Abstract
Cell type-specific DNA methylation patterns are established during mammalian development and maintained in adult somatic cells. Understanding how these patterns of 5-methylcytosine are established and maintained requires the elucidation of mechanisms for both DNA methylation and demethylation. The enzymes involved in the de novo methylation of DNA and the maintenance of the resulting methylation patterns have been fairly well characterized. However, important remaining challenges are to understand how DNA methylation systems function in vivo and in the context of chromatin. In addition, the enzymes and mechanisms for demethylation remain to be elucidated. There is still no consensus as to how active enzymatic demethylation is achieved in mammalian cells, but recent studies implicate base excision repair for genome-wide DNA demethylation in germ cells and early embryos.
Keywords: Chromatin Histone Modification, Chromatin Regulation, Chromatin Structure, DNA Methylation, DNA Methyltransferase, Gene Regulation
Introduction
5-Methylcytosine (5mC)2 in the DNA of mammalian somatic cells is found almost entirely within CpG dinucleotides (1). Approximately 70–80% of cytosine in CpG dyads is methylated on both strands in human somatic cells. In general, CpG methylation is highly prevalent in repetitive sequences and in gene bodies but rare at CpG islands within housekeeping promoters. Recent data have established that, opposite of expectation based on early models (2, 3), the methylation of gene bodies is positively correlated with transcription (4–6). On the other hand, the methylation of promoters and enhancers is consistent with expectation. For these elements, almost all experimental data support the concept that DNA methylation functions to stabilize or lock in the silent state (7). DNA methylation is also involved in several other fundamental processes, such as genomic imprinting, X chromosome inactivation, and suppression of retrotransposon elements (1), and is essential for normal development (8, 9). Consistent with its functional importance, the patterns of DNA methylation are non-random, well regulated, and tissue-specific (10, 11).
Experimental evidence has confirmed, for the most part, the maintenance methylase model (2, 3) outlined in Fig. 1. However, it is now recognized that maintenance methylation is not perfect, and preservation of methylation patterns requires de novo methylation (Ref. 12; see Ref. 13 for a recent review). Although methylation patterns are largely maintained through somatic cell divisions, changes in methylation patterns occur during mammalian development and cell differentiation. In mice, dramatic reprogramming with waves of demethylation and then remethylation occurs in germ cells and early embryos (14). After fertilization, most of the paternal genome is rapidly demethylated before DNA replication begins, indicative of active enzymatic demethylation (15, 16). On the other hand, the maternal genome undergoes apparently passive, replication-dependent demethylation during subsequent cleavage divisions (15). After implantation, a wave of global de novo methylation re-establishes the DNA methylation patterns that will be maintained, in large part, in somatic tissues. Genome-wide demethylation also occurs in primordial germ cells (PGCs) around embryonic days (E) 11.5–12.5 (14, 17), and then de novo methylation establishes a gamete-specific methylation pattern, different for egg and sperm. In addition to these global changes, gene-specific de novo methylation and demethylation occur during lineage-specific differentiation, such as during differentiation of hematopoietic progenitors (18).
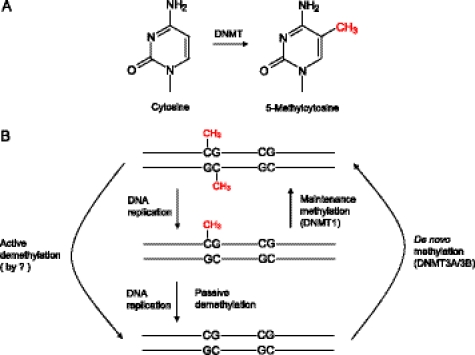
FIGURE 1.
Overview of mechanisms involved in DNA methylation and demethylation in mammals. A, DNMTs catalyze the covalent addition of a methyl group to C-5 of cytosine. B, most of the cytosine methylation occurs within CpG dinucleotides, and a distinction can be made between two DNMT activities: de novo and maintenance methylation. Methylation patterns are established during early development by de novo methyltransferases DNMT3A and DNMT3B and maintained through somatic cell divisions by maintenance methyltransferase DNMT1, which acts preferentially on the hemimethylated CpG sites generated by DNA replication. DNA demethylation can be achieved either passively, by the failure of maintenance methylation after DNA replication, or actively, by replication-independent processes. The enzymes responsible for active demethylation have not been conclusively identified in mammals.
It is clear that understanding how methylation patterns are regulated requires elucidating the mechanisms for de novo DNA methylation and demethylation, as well as maintenance methylation. A recent article by Jones and Liang (13) is recommended for a review of maintenance methylation. Here, we will focus on de novo methylation in the context of chromatin and on the mechanisms potentially involved in active demethylation.
DNA Methylation Machinery
The mammalian DNA (cytosine-5) methyltransferases (DNMTs) that catalyze the transfer of a methyl group from S-adenosyl-l-methionine to cytosine (19) are shown in Fig. 2. Among the three enzymatically active DNMTs, DNMT1 is thought to function as the major maintenance methyltransferase (Fig. 1). This enzyme has a preference for hemimethylated CpG sites, such as those generated by DNA replication (20), and is responsible for copying pre-existing methylation patterns to the newly synthesized strand (21), probably with the help of UHRF1, which also recognizes hemimethylated sites (22, 23). DNMT3A and DNMT3B are de novo methyltransferases active on unmethylated DNA (Fig. 1). Both of them have no preference for hemimethylated CpG substrates in vitro (24, 25) and are responsible for establishing methylation patterns during early development (9). De novo methylation by DNMT3A/3B also contributes to the maintenance of DNA methylation patterns (26, 27), possibly by methylating CpG sites missed by DNMT1 (12, 13).
