Abstract
Changes in chromatin architecture induced by epigenetic mechanisms are essential for normal cellular processes such as gene expression, DNA repair, and cellular division. Compact chromatin presents a barrier to these processes and is highly regulated by epigenetic markers binding to components of the nucleosome. Histone modifications directly influence chromatin dynamics and facilitate recruitment of additional factors such as chromatin remodelers and histone chaperones. One member of this last class of factors, FACT (facilitates chromatin transcription), is categorized as a histone chaperone critical for nucleosome reorganization during replication, transcription, and DNA repair. Significant discoveries regarding the role of histone chaperones and specifically FACT have come over the past dozen years from a number of independent laboratories. Here, we review the structural and biophysical basis for FACT-mediated nucleosome reorganization and discuss up-to-date models for FACT function.
Keywords: Chromatin Histone Modification, Chromatin Regulation, Chromatin Remodeling, Chromatin Structure, Nucleosome, Histone Chaperone
Introduction
Chromatin is a densely packed and tightly regulated nucleoprotein complex that stores the genetic material of a cell in a stable yet readily accessible form. In eukaryotic cells, the repeating core subunit of chromatin is the nucleosome, which is composed of 147 bp of DNA wound around a histone octamer in nearly two superhelical turns (1). Nucleosome assembly follows a stepwise mechanism in which the histone (H3-H4)2 tetramer must bind DNA in position before the two histone H2A-H2B dimers can coordinate with the bound tetramer. Condensation into polynucleosomal arrays is aided by the peripheral linker histone H1 and other nucleosome-associated factors, including histone chaperones (2–4).
Histone chaperones are a diverse family of histone-binding proteins that shield non-nucleosomal histone-DNA interactions. Histone chaperones sequester core histones from DNA until a more energetically favorable nucleosomal arrangement becomes available (5). Histone chaperones, along with ATP-dependent chromatin remodelers, work to deconvolute high order chromatin architecture and reorganize individual nucleosomes to provide accessible DNA templates for cellular machinery. Not surprisingly, histone chaperones must also ensure timely reassembly of chromatin following DNA manipulation. One of these histone chaperones, FACT, is purported to reorganize nucleosomes through destabilization of dimer-tetramer contacts and possibly evicting one H2A-H2B dimer to allow passage of the transcribing RNA polymerase through a nucleosomal template (6, 7).
FACT Function and Architecture
The human (h)2 FACT complex was first identified in 1998 as a factor essential for transcriptional elongation through chromatin (6). Further characterization revealed that hFACT is composed of two subunits (hSpt16 and SSRP1) that are essential for functionality. hFACT has been shown to form stable complexes with the histone H2A-H2B dimer and function through reorganization of nucleosomes within the ORFs of actively transcribed genes (8). Concurrent research on yeast (y) FACT suggests that the yFACT complex has roles in DNA replication in addition to transcriptional regulation (9, 10). As for DNA repair, FACT has been linked to activation of the tumor suppressor protein p53 and to histone variant (H2AX-H2B) exchange in response to induced DNA damage (11, 12). Whether in transcription, replication, or repair, FACT functions by reorganizing nucleosomes through the disruption of core histone-histone and histone-DNA interactions. Additionally, the FACT heterodimer possesses the ability to deposit the H2A-H2B dimer and (H3-H4)2 tetramer onto DNA (13). It is clear that eukaryotic FACT heterocomplexes contribute greatly to chromatin dynamics during critical cellular processes and possess histone chaperone activity.
Two polypeptides first identified as p140 and p80 compose the human form of the heterodimeric FACT complex (14). p140 is better known as Spt16 (suppressor of Ty 16) and is a human homolog of the ySpt16 nuclear protein, which is an essential factor for normal transcription throughout the yeast genome (14). hSpt16 also shares a genetic relationship with a family of transcriptional elongation factors that include Spt4 and transcription factor IIS (14). p80 is identified as a human form of SSRP1 (structure-specific recognition protein 1), which is a homolog of the yPob3 (Pol1-binding protein 3) nuclear protein that has been shown to form heterocomplexes with ySpt16 in vivo (15). In addition, SSRP1 contains an HMG-1 DNA-binding domain implicated in specific binding to nucleosomal DNA (14). Nucleosome reorganization and transcriptional elongation through reconstituted chromatin templates require hSpt16 and SSRP1 bound in a heterodimeric complex (14). In contrast, the functional form of yFACT requires ySpt16, Pob3, and an additional protein subunit (Nhp6) (16–18).
