Abstract
Mitotic inheritance of gene function is obligatory to sustain biological control. Emerging evidence suggests that epigenetic mechanisms are linked to transmission of cell fate, lineage commitment, and maintenance of cellular phenotype in progeny cells. Mechanisms of epigenetic memory include gene silencing by DNA methylation, transcriptional regulation by histone modifications, regulation of gene expression by noncoding small RNA molecules, and retention of regulatory machinery on target gene loci for activation and repression. We will focus on the regulatory implications of epigenetic memory for physiological control and for the onset and progression of disease.
Keywords: Cell Cycle, Epigenetics, Gene Expression, Histone Modification, Nuclear Organization
Introduction
Cell division requires functional changes that include transcriptional silencing by reorganization of regulatory machinery within the nucleus and structural alterations by remodeling of cellular architecture (1, 2). Selective mitotic retention of regulatory signatures is necessary for continuity of function in progeny cells. For example, genes expressed immediately after cell division must be epigenetically marked and poised for transcription. Similarly, genes required exclusively during development and silenced postnatal genes or genes selectively expressed in specific lineages and permanently repressed in others must epigenetically acquire functional memory and retain or relinquish transcriptional status. Equally important, persistence of cell transformation and tumor progression are dependent on continuity of cancer-related gene expression.
Historically, nuclear transplantation studies established retention of competency for pluripotency and epigenetic plasticity (Fig. 1) (3). Genetic control has been shown to be essential but insufficient to accommodate requirements for gene expression (4). Contributions of epigenetic control are evident, and new dimensions are emerging. DNA methylation and histone modifications play key roles in epigenetically transmitting transcriptional memory (5–11). Noncoding small RNA molecules and mitotic retention of gene regulatory machinery are major epigenetic components controlling gene transcription in parent and progeny cells (12–18). These epigenetic mechanisms can be synergistic, antagonistic, or mutually exclusive in regulating the distribution of regulatory information during cell division, development, and differentiation.
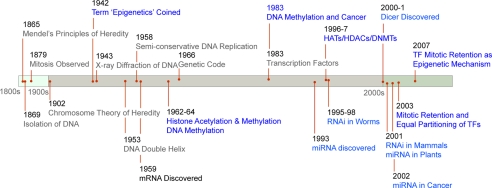
FIGURE 1.
Timeline of important discoveries in genetic and epigenetic regulation of gene expression. A simplified timeline of some key events and discoveries in the fields of genetic and epigenetic gene regulation is presented. Important discoveries in genetic components of gene expression are in black, and the epigenetic events are in blue. The figure was not drawn to scale. HATs, histone acetylases; HDACs, histone deacetylases; DNMTs, DNA methyltransferases; TF, transcription factor; miRNA, microRNA.
We will present an overview of mechanisms operative in cellular epigenetic memory and cohorts of combinatorial and cumulative epigenetic marks that are conveyed through generations for cell fate and lineage commitment. DNA methylation and histone modifications in regulating the interphase gene expression are covered in detail in this minireview series (Fig. 2). We will focus on mitotic inheritance of non-DNA-encoded regulatory parameters in symmetric and asymmetrical cell divisions.
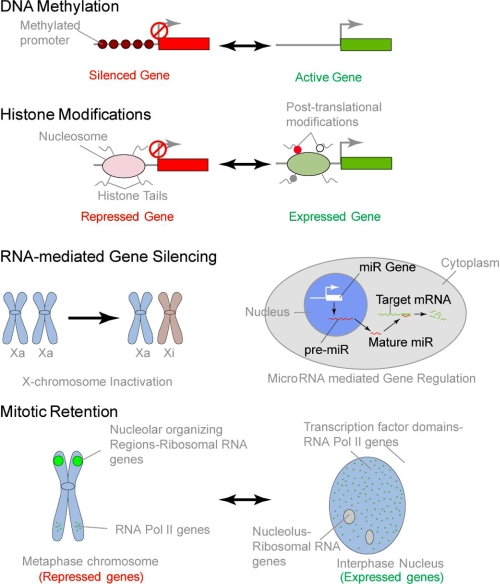
FIGURE 2.
Epigenetic mechanisms of gene expression. Genetic and epigenetic mechanisms contribute to physiological control of mammalian gene expression. Although the genetic information encodes necessary components of gene expression, epigenetic control alters the phenotype without changing the genotype of a cell and provides an additional level of control. Shown here are some well defined epigenetic mechanisms, i.e. DNA methylation, histone modifications, and noncoding RNA-mediated modulation of gene expression. These mechanisms contribute to inheritable transcriptional memory of some of the genes through successive cell divisions and contribute to maintenance of cellular phenotype. Recent studies show that association of transcriptional regulatory machinery with target genes on mitotic chromosomes is a novel epigenetic mechanism that poises genes involved in key cellular processes, such as growth, proliferation, and lineage commitment, for expression in progeny cells. miR, microRNA.