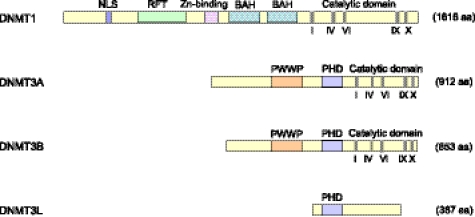
FIGURE 2.
Schematic structure of human DNMTs and DNMT3-like protein. Conserved methyltransferase motifs in the catalytic domain are indicated in Roman numerals. NLS, nuclear localization signal; RFT, replication foci-targeting domain; BAH, bromo-adjacent homology domain; PWWP, a domain containing a conserved proline-tryptophan-tryptophan-proline motif; PHD, a cysteine-rich region containing an atypical plant homeodomain; aa, amino acids. DNMT3L lacks the critical methyltransferase motifs and is catalytically inactive.
Dnmt3a and Dnmt3b have distinct functions during development, as evidenced by the fact that knock-out mice lacking either of them exhibit different defects and die at different developmental stages (9). In germ cells, for example, Dnmt3a, but not Dnmt3b, is essential for de novo methylation of most imprinted loci (28–30). Establishing methylation at most imprinted loci also requires Dnmt3L (28, 31, 32), a protein that was identified on the basis of sequence similarity to the plant homeodomain (PHD) and catalytic domains of Dnmt3a/3b (Fig. 2) (33). Although DNMT3L lacks critical methyltransferase motifs and is catalytically inactive, it can stimulate the activity of DNMT3A/3B (34, 35). Structural analysis indicated that the C-terminal domains of Dnmt3a and Dnmt3L form a tetrameric complex (3L-3a-3a-3L) with two active sites (36), which preferentially methylate two CpGs separated by 8–10 bp in vitro (36, 37). CpG periodicities within the 8–10-bp range have been observed in maternally imprinted loci (36) and in many other regions of the genome (38, 39), but such periodicities do not fully explain why de novo methylation is targeted to specific sequences (39). As will be discussed below, interaction between Dnmt3L and histone H3 tails that are unmethylated at Lys-4 could have a role in targeting methylation to imprinted regions.
In addition to Dnmt3L, a number of other factors are involved in de novo methylation at specific genomic regions. It has been found that the Piwi-interacting small RNA pathway is essential for de novo methylation of retrotransposons in fetal male germ cells, although the underlying mechanism is not clear (40, 41). At the imprinted Gnas locus, transcription across differentially methylated regions is required for the establishment of their maternal methylation marks in oocytes (42). Moreover, as reviewed below, specific histone modifications and histone-modifying enzymes have an important role in establishing DNA methylation patterns in mammals.
De Novo DNA Methylation in the Context of Chromatin
So far, the best studied histone mark linked to DNA methylation is the unmethylated histone H3 Lys-4 (H3K4me0). Genome-wide analyses have revealed a strong inverse correlation between H3K4 methylation and DNA methylation, suggesting that H3K4 methylation may protect DNA from de novo methylation (43, 44). Indeed, the PHD-like domain of DNMT3L interacts with histone H3 tails unmethylated at Lys-4, and this interaction is abolished by methylation at H3K4 (45, 46). DNMT3L also interacts with DNMT3A (34, 35), and both are required for establishing methylation at most imprinted loci in germ cells (30); therefore, it has been proposed that DNMT3L triggers de novo DNA methylation by recruiting DNMT3A2, a germ cell-specific isoform of DNMT3A, to nucleosomes that contain unmethylated H3K4 (36, 45). In support of this hypothesis, knock-out of mouse KDM1B, a histone H3K4 demethylase, results in a significant increase in H3K4 methylation and failure to establish DNA methylation at a subset of imprinted genes in oocytes (47). These findings suggest that demethylation of H3K4 is critical for de novo DNA methylation of some imprinted regions in germ cells.
Interestingly, recent biochemical and structural studies revealed that, even without any accessory proteins, the PHD domain (also known as ADD domain) of DNMT3A can directly interact with H3 tails unmethylated at Lys-4 in vitro (46, 48). In addition, the PWWP domain of DNMT3A was found to specifically interact with H3 tails containing the trimethylated Lys-36 (H3K36me3) in vitro (49). Both interactions increase the activity of DNMT3A2 on chromatin-bound DNA as measured by in vitro assays (48, 49). Thus, it is likely that DNMT3A recognizes specific histone modifications and preferentially methylates associated DNA. Recent genome-wide studies are consistent with this notion; H3K36me3 is located mainly in the bodies of active genes (50, 51), and the distribution of this modification is positively correlated with DNA methylation (50, 52). In addition to H3K4me0 and H3K36me3, histone H4 tails containing symmetrically dimethylated Arg-3 (H4R3me2s) have been reported to interact with the PHD domain of DNMT3A, and this interaction is required for DNA methylation at the human γ-globin promoter (53). However, the DNMT3A-H4R3me2s interaction could not be detected in two recent studies (46, 48); hence, further research is needed to clarify this discrepancy. Although the in vivo roles of these direct interactions between DNMT3A and histone tails remain to be determined, they raise the interesting possibility that DNMT3 methyltransferases themselves can “read” specific histone codes. These histone modifications, many of which may change rapidly, might thus be translated into more long-term stable DNA methylation patterns.