Spt16
All eukaryotic forms of the Spt16 protein are composed of three distinct structurally defined domains in combination with a negatively charged intrinsically disordered C-terminal domain termed here as the N-terminal domain (NTD), the dimerization domain (DD), the middle domain (MD), and C-terminal domain (CTD) (Fig. 1) (11, 19, 20). The Spt16 NTD is highly conserved across all species but has been shown to be nonessential for yeast viability or nucleosome binding. Genetic studies in yeast have revealed a partially redundant role with the MD of the Pob3 protein (21–23). Crystal structures of the ySpt16 NTD coming from two yeast species have been published, and both display an aminopeptidase-like “pita bread” fold (Protein Data Bank codes 3BIP and 3CB5) (Fig. 2, upper panel) (21, 22). Conserved residues on the surface of the Spt16 NTD have been implicated in binding to H3 and H4 N-terminal tails, and additional biophysical experiments have demonstrated a stable physical interaction with the (H3-H4)2 tetramer but not with the H2A-H2B dimer (21). Interestingly, further genetic analysis has provided evidence for a functional relationship between the ySpt16 NTD and the C-terminal “docking domain” extension of H2A, whereas concurrent mutations within both domains cause lethality in yeast (22). Although partially redundant, the Spt16 NTD plays an important role in FACT-mediated nucleosome reorganization, presumably through the destabilization of crucial dimer-tetramer interactions.
FIGURE 1.
Domain architecture and structural alignment of the h/yFACT complexes. The hFACT heterodimeric complex is composed of the hSpt16 (cyan) and SSRP1 (dark green) subunits. Dimerization is accomplished through specific interactions between adjacent DDs in Spt16 and SSRP1/Pob3. The yPob3 (light green) protein is analogous to the SSRP1 protein and combines with ySpt16 and the HMG domain-containing protein Nhp6 to form the yFACT complex. Structural alignments of the h/ySpt16 subunits and SSRP1/Pob3-Nhp6 subunits are based upon previous limited proteolysis experiments and functional analysis of truncated FACT constructs (11, 20, 22, 23).
FIGURE 2.
Structural catalog of the individual domains in FACT with respect to their location in the overall architecture of the complex. Upper panel, the Spt16 subunit of FACT is shown in blue with the NTD displayed as a ribbon diagram from the crystal structure of the ySpt16 NTD (22). The structure reveals an aminopeptidase-like peptide-binding fold suitable for interaction with histone tails. The structures of the remaining domains within h/ySpt16 are presently unknown. Lower panel, the known structures of the Pob3 subunit of FACT are displayed as respective domains in the Pob3 molecule. The NTD/DD and MD present similar PH ligand-binding domains (NTD/DD, single; and MD, double) (Protein Data Bank code 3F5R) (19). The structure of Nhp6a in complex with DNA is also shown (green) and is aligned with the HMG domain from SSRP1 (orange) (29, 30). Structures for remaining SSRP1 domains analogous to those shown for Pob3 are unavailable.
Heterodimerization of FACT complexes is coordinated through the DD of Spt16 and the NTD/DD of SSRP1/Pob3 (Fig. 1) (11). Metazoan and yeast forms of the Spt16 DD are predicted to be partially unfolded and may be stabilized by the neighboring MD, which shares some sequence homology with the MD of SSRP1/Pob3 (29% over a region of 50 residues). The overall fold and structure of both domains remain unknown; however, the sequence homology between the Spt16 MD and the SSRP1/Pob3 MD (structure discussed below) suggests that their folds may be similar.
