity of the genus Nitrobacter. Int. J. Syst. Bacteriol. 42:554-560. [Google Scholar]
- 32.Norton, J. M., J. J. Alzerreca, Y. Suwa, and M. G. Klotz. 2002. Diversity of ammonia monooxygenase operon in autotrophic ammonia-oxidizing bacteria. Arch. Microbiol. 177:139-149. [DOI] [PubMed] [Google Scholar]
- 33.Petit, F., S. Craquelin, J. Guespin-Michel, and C. Buffet-Janvresse. 1999. Nucleic acid extraction from polluted estuarine water for detection of viruses and bacteria by PCR and RT-PCR analysis. Res. Microbiol. 150:143-151. [DOI] [PubMed] [Google Scholar]
- 34.Pommerening-Röser, A., G. Rath, and H. P. Koops. 1996. Phylogenetic diversity within the genus Nitrosomonas. Syst. Appl. Microbiol. 19:344-351. [Google Scholar]
- 35.Porter, K. G., and Y. S. Freig. 1980. Use of DAPI for identifying and counting aquatic microflora. Limnol. Oceanogr. 25:943-948. [Google Scholar]
- 36.Prinĉiĉ, A., I. Mahne, F. Megusar, E. A. Paul, and J. M. Tiedje. 1998. Effects of pH and oxygen and ammonium concentrations on the community structure of nitrifying bacteria from wastewater. Appl. Environ. Microbiol. 64:3584-3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prosser, J. I. 1989. Autotrophic nitrification in bacteria. Adv. Microbiol. Physiol. 30:125-181. [DOI] [PubMed] [Google Scholar]
- 38.Purkhold, U., A. Pommerening-Roser, S. Juretschko, M. C. Schmid, H.-P. Koops, and M. Wagner. 2000. Phylogeny of all recognized species of ammonia oxidizers based on comparative 16S rRNA and amoA sequence analysis: implications for molecular diversity surveys. Appl. Environ. Microbiol. 66:5368-5382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38a.Purkhold, U., M. Wagner, G. Timmermann, A. Pommerening-Roser, and H. P. Koops. 2003. 16S rRNA and amoA-based phylogeny of 12 novelbetaproteobacterial ammonia-oxidizing isolates: extension of the data set and proposal of a new lineage within the nitrosomonads. Int. J. Syst. Evol. Microbiol. 53:1485-1494. [DOI] [PubMed] [Google Scholar]
- 39.Regan, J. M., G. W. Harrington, and D. R. Noguera. 2002. Ammonia- and nitrite-oxidizing bacterial communities in a pilot-scale chloraminated drinking water distribution system. Appl. Environ. Microbiol. 68:73-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rodier, J. 1984. L'analyse de l'eau (eaux naturelles, eaux résiduaires, eau de mer), 7th ed., p. 177. Dunod Edition, Paris, France.
- 41.Romana, L. A., B. Thouvenin, and C. Sammari. 1992. Modèle mathématique sur la nitrification et le cycle de l'oxygène dissous en estuaire de la Seine. Rapport DEL/CCM, AESN, IFREMER, Direction de L'Environnement, Chimie des Contaminants et Modélisation. Hon, Plouzané, France.