In Neurospora crassa, histone H3 Lys-9 trimethylation (H3K9me3) is required for DNA methylation (54). H3K9me3 is recognized by HP1 (heterochromatin protein 1), which directs DNA methylation by recruiting the DIM-2 DNA methyltransferase. In mammals, histone H3K9 methyltransferases, such as G9a and Suv39h, have been implicated in the regulation of DNA methylation, but the mechanism of this process appears to be somewhat different from that in N. crassa. Mouse embryonic stem (ES) cells lacking G9a show a significant reduction of DNA methylation at G9a target promoters, retrotransposons, and major satellite repeats (55, 56). Knock-out of G9a also impairs de novo DNA methylation of a set of embryonic genes during ES cell differentiation (57). Surprisingly, however, DNA methylation appears normal in G9a−/− cells expressing a G9a mutant that is defective in the methylation of H3K9, suggesting that H3K9 methylation per se is not required for DNA methylation (55–57). G9a interacts with Dnmt3a and Dnmt3b, and it has therefore been proposed that G9a mediates de novo methylation by directly recruiting Dnmt3 methyltransferases to the promoters (57). In addition, double knock-out of heterochromatin-associated H3K9 methyltransferases Suv39h1 and Suv39h2 results in loss of methylation at pericentric major satellite repeats in mouse ES cells (58). How Suv39h1 and Suv39h2 contribute to DNA methylation is not clear, although correlative evidence suggests that the in vivo targeting of Dnmt3b to pericentric regions may depend on Suv39h-mediated H3K9 trimethylation (58). Given that SUV39H1 interacts with DNMTs (59), it is possible that, similar to G9a, DNA methylation can be mediated by SUV39H1 itself. The same may also be true for histone H3 Lys-27 (H3K27) methyltransferase EZH2, which interacts with DNMTs and facilitates their binding to EZH2 target promoters (60). It thus seems likely that recruitment of DNMTs by histone methyltransferases may represent one mechanism for targeting DNA methylation to specific genomic regions.
In somatic cells, a strong interaction of nucleosomes with DNMT3A/3B has been observed, but this interaction did not involve binding of DNMT3A/3B to histone H3 and did not require the presence of several known DNMT3A/3B-interacting chromatin proteins, such as EZH2 and HP1α (61). It remains to be determined how this interaction occurs, although it was shown to require an intact nucleosomal structure (61). Interestingly, DNMT3A/3B preferentially bound to nucleosomes associated with highly methylated genomic regions, including the methylated CpG islands and repetitive DNA elements (61). This suggests that de novo methylation in somatic cells is restricted mainly to methylated genomic regions, which is consistent with the proposed role of DNMT3A/3B in restoring methylation at CpG sites missed by DNMT1 during replication (12, 13).
Mechanisms of Active DNA Demethylation
DNA demethylation can be achieved either passively, by simply not methylating the new DNA strand after replication, or actively, by a replication-independent process (Fig. 1B). Passive demethylation probably occurs during mammalian development, e.g. in the maternal genome during pre-implantation growth (15), and it has long been known that inhibition of DNMT1 results in hypomethylated DNA (62). In this minireview, we will focus on active enzymatic demethylation. Considerable evidence supports the existence of genome-wide active demethylation in zygotes (15, 16) and PGCs (14, 17) and locus-specific active demethylation in somatic cells, such as neurons (63) and T lymphocytes (64). However, the mechanism(s) of active demethylation remain poorly understood. A number of mechanisms for the enzymatic removal of the 5-methyl group of 5mC, the 5mC base, or the 5mC nucleotide have been proposed (three are shown in Fig. 3), but, so far, none of them have been conclusively proven. Most early work is controversial and has been reviewed by Ooi and Bestor (65). There is likely to be more than one mechanism. For example, global demethylation may involve mechanisms different from those involved in locus-specific demethylation. Also, the recent discovery of a new modified base, 5-hydroxymethylcytosine (5hmC), in mammalian DNA is likely to have important implications for the mechanisms of active demethylation and open new avenues of research. We will next review several possible mechanisms for active demethylation, with emphasis on recent findings.
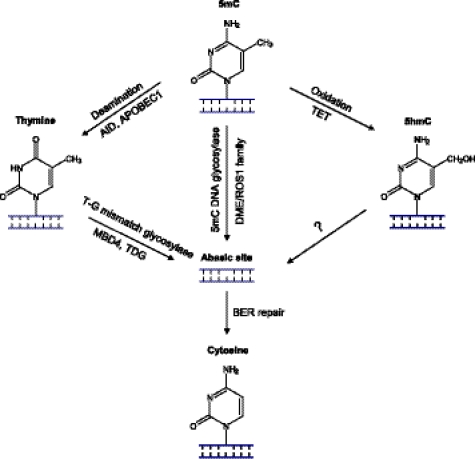
FIGURE 3.
Models for DNA demethylation mechanisms involving BER. In plants, the 5mC base can be directly removed by the DME/ROS1 family of 5mC DNA glycosylases, resulting in an abasic site that is repaired by the BER process. In mammals, no efficient 5mC glycosylases have been conclusively identified, and an alternative pathway initiated by deamination of 5mC has been proposed. Candidate deaminases include AID and APOBEC1, which convert 5mC to thymine. The resulting thymine could be repaired by BER initiated by a T-G mismatch glycosylase such as MBD4 or TDG. Recently, it has been shown that mouse and human TET family proteins can catalyze conversion of 5mC to 5hmC, a new modified base found in mammalian DNA. It is tempting to speculate that 5hmC could be repaired by a BER process, although, so far, no 5hmC DNA glycosylases have been identified.