The CTD of Spt16 is a highly conserved region of the molecule and is similar to regions found in other histone chaperones that are thought to directly interact with the basic histone complexes that form the nucleosome core (Fig. 1) (24). Reported deletion of the CTD in hSpt16 precludes any FACT-nucleosome interaction, prevents transcription through chromatin templates in vitro, and inhibits histone chaperone function (13). The CTD in ySpt16 is especially acidic, with 37 of the 75 residues being a negatively charged aspartic or glutamic acid. In higher eukaryotes such as human and Drosophila, 41 residues are negatively charged over an aligned 75-residue span. However, a C-terminal extension neighboring the acidic CTD contains a large proportion of positively charged arginine and lysine residues. This positively charged extension may provide an extra level of regulation in higher eukaryotes that does not exist in yeast.
SSRP1/Pob3-Nhp6
The hSSRP1 protein contains three well defined domains designated the NTD/DD, the MD, and the HMG-1 domain (Fig. 1) (19, 20). Additionally, two highly charged intrinsically disordered regions surround the HMG-1 domain. As mentioned previously, the SSRP1 homolog consists of two separate proteins: Pob3 and the HMG domain-containing protein Nhp6 (non-histone protein 6) (17). The Pob3 protein includes two structural domains similar to the SSRP1 NTD/DD and MD with a C-terminal intrinsically disordered region (19). The NTDs of SSRP1 and Pob3 have both been implicated as dimerization interfaces with Spt16 through truncation mapping (11, 23). The crystal structure of the first 111 residues of the Pob3 NTD/DD (220 residues total) displays a single pleckstrin homology (PH) domain (Protein Data Bank code 3F5R) (Fig. 2, lower panel). It is unclear whether the remaining 109 residues comprise a second PH domain or another type of fold, but limited proteolysis experiments reveal that this region is structured (19). Alignment of the Pob3 NTD/DD structure with a single PH domain from the pleckstrin crystal structure (Protein Data Bank code 1PLS) gives a root mean square deviation (r.m.s.d.) of 3.27 Å, with the seven-stranded β-barrel and capping helix aligning quite well. In the structure of the Pob3 NTD/DD, an extended loop connects β-strands 3 and 4, and this loop is not present in the pleckstrin structure or sequence. PH domains are characterized by a wide array of ligand binding, including lipids, small peptides, and proteins (19, 26). In the context of FACT, the PH domain of the Pob3 NTD/DD functions as one-half of the Spt16-Pob3 (and possibly SSRP1) dimerization interface.
The MD of Pob3 has also been well characterized structurally, with the crystal structure of this region displaying a double PH domain spanning residues 220–447 of the Pob3 protein (Protein Data Bank code 2GCJ) (Fig. 2, lower panel) (19). The PH domains of the Pob3 MD are both structurally homologous to the single PH domain of the Pob3 NTD/DD, with the second PH domain providing a better alignment (PH2 r.m.s.d. of 1.31 Å compared with PH1 r.m.s.d. of 2.87 Å). Genetic screens have revealed that a highly conserved patch of surface residues, including glutamine 308, may have a role in transcription (Spt− phenotype, abnormal transcriptional initiation site selection) and DNA replication (hydroxyurea sensitivity, a decrease in dNTP production and DNA synthesis) (19). Structural analysis of the Q308K mutant form of the Pob3 MD (Protein Data Bank code 2GCL) revealed a strikingly similar structure to the WT Pob3 MD; thus, the mutant phenotypes are most likely not due to dramatic structural changes within the molecule (19). However, incorporation of a basic residue at this site may disrupt direct interaction with chromatin-related substrates necessary for transcription and replication processes.