DNA Methylation: A Strategy for Transcriptional Silencing
DNA methylation is a well established epigenetic mechanism for both transient and irreversible transcriptional silencing (9–11). Three DNA methyltransferases, designated DNMT1, DNMT3a, and DNMT3b, are responsible for dynamic transcriptional silencing by addition of methyl groups to CpG islands in gene promoters (19, 20). Consequently, compromised binding of transcription factors to gene promoters and/or altered nucleosomal occupancy within the promoter contributes to biological control (21).
DNA methylation permits silenced states of genes to be inherited through multiple cell divisions (9). Gene imprinting and allelic exclusion are examples of developmental gene silencing through successive mitoses. An allele-specific imprinting control region (ICR)3 within the promoter regions of certain genes regulates their monoallelic expression. A differentially methylated domain (DMD) present within the ICR is epigenetically altered by DNA methylation (and histone modifications), which is perpetuated through cell divisions to maintain the silenced state of the gene in progeny cells.
An informative example is the Rasgrf1 (Ras protein-specific guanine nucleotide-releasing factor 1) gene, the product of which is an important component of the Ras pathway (22–24). The ICR of the locus contains a repeated element that is essential and sufficient for the establishment and maintenance of DNA methylation at the DMD for germ line imprinting of the paternal allele. The CCCTC-binding factor, a transcriptional repressor, occupies the DMD on the unmethylated maternal allele and prevents interaction of the enhancer with the promoter, thus silencing the maternal allele (25). Binding of the CCCTC-binding factor to the paternal DMD is prevented by DNA methylation, allowing specific expression of the paternal allele. Thus, DNA methylation plays a critical role in gene imprinting and allelic exclusion at the Rasgrf1 gene locus during development and ensures transmission of “transcriptional memory” through cell divisions.
Although the role of DNA methylation in inheritable silencing of transcriptional states is well understood (9), some key questions remain to be addressed. How are DNA methyltransferases targeted to gene promoters? Recent reports show that H3K9 and H3K27 histone modifications may play a role in directed DNA methylation. It is yet to be resolved whether these histone modifications are a prerequisite for targeting DNA methyltransferases to gene promoters. Equally important is understanding the extent to which enzymatic demethylation and replication-mediated loss of methyl groups are operative in reactivation of genes that are inheritably repressed. The genomic distribution of methylated marks is emerging as a resource for epigenetic signature. It is important to discriminate between methylation signatures that are inheritable through mitoses and methylation patterns confined to a single cell cycle.
Histone Modifications: Determinants of Accessibility
Post-translational modification of nucleosomal histones at the N-terminal tails is a well studied epigenetic mechanism (5–8, 26); histone phosphorylation and acetylation were among the first parameters of epigenetic control (27–29). Histone modifications are dynamic, and enzymes that add or remove post-translational histone modifications have been identified (5–8). Modified histones disrupt contacts between nucleosomes and DNA and render chromatin more accessible for recruitment of non-histone transcriptional regulatory proteins (5–8, 26). Additional complexity is offered by the ordered and sequential nature of histone modifications that either attenuate or accentuate ongoing transcription. Relative turnover of histone modifications, choice of modified residues, and unique functional consequences of each histone modification have led to the proposal of a histone code that governs gene regulatory events. An in-depth discussion of the contributions of histone modifications to context-dependent regulation of gene expression is covered in this minireview series.
The emerging perspective suggests that histone modifications play a role in gene bookmarking during mitosis and in inheritance of chromatin state during differentiation. Histone modifications often work in concert with other epigenetic marks (e.g. DNA methylation) to ensure the inheritance of specific gene expression patterns through cell generations (30–32). Histone modifications have been implicated in a number of epigenetic phenomena, including the inheritable silencing of heterochromatin (33–35). The complexity of heterochromatin formation is reflected by its absence in human embryonic stem cells. In contrast, the sperm heterochromatin is highly compact (33). In each scenario, it is important to maintain the state of heterochromatin. Methylation of Lys-9 of histone H3 plays a crucial role in the transmission of the heterochromatin (34, 35). Once methylated, Lys-9 recruits HP1 (heterochromatin protein 1) to the chromatin. HP1 recruits additional H3K9-methylating activity that modifies nucleosomes on the nascent strand, ensuring the transmission of the H3K9 methylation mark (36). It is likely that other epigenetic modifications and histone variants contribute to inheritable heterochromatin silencing (37). One of the mechanisms that maintain heterochromatin during interphase is the incorporation of a variant of histone H2A (macroH2A) into DNA to be packaged as heterochromatin, but mechanisms that prevent heterochromatin formation in human embryonic stem cells as well as initiate and maintain heterochromatin during and following lineage commitment are minimally understood.
A subset of genes is “marked” by specific histone modifications and/or variants to designate inheritable expression post-mitotically or during differentiation (38). Inheritance of regulatory machinery for myogenesis provides an example in which both histone modifications and variants contribute to epigenetic memory (38, 39). Nuclear transplant experiments have shown that the methylation status of the muscle-specific myoD promoter remains unaltered in non-muscle cell lineages (39). Instead, Gurdon and co-workers have shown that the association of a histone H3 variant (H3.3) with the myoD promoter dictates epigenetic memory. H3.3 is associated with the myoD gene in embryos that display “transcriptional memory” but not in those in which memory has been lost. Mechanistically, blocking the methylation of H3.3 at Lys-4 leads to the loss of transcriptional memory, whereas overexpression of H3.3 contributes to memory. Long-term retention of histone H3.3 with the myoD promoter has been established as a component in lineage-restricted transcriptional memory. Consequently, histone H3.3 stabilizes transcription during development (38, 39).