Only in plants is there firm evidence for the direct removal of the 5mC base, and this is accomplished by a 5mC-specific glycosylase (reviewed in Ref. 66). In Arabidopsis, a family of four 5mC DNA glycosylases (ROS1, DME, DML2, and DML3) that have a preference for 5mC in double-stranded DNA has been identified, and there is strong biochemical and genetic evidence that these enzymes are necessary for active demethylation of specific genes (66). For example, ROS1 is a bifunctional glycosylase with apurinic/apyrimidinic lyase activity, so it first removes the base and then cleaves the abasic site, leaving a nick, which is rapidly repaired (66). This process is like base excision repair (BER), a process well established in mammals for mismatch repair and removal of alkylated bases. Accumulating evidence has suggested a role for BER in active demethylation in mammals, but the enzymes and mechanisms involved in initiating this process appear to be different from those in Arabidopsis.
So far, no mammalian homolog of the DME/ROS1 family of glycosylases has been found, and only weak 5mC glycosylase activity has been reported for thymine DNA glycosylase (TDG) and methyl-CpG-binding domain protein 4 (MBD4) (67, 68). For both of these glycosylases, activity toward 5mC is 30–40 times lower than their T-G mismatch glycosylase activity (67, 68); thus, it is doubtful that they are genuine 5mC DNA glycosylases (69). Consistent with this notion, Mbd4 is not required for global demethylation of the paternal genome in zygotes (70), and Mbd4 knock-out mice are viable and fertile (71). However, its role as a 5mC DNA glycosylase in locus-specific demethylation cannot be completely ruled out. As shown in a recent study (72), hormone-induced phosphorylation of MBD4 significantly stimulates its glycosylase activity toward 5mC, leading to active demethylation of the CYP27B1 gene promoter.
Other proposed mechanisms for active DNA demethylation also involve BER, but only after modification of the 5mC base. A leading candidate mechanism is deamination of 5mC to thymine, followed by BER of the resulting T-G mismatch (Fig. 3, left). Activation-induced cytosine deaminase (AID) and apolipoprotein B mRNA editing enzyme, catalytic polypeptide 1 (APOBEC1) can deaminate 5mC, potentially resulting in T-G mismatches, although these enzymes strongly prefer single-stranded DNA (73). Both enzymes are expressed in mouse oocytes, and Aid expression has also been detected in PGCs, suggesting a possible role in global DNA demethylation (73). These findings led to the hypothesis that demethylation in mammals might be achieved by deamination followed by BER initiated by T-G mismatch glycosylases, such as MBD4 and TDG (73). In support of this hypothesis, studies in zebrafish embryos indicated that overexpression of AID and MBD4 together, but not either alone, can lead to demethylation of DNA (74). In addition, two recent studies have provided evidence that AID is involved in active DNA demethylation in mammals (75, 76). Using heterokaryons made by fusion of mouse ES cells with human fibroblasts, Bhutani et al. (75) demonstrated that AID is required for active demethylation of the OCT4 and NANOG promoters during reprogramming of a fibroblast genome by cell fusion. Another study analyzed genome-wide DNA methylation in PGCs from wild-type and Aid knock-out mice (76). In comparison with their wild-type counterparts, Aid−/− PGCs at E13.5 show higher levels of methylation throughout the genome (76). However, the methylation levels in Aid−/− PGCs are still low, so significant demethylation still occurs even in the absence of AID (76). Importantly, there are no obvious developmental defects in Aid−/− mice, and they are fertile (77). Hence, other factors must also be involved in global demethylation in PGCs, and further studies are needed to determine whether AID plays a major role in this process.
In addition to BER, another major DNA repair pathway, nucleotide excision repair (NER), has been implicated in active demethylation. Using an expression cloning approach, Barreto et al. (78) identified Gadd45a as a protein factor capable of promoting global active demethylation in cultured mammalian cells. The Gadd45a-mediated demethylation was proposed to involve NER based on the observation that it was accompanied by DNA synthesis and required the presence of the NER endonuclease XPG, which directly bound to Gadd45a (78). However, this finding could not be confirmed by another study (79), and no increase in either global or locus-specific methylation was observed in Gadd45a−/− mice (80). Nevertheless, a role for Gadd45 family proteins in locus-specific DNA demethylation has received some support (63, 81). It has been reported that active demethylation at the rRNA gene promoter is mediated by Gadd45a and the NER machinery (81). Another member of the Gadd45 family, Gadd45b, was also found to be required for demethylation of specific gene promoters in mature hippocampal neurons in response to neuronal activity (63). The Gadd45b-dependent DNA demethylation appeared to be highly locus-specific, as significant demethylation occurred at specific regulatory regions of Bdnf and Fgf1, two genes critical for adult neurogenesis, but no global demethylation was observed (63).
Recently, the discoveries of 5hmC in mammalian cells and of enzymes responsible for converting 5mC to 5hmC have suggested new possibilities for demethylation. 5mC has long been thought to be the only naturally occurring modified base in mammalian DNA, but recently, substantial amounts of 5hmC have been detected in mouse Purkinje neurons (82) and in ES cells (83). By searching for mammalian homologs of the trypanosome thymidine hydroxylases, Tahiliani et al. (83) identified three human TET family proteins, TET1, TET2 and TET3, and further demonstrated that TET1 can catalyze conversion of 5mC to 5hmC in vitro and in cultured cells. All three mouse Tet proteins can catalyze a similar reaction, and Tet1 was found to play an important role in ES cell self-renewal and specification of the inner cell mass (84). In mouse ES cells, Tet1 was shown to be required for keeping the Nanog promoter in a hypomethylated state, suggesting a role for Tet1 in regulating DNA methylation (84). It remains unknown whether 5hmC can be an intermediate in active demethylation, but one postulated mechanism (Fig. 3, right) involves BER initiated by a 5hmC-specific DNA glycosylase (83). It is noteworthy that glycosylase activity toward 5hmC has been reported in calf thymus extracts (85).