In higher eukaryotes, the SSRP1 protein contains a C-terminal HMG-1 domain, yet in the yeast version of FACT, the HMG domain is provided separately in the form of the small HMG box protein Nhp6a/b (Fig. 1). HMG domains can readily bind to nucleosomal DNA and may help FACT recognize, bind, and effectively reorganize chromatin (27, 28). NMR solution structures of both the HMG-1 domain of SSRP1 (Drosophila melanogaster) (Protein Data Bank code 1WXL) (29) and the yNhp6a protein (code 1LWM) are available (Fig. 2, lower panel) (30). An additional Nhp6a structure in complex with SRY DNA, which contains a recognition site for HMG box proteins (30), has also been solved using solution NMR (Protein Data Bank code 1J5N) (Fig. 2, lower panel) (30). The Nhp6a-DNA complex structure reveals a characteristic L-shaped HMG fold interacting with the minor groove in the DNA, whereas an extended N-terminal region interacts with the adjacent major groove. A dense network of protein-DNA interactions stabilizes the binding, which includes numerous highly conserved lysine and arginine residues participating in charged interactions with the negatively charged phosphate backbone along with hydrophobic stacking/wedge interactions between select Nhp6a residues and DNA bases (a more detailed characterization is provided in Ref. 30). The structure of the SSRP1 HMG-1 domain is quite similar to the Nhp6a structure, and the proteins align with an r.m.s.d. of 2.35 Å. Electrostatic potential maps show similar charge distributions for the SSRP1 HMG-1 domain, Nhp6a, and other HMG domains of known structure (29). Comparisons between DNA-bound Nhp6a, unbound versions of Nhp6a, and the SSRP1 HMG-1 domain suggest that only small structural changes occur in the protein upon DNA binding; however, analysis of the DNA before and after binding shows that the DNA is considerably underwound, the minor groove is widened, and the DNA displays a significant overall bend when in complex with Nhp6a (30). The presence of HMG domains in all forms of FACT, from yeast to humans, shows that DNA binding is critical to FACT recruitment and nucleosome reorganization.
“Modifying” FACT Function
Histone Modifications
Epigenetic mechanisms share an intimate relationship with factors involved in promoting chromatin dynamics and nucleosome exchange. Modification(s) of nucleosome core histones can have consequences for chromatin architecture (see a comprehensive review in Ref. 31). Many of these epigenetic modifications impart their effect through direct recruitment or blockage of machinery capable of altering chromatin, such as FACT, CHD1, Asf1, Spt6, SWI/SNF, and ACF among many others (31). For example, the ATP-dependent chromatin-remodeling factor CHD1 (chromatin organization modifier, helicase, and DNA-binding domains 1) localizes throughout the coding region of actively transcribing genes through recognition of a trimethylation mark on Lys-4 of histone H3 (H3K4me3) (32–34). CHD1 has also been demonstrated to physically interact with the FACT complex and may work to position FACT on nucleosomes within ORFs of active genes (35, 36). Another epigenetic marker, H2BK120ub1 (histone H2B monoubiquitination at lysine 120), is associated with transcriptional elongation and is dependent upon the PAF (RNA polymerase-associating factor) complex (37–39). Yeast genetic analysis has revealed that the PAF complex functionally interacts with FACT, and subsequent in vitro experiments confirmed a robust physical interaction (40, 41). Furthermore, the PAF complex and H2B monoubiquitination enhance FACT-mediated transcription in a reconstituted chromatin system in vitro (40). Interestingly, H2A monoubiquitination (H2AK119ub1) by 2A-HUB (histone H2A-homologous to ubiquitin) represses transcriptional initiation through the apparent blockage of FACT binding to the GAL4 promoter (42). FACT has also been implicated in DNA repair processes and the exchange of histone variants. The histone H2A variant H2AX is phosphorylated at Ser-139 (H2AXS139phos) by DNA-dependent protein kinase in response to DNA damage (43–45). Accordingly, FACT-mediated H2AX exchange is facilitated by phosphorylation of H2AX. It is hypothesized that changes to the overall nucleosome structure as a result of H2AX phosphorylation allow more efficient exchange by FACT (12). A diverse set of epigenetic mechanisms influence FACT function through disruption of nucleosome structure and stability. This list is certain to expand as research continues to uncover additional histone modifications and their effects on chromatin.
Modifications to FACT
In addition to the extensive collection of histone modifications that influence FACT activity, other factors physically alter the FACT complex as a response to specific cellular conditions. Direct modification of FACT may provide an intrinsic regulation method for nucleosome reorganization. One example is the poly(ADP-ribosyl)ation of the Spt16 subunit of FACT by PARP1 (poly(ADP-ribose) polymerase 1) in response to genotoxic stress in vivo and the inability of poly(ADP-ribosyl)ated FACT to bind nucleosomes in vitro (46). Furthermore, PARP1-mediated poly(ADP-ribosyl)ation of FACT has been shown to inhibit H2AX-H2B exchange during DNA damage repair (12). Modifications to the SSRP1 subunit of FACT have also been demonstrated to alter FACT activity levels. Direct phosphorylation of SSRP1 can occur through interaction with CK2 (casein kinase 2) (11, 47), and this modification inhibits nucleosomal DNA binding (20). Phosphorylation of SSRP1 may act to create a storage pool of inactive FACT that can be rapidly activated to facilitate changes in vivo. Physical modification of the Spt16 and SSRP1 subunits efficiently provides an additional level of control over FACT-induced nucleosome reorganization and chromatin dynamics.