These observations indicate that mechanisms exist during mitosis to maintain open chromatin architecture on selective genes for expression following cell division. However, the extent to which the histone code that operates in interphase is retained during mitosis remains to be determined. Important open-ended questions relate to the existence of a mitotic histone code. Are both histone modifications and histone variants involved in mitotic gene bookmarking? Does the entire cohort of histone modifications persist? Is the stoichiometry of histone protein variants present in nucleosomes on a gene locus a critical determinant for epigenetic transmission of transcriptional state? Does genomic and biological context determine whether a gene will be bookmarked? What components of enzymology are involved?
Noncoding RNA Molecules
Noncoding RNAs range from very small (21–25-nucleotide microRNAs) to very large, such as the Air transcript, which exceeds 100 kb in length (40–44). The presence of RNAi in lower eukaryotes and microRNAs in higher eukaryotes indicates biological roles for noncoding RNA; these include transcriptional regulation and translational inhibition that are accomplished through direct base pairing with the DNA or RNA target, by mimicking the structure of other nucleic acids, or by functioning as a component of an RNA-protein complex (41–44).
A well studied example of RNA involvement in heritable epigenetic regulation is X chromosome inactivation by Xist RNA in mammals (45–50). Xist RNA is a large, noncoding, and alternatively spliced RNA that associates with the X chromosome from which it is transcribed (45). Xist RNA is essential for inactivation of one copy of the X chromosome when more than one copy is present (48). Inactivation of the paternal X chromosome occurs in extraembryonic tissues, and consistent with this, the paternal Xist copy is expressed preferentially in female preimplantation embryos (49). Xist RNA coats the X chromosome that is silenced and, together with recruitment of the chromatin-remodeling and DNA methylation machinery, leads to stable inheritable silencing of one copy of the X chromosome (50). A complete understanding of mechanisms that mediate, preserve, and propagate X chromosome inactivation will require additional experimental evidence. Histone modifications also contribute to X chromosome inactivation (47), but specific roles of these modifications are not well established. Adapter proteins that integrate independent epigenetic mechanisms for X chromosome inactivation are not known. It also remains to be established whether all RNA-mediated silencing is heritable and the extent to which RNA control of gene expression is reversible or permanent.
Mitotic Retention of Gene Regulatory Machinery: Sustaining the Family Legacy
Mitotic remodeling of cellular and nuclear architecture includes dynamic relocalization of transcription factors (e.g. Ets1, Oct2, B-Myb, and Sp factors) and transient degradation of key regulatory proteins (e.g. cyclins) (1, 2, 51–54). After mitosis, the structural and functional integrity of the cell's regulatory machinery must be re-established to accommodate cell cycle and growth control as well as phenotype. Enzymatically and chemically sensitive sites on mitotic chromosomes mark active genes (18, 55), indicating that some regulatory complexes remain bound to the condensed chromatin for rapid reactivation of genes following mitosis. Mitotic retention of the transcription factor IID complex at gene promoters supports a “bookmarking” mechanism to resume active transcription upon exit from mitosis (17, 56). Recent evidence that phenotypic transcription factors are associated with target gene loci on mitotic chromosomes adds a tissue-specific dimension to this concept (57–61).
The tandemly organized and developmentally regulated globin gene loci exhibit DNase I-hypersensitive sites that are linked to erythroid lineage-restricted gene expression (62, 63). These hypersensitive sites are retained for >20 generations (64). Mechanistically, NF-E2, a globin gene regulator, remains bound to mitotic chromosomes (65), but the GATA1 erythroid transcription factor dissociates from condensed chromatin (65, 66). NF-E2 recruits chromatin-remodeling factors that include TAFII130 and CBP (cAMP-responsive element-binding protein-binding protein) to support re-expression of the globin gene post-mitotically (65). Thus, NF-E2 supports the persistent hypersensitive state of globin genes. This is complemented by active histone modifications (e.g. H3 acetylation and H3 Lys-4 dimethylation and Lys-79 dimethylation) in distal regulatory domains of globin gene loci that are transcriptionally competent (65). Collectively, an “epigenetic memory” mechanism mediates efficient and lineage-restricted reactivation of globin gene transcription following mitosis.