- 42.Rotthauwe, J. H., K. P. Witzel, and W. Liesack. 1997. The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl. Environ. Microbiol. 63:4704-4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sanden, B., and G. Dalhammar. 2000. Application of an amperometric immunosensor for the enumeration of Nitrobacter in activated sludge. Appl. Microbiol. Biotechnol. 54:413-417. [DOI] [PubMed] [Google Scholar]
- 44.Schmid, M., U. Twachtmann, M. Klein, M. Strous, S. Juretschko, M. Jetten, J. W. Metzger, K. H. Schleifer, and M. Wagner. 2000. Molecular evidence for genus level diversity of bacteria capable of catalyzing anaerobic ammonium oxidation. Syst. Appl. Microbiol. 23:93-106. [DOI] [PubMed] [Google Scholar]
- 45.Schramm, A., L. H. Larsen, N. P. Revsbech, N. B. Ramsing, R. Amann, and K. H. Schleifer. 1996. Structure and function of a nitrifying biofilm as determined by in situ hybridization and the use of microelectrodes. Appl. Environ. Microbiol. 62:4641-4647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schramm, A., D. De Beer, M. Wagner, and R. Amann. 1998. Identification and activities in situ of Nitrosospira and Nitrospira spp. as dominant populations in a nitrifying fluidized bed reactor. Appl. Environ. Microbiol. 64:3480-3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schramm, A., D. de Beer, J. C. van den Heuvel, S. Ottengraf, and R. Amann. 1999. Microscale distribution of populations and activities of Nitrosospira and Nitrospira spp. along a macroscale gradient in a nitrifying bioreactor: quantification by in situ hybridization and the use of microsensors. Appl. Environ. Microbiol. 65:3690-3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Servais, P., and J. Garnier. 1993. Contribution of heterotrophic bacterial production to the carbon budget of the River Seine (France). Microb. Ecol. 25:19-33. [DOI] [PubMed] [Google Scholar]
- 49.Servais, P., J. Garnier, N. Demarteau, N. Brion, and G. Billen. 1999. Supply of organic matter and bacteria to aquatic ecosystems through wastewater effluents. Water Res. 33:3521-3531. [Google Scholar]
- 50.Slavyck, G., and J. J. McIsaac. 1972. Comparison of two automated ammonium methods in a region of coastal upwelling. Deep Sea Res. 19:1-4. [Google Scholar]
- 51.Somville, M. 1978. A method for the measurement of nitrification rates in water. Water Res. 12:843-848. [Google Scholar]
- 52.Speksnijder, A. G. C. L., G. A. Kowalchuk, K. Roest, and H. J. Laanbroek. 1998. Recovery of a Nitrosomonas-like 16S rDNA sequence group from freshwater habitats. Syst. Appl. Microbiol. 21:321-330. [DOI] [PubMed] [Google Scholar]
- 53.Stehr, G., B. Bottcher, P. Dittberner, G. Rath, and H.-P Koops. 1995. The ammonia-oxidizing nitrifying population of the River Elbe estuary. FEMS Microbiol. Ecol. 17:177-186. [Google Scholar]
- 54.Stephen, J. R., G. A. Kowalchuk, M.-A. V. Bruns, A. E. McCaig, C. J. Phillips, T. M. Embley, and J. I. Prosser. 1998. Analysis of β-subgroup proteobacterial ammonia oxidizer populations in soil by denaturing gradient gel electrophoresis analysis and hierarchical phylogenetic probing. Appl. Environ. Microbiol. 64:2958-2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stephen, J. R., Y. J. Chang, S. J. MacNaughton, G. A. Kowalchuk, K. T. Leung, C. A. Flemming, and D. C. White. 1999. Effect of toxic metals on indigenous soil beta-subgroup proteobacterium ammonia oxidizer community structure and protection against toxicity by inoculated metal-resistant bacteria. Appl. Environ. Microbiol. 65:95-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Strom, P. F., V. A. Matulewich, and M. S. Finstein. 1976. Concentrations of nitrifying bacteria in sewages, effluents, and a receiving stream and resistance of these organisms to chlorination. Appl. Environ. Microbiol. 31:731-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Suwa, Y., T. Sumino, and K. Noto. 1997. Phylogenetic relationships of activated sludge isolates of ammonia oxidizers with different sensitivities to ammonium sulfate. J. Gen. Appl. Microbiol. 43:373-379. [DOI] [PubMed] [Google Scholar]
- 58.Teske, A., E. Alm, J. M. Regan, S. Toze, B. E. Rittmann, and D. A. Stahl. 1994. Evolutionary relationships among ammonia- and nitrite-oxidizing bacteria. J. Bacteriol. 176:6623-6630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Utåker, J. B., and I. F. Nes. 1998. A qualitative evaluation of the published oligonucleotides specific for the 16S rRNA gene sequences of ammonia-oxidizing bacteria. Syst. Appl. Microbiol. 21:72-88. [DOI] [PubMed] [Google Scholar]
- 61.Wagner, M., G. Rath, R. Amann, H.-P. Koops, J. Flood, and R. Amann. 1996. In situ analysis of nitrifying bacteria in sewage treatment plants. Water Sci. Technol. 34:237-244. [Google Scholar]
- 62.Watson, S. W., E. Bock, H. Harms, H.-P. Koops, and A. B. Hooper. 1989. Nitrifying bacteria, p. 1808-1834. In J. T. Staley, M. P. Bryant, N. Pfenning, and J. G. Holt (ed.), Bergey's manual of systematic bacteriology. Williams and Wilkins, Baltimore, Md.