FACT Functional Models
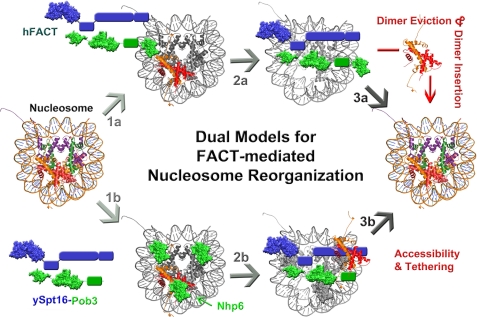
Many questions remain concerning FACT function and the mechanisms by which FACT performs its many functions in vivo. For instance, how is the FACT heterocomplex recruited to specific sites in chromatin, and by what means is FACT turned “on” and “off?” Here, we focus on the proposed mechanistic models for this histone chaperone and on direct nucleosome reorganization functions of FACT. Two major models exist in the literature for FACT-mediated nucleosome reorganization. The “dimer eviction model” proposes that the FACT complex utilizes its histone chaperone function to actively displace a single H2A-H2B dimer from a nucleosome to promote DNA accessibility (13, 14, 48). The “global accessibility/non-eviction model” suggests that FACT-induced H2A-H2B dimer displacement is a nonessential byproduct of FACT action and not essential for nucleosome reorganization (7, 49). This model depicts FACT loosening internal contacts within the nucleosome and in effect changing its dynamic nature to allow sufficient access to the DNA template.
Dimer Eviction Model
The dimer eviction model describes the mechanistic details of FACT function based heavily upon the proposed histone chaperone utility of FACT. This model proceeds through three basic steps (Fig. 3): step 1a, FACT binding nucleosomes at a near 1:1 stoichiometry through the acidic Spt16 CTD; step 2a, FACT-mediated histone H2A-H2B dimer displacement; and step 3a, reinsertion of the H2A-H2B dimer after cellular factors such as RNA polymerase II have performed their functions. The FACT heterocomplex has been shown to readily bind nucleosomes, and in vitro chromatin transcription assays revealed that maximal FACT activity occurs when the FACT/nucleosome ratio is near 1:1 (14, 16, 20). The H2A-H2B dimer eviction step in this process (step 2a) is supported by evidence that FACT binds not only nucleosomes but also the H2A-H2B dimer in vitro (14). FACT-mediated hexasome formation occurs as a result of transcriptional elongation through chromatin templates (13). In accordance, FACT activity is obstructed by covalent cross-linking of the nucleosome core histones (14). Supporting research also shows that H2A-H2B dimer dissociation from nucleosomes is intimately associated with transcriptional initiation (50), transcriptional elongation (51, 52), replication (53), and DNA damage repair (12). Finally, FACT may assist in reassembly of the nucleosome based upon the histone chaperone activity of FACT (13), and solid evidence showing that disruption of FACT activity inhibits nucleosome reformation after passage of the RNA polymerase (54, 55) supports this notion. In all, the dimer eviction model for FACT-mediated nucleosome disassembly and reconstruction combines numerous lines of evidence that strongly suggests that H2A-H2B dimer dissociation occurs concomitantly with critical stages in DNA metabolic processes (i.e. transcription, replication, and repair) and that FACT activity is critical to nucleosome stabilization subsequent to these actions.
FIGURE 3.