Differentiation of mesenchymal progenitor cells into osteoblasts, adipocytes, or myoblasts is dictated by specific sets of master transcription factors (i.e. RUNX2 in osteoblasts, CCAAT-enhancer-binding protein α in adipocytes, and MyoD and myogenin in myoblasts) (67–69). In interphase, these proteins reside in distinct nuclear microenvironments where they interact with coregulatory proteins as well as cognate DNA elements to carry out biological functions (70). Studies from our group and others have shown that these regulatory proteins are also associated with phenotypic target genes on mitotic chromosomes in their respective lineages (57–61). During mitosis, these proteins localize to nucleolar organizing regions, the precursors of interphase nucleoli, and occupy RNA polymerase (Pol) I-transcribed ribosomal RNA genes as well as RNA Pol II-transcribed phenotypic genes that are involved in control of the cell cycle and differentiation. Post-mitotically, phenotype regulatory proteins are equally partitioned to progeny cells and, consistent with their antiproliferative and differentiation-promoting roles, down-regulate rRNA gene transcription, which is intimately linked to cell growth, thus coordinating cell proliferation and growth with lineage maintenance (57–61). The occupancy and regulation of genes that are transcribed by both RNA Pol I and Pol II by these proteins during interphase and mitosis reflect coordinate genetic and epigenetic control of cell proliferation, growth, and differentiation.
Asymmetric Partitioning of Transcription Factors: Separate but Not Equal
Lineage progenitors and stem cells must undergo asymmetric cell division to sustain the availability of a progenitor pool for two important cell types: pluripotent cells and lineage-committed cells (71, 72). Most informative observations regarding the asymmetric cell division have come from lower eukaryotes (73–75). For example, in budding yeast, the Ace2 chromatin-remodeling factor is asymmetrically accumulated in the nucleus of the daughter cell (76). Ace2, which is translated in G2 cells, is retained in the cytoplasm. In late mitosis, a dephosphorylation event leads to unmasking of the nuclear localization signal and nuclear translocation of the protein. Once in the daughter nucleus, Ace2 is phosphorylated by the daughter cell-specific Cbk1 kinase, which results in masking the export signal and the daughter cell-specific nuclear retention of Ace2 (76). It remains to be explored whether the daughter cell-specific retention of Ace2 results in resumption of gene expression that is restricted to the progeny.
Asymmetric cell division has also been observed during Drosophila neurogenesis, in which the progenitor undergoes several rounds of asymmetric division (74). Each division produces a large cell that remains a neuroblast and a smaller ganglion mother cell that eventually differentiates into neurons and glia of the central nervous system. In parallel, the sensory organ precursor also undergoes asymmetric cell divisions to give rise to five distinct cell types of the peripheral nervous system. In both these progenitors, Numb, an attenuator of Notch signaling, and the Prospero transcription factor distribute asymmetrically to only one of the daughter nuclei (77–79). This asymmetry is achieved through the actions of a protein complex that localizes to the apical cortex of dividing neuroblasts and regulates orientation of the mitotic spindle, thus controlling the polarity of these proteins.
Similar mechanisms may be operative in mammalian cells for maintenance of stem/progenitor cell pools while giving rise to distinct cell types from a single progenitor. However, experimental evidence to establish the existence of asymmetrical distribution of regulatory complexes in mammals is lacking. It also remains to be determined how phenotypic properties of progeny cells that arise from an asymmetric cell division are established. A principal limitation is the lack of an in vitro model system in which in vivo mechanisms that underlie asymmetric distribution can be recapitulated.
Mitotic retention of gene regulatory machinery, particularly on chromosomes, offers a new dimension to epigenetic regulation during cell division, both symmetric and asymmetric. Together with DNA methylation and histone modifications, transcription factor association with mitotic gene loci or their retention during mitosis provides architectural epigenetic signatures, with several regulatory implications. For example, transcription factor association may “tag” genes for rapid reactivation post-mitotically. Another implication is that mitotic association of chromatin-remodeling proteins (e.g. Ace2) with gene loci may render genes more accessible for regulatory proteins in G1 cells. Additionally, mitotic association of phenotypic regulatory proteins with genes in an allele-specific manner in omnipotent and pluripotent cells can facilitate asymmetric distribution of transcription factors to cells destined to commit to a particular lineage. In committed cells, equal partitioning of phenotypic transcription factors can then lead to maintenance of the lineage. Symmetric distribution of regulatory factors has been demonstrated (57), but whether this is the rule or exception must be established. Other key questions remain. How are genes that do not retain cognate regulatory factors during mitosis marked for activation or suppression in progeny cells? How are genes epigenetically regulated in a selective manner during asymmetric cell division? Are all, some, or none of the coregulatory proteins of a particular transcription factor present at mitotic gene loci? Despite open-ended questions, this epigenetic mechanism offers a flexible yet cell type-specific and context-dependent dimension to gene regulation.
Epigenetic Memory Sustains the Disease Phenotypes
Cancer development depends not only on genetic alterations but also on an abnormal cellular memory or epigenetic changes that convey heritable gene expression patterns critical for tumor initiation and progression (80–83). These aberrant epigenetic mechanisms result in global changes in chromatin packaging and in localized gene promoter changes that influence the transcription of genes important to cancer.