BER Activity in Mammalian Germ Cells and Zygotes
The possible mechanisms of active demethylation, as reviewed above, were proposed based mainly on studies using in vitro assays and cultured cells. It is only recently that attempts have been made to directly investigate the molecular basis of global active demethylation in zygotes and PGCs.
Two recent studies have provided several lines of evidence to support a role of BER in active demethylation of the paternal genome in mouse zygotes (86, 87). First, high levels of BER components PARP1, APE1, and XRCC1 were seen in zygotic pronuclei. In contrast, NER components ERCC1 and chromatin-bound XPA were almost undetectable throughout zygotic development (86). Second, XRCC1, a single-strand break (SSB) sensor protein, bound only to chromatin in the paternal pronucleus, but not in the maternal pronucleus, and the presence of chromatin-bound XRCC1 coincided with the timing of active DNA demethylation (86). Another SSB sensor protein, PARP1, was also detected predominantly in the paternal pronucleus (86, 87). Third, a modified nick translation assay detected SSBs only in the paternal DNA at early pronuclear stage 3, in which active demethylation occurs (87). Finally, inhibition of BER resulted in a significantly higher level of DNA methylation in the paternal genome, indicating a critical role of BER in active demethylation in zygotes (86). Global demethylation in PGCs may also be mechanistically linked to the BER pathway, as it has been shown that expression of BER factors, such as Parp1, Ape1, and Xrcc1, is up-regulated in PGCs at E11.5, coincident with the time when genome-wide DNA demethylation occurs (86).
These recent studies suggest a link between BER and active demethylation in PGCs and zygotes. If so, how is BER triggered during this process? BER triggered by AID-mediated deamination of 5mC is not likely to play a major role in demethylation during gametogenesis and early development because Aid−/− mice are normal and fertile; hence, it is possible that BER is initiated by other mechanisms. Recently, it was found that Tet1 is significantly expressed in E11.5 PGCs (86), and Tet1 protein is present in the nuclei of one-cell embryos (84). Therefore, it will be of great interest to investigate whether 5hmC is present in the paternal genome in zygotes and in PGCs at a time when global demethylation occurs. As mentioned previously, 5hmC has been proposed to be a direct target for BER, although, so far, no 5hmC-specific glycosylases have been identified.
Further research is needed to firmly establish the role of BER in genome-wide DNA demethylation. However, irrespective of whether BER is involved, it is conceivable that global demethylation is a highly regulated process that involves many factors. To identify molecular components required for paternal genome demethylation in zygotes, Okada et al. (88) recently developed a novel assay using GFP fused to a protein that has high affinity for unmethylated CpG. This allowed them to monitor paternal genome demethylation by live-cell imaging. Using this assay together with siRNA knockdown, they screened a dozen candidate genes and found that knockdown of Elp3, a component of the elongator complex, impaired DNA demethylation in paternal pronuclei. Knockdown of two other elongator subunits, Elp1 and Elp4, also impaired demethylation, suggesting that the entire transcription elongator complex may be required for the demethylation process. Although the molecular mechanism for elongator complex-mediated DNA demethylation has yet to be elucidated, Elp3 has a Fe-S radical S-adenosylmethionine (SAM) domain, suggesting an oxidative mechanism with 5hmC as an intermediate (89).
Future Directions
It has become increasingly clear that histone modifications and chromatin-associated factors can have a profound effect on the establishment and maintenance of DNA methylation. One theme emerging from recent in vitro studies is that DNMT3A methyltransferase can directly interact with specifically modified histone tails. It will be of interest to examine whether such interactions could modulate the enzymatic activity of DNMT3A and/or whether they are involved in the recruitment of DNMT3A. A challenge for future research will be to understand at a mechanistic level how de novo DNA methylation occurs within a chromatin context in vivo. On the other hand, research on active DNA demethylation is gaining momentum. Although there is still no consensus on the mechanisms of active demethylation in mammals, emerging evidence suggests that it may involve BER. As definitive proof for BER involvement requires genetic evidence, it will be critically important in future studies to identify the involved BER factors and use knock-out mice to examine their roles in active demethylation. In addition, it is likely that the newly discovered 5hmC base in mammalian DNA will be the subject of intense study in the coming years, and it will be of particular interest to determine whether it plays a role in the DNA demethylation process.
This is the first article in the Thematic Minireview Series on Epigenetics. This minireview will be reprinted in the 2011 Minireview Compendium, which will be available in January, 2012.
- 5mC
- 5-methylcytosine
- PGC
- primordial germ cell
- E
- embryonic day
- DNMT
- DNA methyltransferase
- PHD
- plant homeodomain
- ES
- embryonic stem
- 5hmC
- 5-hydroxymethylcytosine
- BER
- base excision repair
- TDG
- thymine DNA glycosylase
- AID
- activation-induced cytosine deaminase
- NER
- nucleotide excision repair
- SSB
- single-strand break.