Dual models for FACT-mediated nucleosome reorganization. The dimer eviction model proceeds through the schematic in steps 1a, 2a, and 3a. In step 1a, the FACT complex binds to the nucleosome, which is thought to be dependent upon the acidic CTD of Spt16 (13) and the N-terminal tails of the core histones (7). Next (step 2a), the FACT complex binds and evicts a single H2A-H2B dimer, creating a hexasome structure. The last step (step 3a) reinserts the evicted H2A-H2B dimer (after the cellular machinery has performed the necessary function, i.e. transcription, replication, or DNA repair) to restore a complete nucleosome. The global accessibility/non-eviction model progresses through steps 1b, 2b, and 3b. This model is also shown using yFACT, which utilizes numerous copies of Nhp6 to recognize and bind nucleosomes in the first step (step 1b) (18). In step 2b, the ySpt16-Pob3 complex is then recruited to the Nhp6-bound nucleosome, and binding induces gross conformational changes and accessibility within the nucleosome but without H2A-H2B dimer eviction (7). The H2A-H2B dimer remains tethered to the nucleosome via FACT, which facilitates reinsertion after relevant cellular processes have concluded (step 3b). Although the latter model utilizes yFACT, the overall mechanism driving global accessibility without dimer eviction could also be applicable to metazoan forms of FACT.
Global Accessibility/Non-eviction Model
Is H2A-H2B dimer eviction the critical activity for or an ancillary consequence of FACT-mediated nucleosome reorganization? The global accessibility/non-eviction model endorses the latter notion based primarily on the finding that yFACT can promote hydroxyl radical and nuclease accessibility without H2A-H2B dimer displacement in vitro while facilitating transcriptional activation at the GAL1–10 promoter without significant dimer loss in vivo (7). This fairly recent model, which was first proposed in 2008 (49), suggests the progression through three discrete steps depicted in Fig. 3: step 1b, Nhp6 binding to the nucleosome triggers small changes and recruits ySpt16-Pob3; step 2b, yFACT binding induces larger reorganization events by tethering nucleosome components; and step 3b, a wholly intact nucleosome is restored (Fig. 3) (7). Several key studies have not only linked Nhp6 to comprehensive yFACT function (10, 16, 17) but have also shown that multiple copies of Nhp6 are required for yFACT recruitment to nucleosomes (18). The HMG box domain of Nhp6 permits nucleosome binding in the absence of the ySpt16-Pob3 complex. In addition, complete yFACT (ySpt16-Pob3 + Nhp6) binding to nucleosomes produces large-scale transformations that allow global accessibility of the nucleosome but do not correspond with H2A-H2B dimer displacement (7, 56). Although it has been shown that yFACT can cause H2A-H2B dimer displacement in vitro, the amount of H2A-H2B dimer loss does not correlate with the level of nucleosome accessibility (7). In addition, transcription activation at the GAL1–10 promoter occurs with minimal loss of the H2A-H2B dimer or (H3-H4)2 tetramer (7). These results support the notion that the nucleosomal elements are continuously tethered between FACT and a non-canonical form of the nucleosome during times of DNA accessibility. Finally, FACT must be able to reverse action and reestablish a canonical nucleosome after DNA-linked processes have occurred. A tightly regulated equilibrium between the accessible and non-accessible forms of the nucleosome fits well with the idea that additional chromatin-modulating factors may bind to the accessible form to further promote cellular processes and are released upon or inhibited by the non-accessible form (57). It is also interesting to speculate how epigenetic markers (i.e. histone modifications) might influence the “equilibrium” proposed in this model given that these marks are known to recruit/inhibit chromatin-modulating factors to chromatin (31).
Conclusions and Perspectives
Exclusive or Combinatorial Model(s)
Are the potential mechanisms outlined above mutually exclusive, or can aspects of both models be correct under certain circumstances? Another view could be that yeast and metazoan forms of FACT may utilize related yet different mechanisms. Some aspects of the two models, which pertain to the h/yFACT architecture, are undoubtedly different, such as the role Nhp6 plays in yFACT recruitment to nucleosomes. Binding of multiple Nhp6 subunits to the nucleosome is required to bring in the Spt16-Pob3 complex and form the complete yFACT complex (18). This first critical step in nucleosome reorganization is different for higher eukaryotes that do not possess a separate HMG domain-containing protein. For these versions of FACT, the conserved CTD of Spt16 is essential for nucleosome binding and overall FACT function (13). The HMG-1 domain contained within the SSRP1 molecule still plays an important role in FACT-nucleosome binding as shown through reduced binding of nucleosomes by Spt16 alone when compared with the FACT heterodimer (13). The idea that FACT from different species may operate through opposing mechanisms is also supported by recent evidence suggesting that the source of histone subunits within the nucleosomes directly affects stability and dynamics (7).