The hematopoietic master regulator RUNX1 provides an example of deregulated epigenetic control during mitosis. RUNX1 is required for definitive hematopoiesis and is frequently translocated in hematological malignancies (84). One of the most prevalent chromosomal translocations involving the RUNX1 gene is the 8;21 translocation, present in >15% of all patients with acute myelogenous leukemia (85–87). t(8;21) results in expression of a chimeric protein, AML1-ETO, from the RUNX1 gene locus. AML1-ETO shares the DNA-binding domain of the normal RUNX1 protein but lacks the C terminus, which is replaced by the nearly full-length ETO protein (85–87). AML1-ETO localizes to distinct nuclear microenvironments, interferes with RUNX1 function, and results in a blast-like phenotype of myeloid progenitors (88–90). RUNX1 associates with mitotic chromosomes and is equally distributed to the progeny cells (91). RUNX1 also occupies rRNA gene promoters during interphase and down-regulates the expression of pre-rRNA transcripts. However, during mitosis and interphase in leukemic cells that express AML1-ETO from one allele, rRNA genes are occupied by the chimeric protein instead. AML1-ETO occupancy of rRNA genes up-regulates rRNA genes concomitant with the growth advantage of leukemic cells (91). This example provides mechanistic insights into inheritable progression and maintenance of a disease phenotype and offers novel opportunities to specifically target AML1-ETO during mitosis.
Targeting the Inherited Epigenome in Cancer: Limitations and Promises
Genome-wide screens of histone modifications and DNA methylation have provided an unbiased means to define diagnostic epigenetic signatures for tumor type and have offered promising therapeutic targets (80–83). Drugs that inhibit DNA methyltransferase activity (such as azacytidine (5-azacytidine), decitabine (5-aza-2-deoxycytidine), fazarabine (1-β-d-arabinofuranosyl-5-azacytosine), and dihydro-5-azacytidine) or histone deacetylase function (e.g. hydroxamates such as suberoylanilide hydroxamic acid, a cyclic peptide-benzamide such as MS-275, and aliphatic acids such as phenylbutyrate) are in clinical trials (92). Global effects of these inhibitors that may impede normal cellular functions are a limitation. A confounding consideration is plasticity that may be a function of DNA hemimethylation, which can influence the composite epigenetic signature as well as the optimal approach for drug design. How is the enzymology targeted specifically to these silenced genes? Are certain histone modifications unique to gene loci of oncogenes and tumor suppressors? Consistent with these concerns, it has become clear over the past decade or so that targeting DNA methylation or histone modifications alone is not sufficient for therapeutic purposes. Because these mechanisms are cumulative and context-dependent, it is likely that they function in concert with each other or with other epigenetic mechanisms to contribute to cancer phenotype. Consequently, the strategy for effective therapy may be complex and multifactorial.
Small noncoding RNA molecules and their relatively specific inhibition of gene expression offer attractive options for use as diagnostic markers and for targeting selective gene sets. Although some studies have identified roles of small RNA molecules as epigenetic carriers, additional studies are needed to define their activity as a principal epigenetic mechanism.
In addition to its contribution to physiologically responsive gene expression during development and tissue remodeling, retention of regulatory proteins with gene loci during cell division can be translated to targeted therapy with enhanced specificity and reduced off-target effects. In contrast, global inhibition of histone modification or DNA methylation does not provide a viable option for selectivity. During mitosis, minimal components of transcriptional regulatory complexes may be present, thus unmasking epitopes to generate a druggable target. Such a mechanism can favorably influence pharmacological kinetics, i.e. the minimal drug concentration will be required. In addition, mitotic association of regulatory proteins, combined with global genome-wide assessment of histone modifications and DNA methylation, may provide a diagnostic “epigenetic signature.”
Concluding Remarks: Illuminating the Dark Side of Gene Regulation by Understanding Epigenetic Mechanisms
With increased capabilities for genome-wide expression profiling and accruing signatures for gene expression that are linked to biological control and pathology, gaps in our ability to mechanistically account for transcriptional activation, suppression, and persistence of expression status during cell division have become evident. Elucidation of control that is outside of DNA sequences can provide additional levels of regulation. Epigenetic mechanisms that include DNA methylation, histone modifications, noncoding RNA molecules, and mitotic retention of transcription factors individually and as combinatorial signatures have been shown to be informative in predicting the biological outcome and in diagnosing the diseased state. The definition of epigenetic control is evolving by necessity to accommodate recognition for an expanding repertoire of non-DNA-encoded information that establishes and sustains regulatory machinery. Understanding the control of these non-genomic but inheritable parameters can yield novel dimensions to regulation of cell fate and lineage commitment. In turn, these observations will provide important insights into development, differentiation, and tissue remodeling, as well as an appreciation for regulatory mechanisms that are compromised with the onset and progression of disease. Finally, although abrogation of epigenetic mechanisms in disease can result in dysfunction similar to that caused by point mutations or chromosomal translocations, reversibility renders them viable therapeutic targets.
Acknowledgment
We thank Patricia Jamieson for assistance in preparation of this article.
This work was supported, in whole or in part, by National Institutes of Health Grants P01 AR048818, P01 CA082834, and 5 P30 DK32520. This is the second article in the Thematic Minireview Series on Epigenetics. This minireview will be reprinted in the 2011 Minireview Compendium, which will available in January, 2012.
- ICR
- imprinting control region
- DMD
- differentially methylated domain
- Pol
- polymerase.