REFERENCES
- 1. Bird A. (2002) Genes Dev. 16, 6–21 [DOI] [PubMed] [Google Scholar]
- 2. Holliday R., Pugh J. E. (1975) Science 187, 226–232 [PubMed] [Google Scholar]
- 3. Riggs A. D. (1975) Cytogenet. Cell Genet. 14, 9–25 [DOI] [PubMed] [Google Scholar]
- 4. Laurent L., Wong E., Li G., Huynh T., Tsirigos A., Ong C. T., Low H. M., Kin Sung K. W., Rigoutsos I., Loring J., Wei C. L. (2010) Genome Res. 20, 320–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lister R., Pelizzola M., Dowen R. H., Hawkins R. D., Hon G., Tonti-Filippini J., Nery J. R., Lee L., Ye Z., Ngo Q. M., Edsall L., Antosiewicz-Bourget J., Stewart R., Ruotti V., Millar A. H., Thomson J. A., Ren B., Ecker J. R. (2009) Nature 462, 315–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rauch T. A., Wu X., Zhong X., Riggs A. D., Pfeifer G. P. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 671–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Razin A., Riggs A. D. (1980) Science 210, 604–610 [DOI] [PubMed] [Google Scholar]
- 8. Li E., Bestor T. H., Jaenisch R. (1992) Cell 69, 915–926 [DOI] [PubMed] [Google Scholar]
- 9. Okano M., Bell D. W., Haber D. A., Li E. (1999) Cell 99, 247–257 [DOI] [PubMed] [Google Scholar]
- 10. Eckhardt F., Lewin J., Cortese R., Rakyan V. K., Attwood J., Burger M., Burton J., Cox T. V., Davies R., Down T. A., Haefliger C., Horton R., Howe K., Jackson D. K., Kunde J., Koenig C., Liddle J., Niblett D., Otto T., Pettett R., Seemann S., Thompson C., West T., Rogers J., Olek A., Berlin K., Beck S. (2006) Nat. Genet. 38, 1378–1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Illingworth R., Kerr A., Desousa D., Jørgensen H., Ellis P., Stalker J., Jackson D., Clee C., Plumb R., Rogers J., Humphray S., Cox T., Langford C., Bird A. (2008) PLoS Biol. 6, e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Riggs A. D., Xiong Z. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 4–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jones P. A., Liang G. (2009) Nat. Rev. Genet. 10, 805–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Morgan H. D., Santos F., Green K., Dean W., Reik W. (2005) Hum. Mol. Genet. 14, R47–R58 [DOI] [PubMed] [Google Scholar]
- 15. Mayer W., Niveleau A., Walter J., Fundele R., Haaf T. (2000) Nature 403, 501–502 [DOI] [PubMed] [Google Scholar]
- 16. Oswald J., Engemann S., Lane N., Mayer W., Olek A., Fundele R., Dean W., Reik W., Walter J. (2000) Curr. Biol. 10, 475–478 [DOI] [PubMed] [Google Scholar]
- 17. Hajkova P., Erhardt S., Lane N., Haaf T., El-Maarri O., Reik W., Walter J., Surani M. A. (2002) Mech. Dev. 117, 15–23 [DOI] [PubMed] [Google Scholar]
- 18. Ji H., Ehrlich L. I., Seita J., Murakami P., Doi A., Lindau P., Lee H., Aryee M. J., Irizarry R. A., Kim K., Rossi D. J., Inlay M. A., Serwold T., Karsunky H., Ho L., Daley G. Q., Weissman I. L., Feinberg A. P. (2010) Nature 467, 338–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Goll M. G., Bestor T. H. (2005) Annu. Rev. Biochem. 74, 481–514 [DOI] [PubMed] [Google Scholar]
- 20. Pradhan S., Bacolla A., Wells R. D., Roberts R. J. (1999) J. Biol. Chem. 274, 33002–33010 [DOI] [PubMed] [Google Scholar]
- 21. Chen T., Li E. (2004) Curr. Top. Dev. Biol. 60, 55–89 [DOI] [PubMed] [Google Scholar]
- 22. Bostick M., Kim J. K., Estève P. O., Clark A., Pradhan S., Jacobsen S. E. (2007) Science 317, 1760–1764 [DOI] [PubMed] [Google Scholar]
- 23. Sharif J., Muto M., Takebayashi S., Suetake I., Iwamatsu A., Endo T. A., Shinga J., Mizutani-Koseki Y., Toyoda T., Okamura K., Tajima S., Mitsuya K., Okano M., Koseki H. (2007) Nature 450, 908–912 [DOI] [PubMed] [Google Scholar]
- 24. Gowher H., Jeltsch A. (2001) J. Mol. Biol. 309, 1201–1208 [DOI] [PubMed] [Google Scholar]
- 25. Okano M., Xie S., Li E. (1998) Nat. Genet. 19, 219–220 [DOI] [PubMed] [Google Scholar]
- 26. Chen T., Ueda Y., Dodge J. E., Wang Z., Li E. (2003) Mol. Cell. Biol. 23, 5594–5605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liang G., Chan M. F., Tomigahara Y., Tsai Y. C., Gonzales F. A., Li E., Laird P. W., Jones P. A. (2002) Mol. Cell. Biol. 22, 480–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kaneda M., Okano M., Hata K., Sado T., Tsujimoto N., Li E., Sasaki H. (2004) Nature 429, 900–903 [DOI] [PubMed] [Google Scholar]
- 29. Kato Y., Kaneda M., Hata K., Kumaki K., Hisano M., Kohara Y., Okano M., Li E., Nozaki M., Sasaki H. (2007) Hum. Mol. Genet. 16, 2272–2280 [DOI] [PubMed] [Google Scholar]
- 30. Sasaki H., Matsui Y. (2008) Nat. Rev. Genet. 9, 129–140 [DOI] [PubMed] [Google Scholar]
- 31. Bourc'his D., Xu G. L., Lin C. S., Bollman B., Bestor T. H. (2001) Science 294, 2536–2539 [DOI] [PubMed] [Google Scholar]
- 32. Hata K., Okano M., Lei H., Li E. (2002) Development 129, 1983–1993 [DOI] [PubMed] [Google Scholar]