REFERENCES
- 1. Gottesfeld J. M., Forbes D. J. (1997) Trends Biochem. Sci. 22, 197–202 [DOI] [PubMed] [Google Scholar]
- 2. Sullivan M., Morgan D. O. (2007) Nat. Rev. Mol. Cell Biol. 8, 894–903 [DOI] [PubMed] [Google Scholar]
- 3. Gurdon J. B. (1964) Adv. Morphog. 4, 1–43 [DOI] [PubMed] [Google Scholar]
- 4. Goldberg A. D., Allis C. D., Bernstein E. (2007) Cell 128, 635–638 [DOI] [PubMed] [Google Scholar]
- 5. Lee K. K., Workman J. L. (2007) Nat. Rev. Mol. Cell Biol. 8, 284–295 [DOI] [PubMed] [Google Scholar]
- 6. Mellor J., Dudek P., Clynes D. (2008) Curr. Opin. Genet. Dev. 18, 116–122 [DOI] [PubMed] [Google Scholar]
- 7. Jenuwein T., Allis C. D. (2001) Science 293, 1074–1080 [DOI] [PubMed] [Google Scholar]
- 8. Berger S. L. (2007) Nature 447, 407–412 [DOI] [PubMed] [Google Scholar]
- 9. Lande-Diner L., Cedar H. (2005) Nat. Rev. Genet. 6, 648–654 [DOI] [PubMed] [Google Scholar]
- 10. Fazzari M. J., Greally J. M. (2004) Nat. Rev. Genet. 5, 446–455 [DOI] [PubMed] [Google Scholar]
- 11. Jones P. A., Baylin S. B. (2007) Cell 128, 683–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Costa F. F. (2008) Gene 410, 9–17 [DOI] [PubMed] [Google Scholar]
- 13. Filipowicz W., Bhattacharyya S. N., Sonenberg N. (2008) Nat. Rev. Genet. 9, 102–114 [DOI] [PubMed] [Google Scholar]
- 14. Goodrich J. A., Kugel J. F. (2006) Nat. Rev. Mol. Cell Biol. 7, 612–616 [DOI] [PubMed] [Google Scholar]
- 15. Moazed D. (2009) Nature 457, 413–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zaidi S. K., Young D. W., Montecino M. A., Lian J. B., van Wijnen A. J., Stein J. L., Stein G. S. (2010) Nat. Rev. Genet. 11, 583–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sarge K. D., Park-Sarge O. K. (2005) Trends Biochem. Sci. 30, 605–610 [DOI] [PubMed] [Google Scholar]
- 18. John S., Workman J. L. (1998) BioEssays 20, 275–279 [DOI] [PubMed] [Google Scholar]
- 19. Gold M., Hurwitz J., Anders M. (1963) Biochem. Biophys. Res. Commun. 11, 107–114 [DOI] [PubMed] [Google Scholar]
- 20. Tucker K. L., Talbot D., Lee M. A., Leonhardt H., Jaenisch R. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 12920–12925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Edwards C. A., Ferguson-Smith A. C. (2007) Curr. Opin. Cell Biol. 19, 281–289 [DOI] [PubMed] [Google Scholar]
- 22. Yoon B. J., Herman H., Sikora A., Smith L. T., Plass C., Soloway P. D. (2002) Nat. Genet. 30, 92–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Martegani E., Vanoni M., Zippel R., Coccetti P., Brambilla R., Ferrari C., Sturani E., Alberghina L. (1992) EMBO J. 11, 2151–2157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bernards A., Settleman J. (2005) Growth Factors 23, 143–149 [DOI] [PubMed] [Google Scholar]
- 25. Yoon B., Herman H., Hu B., Park Y. J., Lindroth A., Bell A., West A. G., Chang Y., Stablewski A., Piel J. C., Loukinov D. I., Lobanenkov V. V., Soloway P. D. (2005) Mol. Cell. Biol. 25, 11184–11190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ruthenburg A. J., Li H., Patel D. J., Allis C. D. (2007) Nat. Rev. Mol. Cell Biol. 8, 983–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Stevely W. S., Stocken L. A. (1966) Biochem. J. 100, 20C–21C [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gutierrez R. M., Hnilica L. S. (1967) Science 157, 1324–1325 [DOI] [PubMed] [Google Scholar]
- 29. Allfrey V. G., Faulkner R., Mirsky A. E. (1964) Proc. Natl. Acad. Sci. U.S.A. 51, 786–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Stancheva I. (2005) Biochem. Cell Biol. 83, 385–395 [DOI] [PubMed] [Google Scholar]
- 31. Rice J. C., Allis C. D. (2001) Curr. Opin. Cell Biol. 13, 263–273 [DOI] [PubMed] [Google Scholar]
- 32. Zhang Y., Reinberg D. (2001) Genes Dev. 15, 2343–2360 [DOI] [PubMed] [Google Scholar]
- 33. Grewal S. I., Jia S. (2007) Nat. Rev. Genet. 8, 35–46 [DOI] [PubMed] [Google Scholar]
- 34. Richards E. J., Elgin S. C. (2002) Cell 108, 489–500 [DOI] [PubMed] [Google Scholar]
- 35. Krauss V. (2008) Genetica 133, 93–106 [DOI] [PubMed] [Google Scholar]
- 36. Eskeland R., Eberharter A., Imhof A. (2007) Mol. Cell. Biol. 27, 453–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ladurner A. G. (2003) Mol. Cell 12, 1–3 [DOI] [PubMed] [Google Scholar]
- 38. Ng R. K., Gurdon J. B. (2008) Cell Cycle 7, 1173–1177 [DOI] [PubMed] [Google Scholar]
- 39. Ng R. K., Gurdon J. B. (2008) Nat. Cell Biol. 10, 102–109 [DOI] [PubMed] [Google Scholar]
- 40. Eliceiri G. L. (1974) Cell 3, 11–14 [DOI] [PubMed] [Google Scholar]
- 41. Fire A., Xu S., Montgomery M. K., Kostas S. A., Driver S. E., Mello C. C. (1998) Nature 391, 806–811 [DOI] [PubMed] [Google Scholar]
- 42. Ambros V. (2001) Cell 107, 823–826 [DOI] [PubMed] [Google Scholar]
- 43. McManus M. T., Sharp P. A. (2002) Nat. Rev. Genet. 3, 737–747 [DOI] [PubMed] [Google Scholar]
- 44. Croce C. M., Calin G. A. (2005) Cell 122, 6–7 [DOI] [PubMed] [Google Scholar]
- 45. Brown C. J., Hendrich B. D., Rupert J. L., Lafrenière R. G., Xing Y., Lawrence J., Willard H. F. (1992) Cell 71, 527–542 [DOI] [PubMed] [Google Scholar]
- 46. Brockdorff N., Ashworth A., Kay G. F., Cooper P., Smith S., McCabe V. M., Norris D. P., Penny G. D., Patel D., Rastan S. (1991) Nature 351, 329–331 [DOI] [PubMed] [Google Scholar]
- 47. Heard E., Clerc P., Avner P. (1997) Annu. Rev. Genet. 31, 571–610 [DOI] [PubMed] [Google Scholar]
- 48. Penny G. D., Kay G. F., Sheardown S. A., Rastan S., Brockdorff N. (1996) Nature 379, 131–137 [DOI] [PubMed] [Google Scholar]
- 49. Johnston C. M., Nesterova T. B., Formstone E. J., Newall A. E., Duthie S. M., Sheardown S. A., Brockdorff N. (1998) Cell 94, 809–817 [DOI] [PubMed] [Google Scholar]
- 50. Clemson C. M., McNeil J. A., Willard H. F., Lawrence J. B. (1996) J. Cell Biol. 132, 259–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Spencer C. A., Kruhlak M. J., Jenkins H. L., Sun X., Bazett-Jones D. P. (2000) J. Cell Biol. 150, 13–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. He S., Davie J. R. (2006) J. Cell Sci. 119, 1063–1070 [DOI] [PubMed] [Google Scholar]
- 53. Martínez-Balbás M. A., Dey A., Rabindran S. K., Ozato K., Wu C. (1995) Cell 83, 29–38 [DOI] [PubMed] [Google Scholar]
- 54. Pines J. (2006) Trends Cell Biol. 16, 55–63 [DOI] [PubMed] [Google Scholar]
- 55. Xin L., Zhou G. L., Song W., Wu X. S., Wei G. H., Hao D. L., Lv X., Liu D. P., Liang C. C. (2007) BMC Mol. Biol. 8, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Christova R., Oelgeschläger T. (2002) Nat. Cell Biol. 4, 79–82 [DOI] [PubMed] [Google Scholar]
- 57. Zaidi S. K., Young D. W., Pockwinse S. M., Javed A., Lian J. B., Stein J. L., van Wijnen A. J., Stein G. S. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 14852–14857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Young D. W., Hassan M. Q., Pratap J., Galindo M., Zaidi S. K., Lee S. H., Yang X., Xie R., Javed A., Underwood J. M., Furcinitti P., Imbalzano A. N., Penman S., Nickerson J. A., Montecino M. A., Lian J. B., Stein J. L., van Wijnen A. J., Stein G. S. (2007) Nature 445, 442–446 [DOI] [PubMed] [Google Scholar]
- 59. Young D. W., Hassan M. Q., Yang X. Q., Galindo M., Javed A., Zaidi S. K., Furcinitti P., Lapointe D., Montecino M., Lian J. B., Stein J. L., van Wijnen A. J., Stein G. S. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 3189–3194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ali S. A., Zaidi S. K., Dacwag C. S., Salma N., Young D. W., Shakoori A. R., Montecino M. A., Lian J. B., van Wijnen A. J., Imbalzano A. N., Stein G. S., Stein J. L. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 6632–6637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Tang Q. Q., Lane M. D. (1999) Genes Dev. 13, 2231–2241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Cao A., Moi P. (2002) Pediatr. Res. 51, 415–421 [DOI] [PubMed] [Google Scholar]