- 33. Aapola U., Kawasaki K., Scott H. S., Ollila J., Vihinen M., Heino M., Shintani A., Kawasaki K., Minoshima S., Krohn K., Antonarakis S. E., Shimizu N., Kudoh J., Peterson P. (2000) Genomics 65, 293–298 [DOI] [PubMed] [Google Scholar]
- 34. Chen Z. X., Mann J. R., Hsieh C. L., Riggs A. D., Chédin F. (2005) J. Cell. Biochem. 95, 902–917 [DOI] [PubMed] [Google Scholar]
- 35. Suetake I., Shinozaki F., Miyagawa J., Takeshima H., Tajima S. (2004) J. Biol. Chem. 279, 27816–27823 [DOI] [PubMed] [Google Scholar]
- 36. Jia D., Jurkowska R. Z., Zhang X., Jeltsch A., Cheng X. (2007) Nature 449, 248–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jurkowska R. Z., Anspach N., Urbanke C., Jia D., Reinhardt R., Nellen W., Cheng X., Jeltsch A. (2008) Nucleic Acids Res. 36, 6656–6663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ferguson-Smith A. C., Greally J. M. (2007) Nature 449, 148–149 [DOI] [PubMed] [Google Scholar]
- 39. Glass J. L., Fazzari M. J., Ferguson-Smith A. C., Greally J. M. (2009) Mamm. Genome 20, 633–643 [DOI] [PubMed] [Google Scholar]
- 40. Aravin A. A., Sachidanandam R., Bourc'his D., Schaefer C., Pezic D., Toth K. F., Bestor T., Hannon G. J. (2008) Mol. Cell 31, 785–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kuramochi-Miyagawa S., Watanabe T., Gotoh K., Totoki Y., Toyoda A., Ikawa M., Asada N., Kojima K., Yamaguchi Y., Ijiri T. W., Hata K., Li E., Matsuda Y., Kimura T., Okabe M., Sakaki Y., Sasaki H., Nakano T. (2008) Genes Dev. 22, 908–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chotalia M., Smallwood S. A., Ruf N., Dawson C., Lucifero D., Frontera M., James K., Dean W., Kelsey G. (2009) Genes Dev. 23, 105–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Meissner A., Mikkelsen T. S., Gu H., Wernig M., Hanna J., Sivachenko A., Zhang X., Bernstein B. E., Nusbaum C., Jaffe D. B., Gnirke A., Jaenisch R., Lander E. S. (2008) Nature 454, 766–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Weber M., Hellmann I., Stadler M. B., Ramos L., Pääbo S., Rebhan M., Schübeler D. (2007) Nat. Genet. 39, 457–466 [DOI] [PubMed] [Google Scholar]
- 45. Ooi S. K., Qiu C., Bernstein E., Li K., Jia D., Yang Z., Erdjument-Bromage H., Tempst P., Lin S. P., Allis C. D., Cheng X., Bestor T. H. (2007) Nature 448, 714–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Otani J., Nankumo T., Arita K., Inamoto S., Ariyoshi M., Shirakawa M. (2009) EMBO Rep. 10, 1235–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ciccone D. N., Su H., Hevi S., Gay F., Lei H., Bajko J., Xu G., Li E., Chen T. (2009) Nature 461, 415–418 [DOI] [PubMed] [Google Scholar]
- 48. Zhang Y., Jurkowska R., Soeroes S., Rajavelu A., Dhayalan A., Bock I., Rathert P., Brandt O., Reinhardt R., Fischle W., Jeltsch A. (2010) Nucleic Acids Res. 38, 4246–4253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Dhayalan A., Rajavelu A., Rathert P., Tamas R., Jurkowska R. Z., Ragozin S., Jeltsch A. (2010) J. Biol. Chem. 285, 26114–26120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hodges E., Smith A. D., Kendall J., Xuan Z., Ravi K., Rooks M., Zhang M. Q., Ye K., Bhattacharjee A., Brizuela L., McCombie W. R., Wigler M., Hannon G. J., Hicks J. B. (2009) Genome Res. 19, 1593–1605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Mikkelsen T. S., Ku M., Jaffe D. B., Issac B., Lieberman E., Giannoukos G., Alvarez P., Brockman W., Kim T. K., Koche R. P., Lee W., Mendenhall E., O'Donovan A., Presser A., Russ C., Xie X., Meissner A., Wernig M., Jaenisch R., Nusbaum C., Lander E. S., Bernstein B. E. (2007) Nature 448, 553–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ball M. P., Li J. B., Gao Y., Lee J. H., LeProust E. M., Park I. H., Xie B., Daley G. Q., Church G. M. (2009) Nat. Biotechnol. 27, 361–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhao Q., Rank G., Tan Y. T., Li H., Moritz R. L., Simpson R. J., Cerruti L., Curtis D. J., Patel D. J., Allis C. D., Cunningham J. M., Jane S. M. (2009) Nat. Struct. Mol. Biol. 16, 304–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Rountree M. R., Selker E. U. (2010) Heredity 105, 38–44 [DOI] [PubMed] [Google Scholar]
- 55. Dong K. B., Maksakova I. A., Mohn F., Leung D., Appanah R., Lee S., Yang H. W., Lam L. L., Mager D. L., Schübeler D., Tachibana M., Shinkai Y., Lorincz M. C. (2008) EMBO J. 27, 2691–2701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Tachibana M., Matsumura Y., Fukuda M., Kimura H., Shinkai Y. (2008) EMBO J. 27, 2681–2690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Epsztejn-Litman S., Feldman N., Abu-Remaileh M., Shufaro Y., Gerson A., Ueda J., Deplus R., Fuks F., Shinkai Y., Cedar H., Bergman Y. (2008) Nat. Struct. Mol. Biol. 15, 1176–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lehnertz B., Ueda Y., Derijck A. A., Braunschweig U., Perez-Burgos L., Kubicek S., Chen T., Li E., Jenuwein T., Peters A. H. (2003) Curr. Biol. 13, 1192–1200 [DOI] [PubMed] [Google Scholar]
- 59. Fuks F., Hurd P. J., Deplus R., Kouzarides T. (2003) Nucleic Acids Res. 31, 2305–2312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Viré E., Brenner C., Deplus R., Blanchon L., Fraga M., Didelot C., Morey L., Van Eynde A., Bernard D., Vanderwinden J. M., Bollen M., Esteller M., Di Croce L., de Launoit Y., Fuks F. (2006) Nature 439, 871–874 [DOI] [PubMed] [Google Scholar]
- 61. Jeong S., Liang G., Sharma S., Lin J. C., Choi S. H., Han H., Yoo C. B., Egger G., Yang A. S., Jones P. A. (2009) Mol. Cell. Biol. 29, 5366–5376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Jones P. A., Taylor S. M. (1980) Cell 20, 85–93 [DOI] [PubMed] [Google Scholar]
- 63. Ma D. K., Jang M. H., Guo J. U., Kitabatake Y., Chang M. L., Pow-Anpongkul N., Flavell R. A., Lu B., Ming G. L., Song H. (2009) Science 323, 1074–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Bruniquel D., Schwartz R. H. (2003) Nat. Immunol. 4, 235–240 [DOI] [PubMed] [Google Scholar]
- 65. Ooi S. K., Bestor T. H. (2008) Cell 133, 1145–1148 [DOI] [PubMed] [Google Scholar]
- 66. Zhu J. K. (2009) Annu. Rev. Genet. 43, 143–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Zhu B., Zheng Y., Angliker H., Schwarz S., Thiry S., Siegmann M., Jost J. P. (2000) Nucleic Acids Res. 28, 4157–4165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Zhu B., Zheng Y., Hess D., Angliker H., Schwarz S., Siegmann M., Thiry S., Jost J. P. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 5135–5139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Cortázar D., Kunz C., Saito Y., Steinacher R., Schär P. (2007) DNA Repair 6, 489–504 [DOI] [PubMed] [Google Scholar]
- 70. Santos F., Dean W. (2004) Reproduction 127, 643–651 [DOI] [PubMed] [Google Scholar]
- 71. Millar C. B., Guy J., Sansom O. J., Selfridge J., MacDougall E., Hendrich B., Keightley P. D., Bishop S. M., Clarke A. R., Bird A. (2002) Science 297, 403–405 [DOI] [PubMed] [Google Scholar]
- 72. Kim M. S., Kondo T., Takada I., Youn M. Y., Yamamoto Y., Takahashi S., Matsumoto T., Fujiyama S., Shirode Y., Yamaoka I., Kitagawa H., Takeyama K., Shibuya H., Ohtake F., Kato S. (2009) Nature 461, 1007–1012 [DOI] [PubMed] [Google Scholar]
- 73. Morgan H. D., Dean W., Coker H. A., Reik W., Petersen-Mahrt S. K. (2004) J. Biol. Chem. 279, 52353–52360 [DOI] [PubMed] [Google Scholar]
- 74. Rai K., Huggins I. J., James S. R., Karpf A. R., Jones D. A., Cairns B. R. (2008) Cell 135, 1201–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Bhutani N., Brady J. J., Damian M., Sacco A., Corbel S. Y., Blau H. M. (2010) Nature 463, 1042–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Popp C., Dean W., Feng S., Cokus S. J., Andrews S., Pellegrini M., Jacobsen S. E., Reik W. (2010) Nature 463, 1101–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Muramatsu M., Kinoshita K., Fagarasan S., Yamada S., Shinkai Y., Honjo T. (2000) Cell 102, 553–563 [DOI] [PubMed] [Google Scholar]
- 78. Barreto G., Schäfer A., Marhold J., Stach D., Swaminathan S. K., Handa V., Döderlein G., Maltry N., Wu W., Lyko F., Niehrs C. (2007) Nature 445, 671–675 [DOI] [PubMed] [Google Scholar]
- 79. Jin S. G., Guo C., Pfeifer G. P. (2008) PLoS Genet. 4, e1000013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Engel N., Tront J. S., Erinle T., Nguyen N., Latham K. E., Sapienza C., Hoffman B., Liebermann D. A. (2009) Epigenetics 4, 98–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Schmitz K. M., Schmitt N., Hoffmann-Rohrer U., Schäfer A., Grummt I., Mayer C. (2009) Mol. Cell 33, 344–353 [DOI] [PubMed] [Google Scholar]
- 82. Kriaucionis S., Heintz N. (2009) Science 324, 929–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Tahiliani M., Koh K. P., Shen Y., Pastor W. A., Bandukwala H., Brudno Y., Agarwal S., Iyer L. M., Liu D. R., Aravind L., Rao A. (2009) Science 324, 930–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Ito S., D'Alessio A. C., Taranova O. V., Hong K., Sowers L. C., Zhang Y. (2010) Nature 466, 1129–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Cannon S. V., Cummings A., Teebor G. W. (1988) Biochem. Biophys. Res. Commun. 151, 1173–1179 [DOI] [PubMed] [Google Scholar]
- 86. Hajkova P., Jeffries S. J., Lee C., Miller N., Jackson S. P., Surani M. A. (2010) Science 329, 78–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Wossidlo M., Arand J., Sebastiano V., Lepikhov K., Boiani M., Reinhardt R., Schöler H., Walter J. (2010) EMBO J. 29, 1877–1888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Okada Y., Yamagata K., Hong K., Wakayama T., Zhang Y. (2010) Nature 463, 554–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Wu S. C., Zhang Y. (2010) Nat. Rev. Mol. Cell Biol. 11, 607–620 [DOI] [PMC free article] [PubMed] [Google Scholar]