- 63. McGhee J. D., Wood W. I., Dolan M., Engel J. D., Felsenfeld G. (1981) Cell 27, 45–55 [DOI] [PubMed] [Google Scholar]
- 64. Groudine M., Weintraub H. (1982) Cell 30, 131–139 [DOI] [PubMed] [Google Scholar]
- 65. Martin D. I., Orkin S. H. (1990) Genes Dev. 4, 1886–1898 [DOI] [PubMed] [Google Scholar]
- 66. Whitelaw E., Tsai S. F., Hogben P., Orkin S. H. (1990) Mol. Cell. Biol. 10, 6596–6606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Lian J. B., Javed A., Zaidi S. K., Lengner C., Montecino M., van Wijnen A. J., Stein J. L., Stein G. S. (2004) Crit. Rev. Eukaryot. Gene Expr. 14, 1–41 [PubMed] [Google Scholar]
- 68. Weintraub H., Davis R., Tapscott S., Thayer M., Krause M., Benezra R., Blackwell T. K., Turner D., Rupp R., Hollenberg S. (1991) Science 251, 761–766 [DOI] [PubMed] [Google Scholar]
- 69. MacDougald O. A., Lane M. D. (1995) Annu. Rev. Biochem. 64, 345–373 [DOI] [PubMed] [Google Scholar]
- 70. Zaidi S. K., Young D. W., Javed A., Pratap J., Montecino M., van Wijnen A., Lian J. B., Stein J. L., Stein G. S. (2007) Nat. Rev. Cancer 7, 454–463 [DOI] [PubMed] [Google Scholar]
- 71. Roegiers F., Jan Y. N. (2004) Curr. Opin. Cell Biol. 16, 195–205 [DOI] [PubMed] [Google Scholar]
- 72. Morrison S. J., Kimble J. (2006) Nature 441, 1068–1074 [DOI] [PubMed] [Google Scholar]
- 73. Thorpe P. H., Bruno J., Rothstein R. (2008) Cold Spring Harbor Symp. Quant. Biol. 73, 81–88 [DOI] [PubMed] [Google Scholar]
- 74. Januschke J., Gonzalez C. (2008) Oncogene 27, 6994–7002 [DOI] [PubMed] [Google Scholar]
- 75. Cowan C. R., Hyman A. A. (2004) Annu. Rev. Cell Dev. Biol. 20, 427–453 [DOI] [PubMed] [Google Scholar]
- 76. Colman-Lerner A., Chin T. E., Brent R. (2001) Cell 107, 739–750 [DOI] [PubMed] [Google Scholar]
- 77. Knoblich J. A., Jan L. Y., Jan Y. N. (1995) Nature 377, 624–627 [DOI] [PubMed] [Google Scholar]
- 78. Rhyu M. S., Jan L. Y., Jan Y. N. (1994) Cell 76, 477–491 [DOI] [PubMed] [Google Scholar]
- 79. Hirata J., Nakagoshi H., Nabeshima Y., Matsuzaki F. (1995) Nature 377, 627–630 [DOI] [PubMed] [Google Scholar]
- 80. Esteller M. (2008) N. Engl. J. Med. 358, 1148–1159 [DOI] [PubMed] [Google Scholar]
- 81. Das P. M., Singal R. (2004) J. Clin. Oncol. 22, 4632–4642 [DOI] [PubMed] [Google Scholar]
- 82. Herranz M., Esteller M. (2006) Clin. Transl. Oncol. 8, 242–249 [DOI] [PubMed] [Google Scholar]
- 83. Ting A. H., McGarvey K. M., Baylin S. B. (2006) Genes Dev. 20, 3215–3231 [DOI] [PubMed] [Google Scholar]
- 84. Speck N. A., Gilliland D. G. (2002) Nat. Rev. Cancer 2, 502–513 [DOI] [PubMed] [Google Scholar]
- 85. Erickson P., Gao J., Chang K. S., Look T., Whisenant E., Raimondi S., Lasher R., Trujillo J., Rowley J., Drabkin H. (1992) Blood 80, 1825–1831 [PubMed] [Google Scholar]
- 86. Nucifora G., Birn D. J., Erickson P., Gao J., LeBeau M. M., Drabkin H. A., Rowley J. D. (1993) Blood 81, 883–888 [PubMed] [Google Scholar]
- 87. Meyers S., Downing J. R., Hiebert S. W. (1993) Mol. Cell. Biol. 13, 6336–6345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Peterson L. F., Zhang D. E. (2004) Oncogene 23, 4255–4262 [DOI] [PubMed] [Google Scholar]
- 89. Zeng C., van Wijnen A. J., Stein J. L., Meyers S., Sun W., Shopland L., Lawrence J. B., Penman S., Lian J. B., Stein G. S., Hiebert S. W. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 6746–6751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. McNeil S., Zeng C., Harrington K. S., Hiebert S., Lian J. B., Stein J. L., van Wijnen A. J., Stein G. S. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 14882–14887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Bakshi R., Zaidi S. K., Pande S., Hassan M. Q., Young D. W., Montecino M., Lian J. B., van Wijnen A. J., Stein J. L., Stein G. S. (2008) J. Cell Sci. 21, 3981–3990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Bruserud Ø., Stapnes C., Ersvaer E., Gjertsen B. T., Ryningen A. (2007) Curr. Pharm. Biotechnol. 8, 388–400 [DOI] [PubMed] [Google Scholar]