Although there are discrepancies between the nucleosome-binding mode of yeast and metazoan FACT, both forms can stably bind nucleosomes and function to permit cellular access to chromatin templates. Thus, the main disparity between the two previously discussed models stems from H2A-H2B dimer eviction and its role as an essential step or a by-product of FACT function. Exchange of the histone variant H2AX is tightly regulated via phosphorylation and FACT (12). This implies that the histone chaperone activity of FACT is responsible for physical exchange of histones within select nucleosomes during DNA damage repair. Could FACT exert its histone chaperone activity to facilitate histone variant exchange but function differently during transcriptional elongation, DNA replication, and/or stress conditions? Conditional functionality in FACT has been shown to occur during replication stress through the dependence upon normally nonessential domains in the Spt16 subunit (23). Moreover, the effects of histone modifications such as the phosphorylation of H2AX regulating dimer exchange by FACT (12) may contribute to possible varying functionality of the FACT complex. Additional research will be required to evaluate whether or not FACT can exert varying degrees of nucleosome reorganization to suit specific processes in vivo.
Future Directions
The present mechanistic ambiguity surrounding FACT-mediated chromatin dynamics offers a wide array of opportunities for future research. For instance, a quantitative analysis of the numerous purported FACT-nucleosome interactions would help to clarify mechanistic details of FACT-dependent nucleosome reorganization. A better understanding of the roles performed by the individual FACT domains could determine whether the domains work cooperatively or independently to achieve nucleosome reorganization and/or H2A-H2B dimer eviction. In addition, do components of the nucleosome and other chromatin-associated elements compete for interaction with FACT, and if so, do these molecules readily exchange depending upon cellular conditions? Analogous studies with the histone chaperone Nap1 have clearly demonstrated that Nap1 works to control non-nucleosomal histone-DNA interactions until conditions for nucleosome formation are favorable (58). FACT may function in a similar matter based upon recent reports that FACT prevents the accumulation of free histones evicted from chromatin and that seizure of these histones prevents a cell cycle delay in G1 (25). Finally, the holy grail in the biophysical characterization of FACT-mediated nucleosome reorganization would be a crystal or solution structure of a FACT-nucleosome complex. High resolution structural details of the FACT-nucleosome interaction have the potential to dramatically increase the understanding of FACT function and could help design future studies aimed at further mechanistic characterization.
This work was supported, in whole or in part, by National Institutes of Health Grant 1P01GM088409-01A1. This work was also supported by the Howard Hughes Medical Institute (to D. D. W. and K. L.). This is the fourth article in the Thematic Minireview Series on Epigenetics. This minireview will be reprinted in the 2011 Minireview Compendium, which will be available in January, 2012.
- h
- human
- y
- yeast
- NTD
- N-terminal domain
- DD
- dimerization domain
- MD
- middle domain
- CTD
- C-terminal domain
- PH
- pleckstrin homology
- r.m.s.d.
- root mean square deviation.
REFERENCES
- 1. Luger K., Mäder A. W., Richmond R. K., Sargent D. F., Richmond T. J. (1997) Nature 389, 251–260 [DOI] [PubMed] [Google Scholar]
- 2. Tremethick D. J. (2007) Cell 128, 651–654 [DOI] [PubMed] [Google Scholar]
- 3. Woodcock C. L., Ghosh R. P. (2010) Cold Spring Harbor Perspect. Biol. 2:a000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. McBryant S. J., Lu X., Hansen J. C. (2010) Cell Res. 20, 519–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ransom M., Dennehey B. K., Tyler J. K. (2010) Cell 140, 183–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Orphanides G., LeRoy G., Chang C. H., Luse D. S., Reinberg D. (1998) Cell 92, 105–116 [DOI] [PubMed] [Google Scholar]
- 7. Xin H., Takahata S., Blanksma M., McCullough L., Stillman D. J., Formosa T. (2009) Mol. Cell 35, 365–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Orphanides G., Reinberg D. (2000) Nature 407, 471–475 [DOI] [PubMed] [Google Scholar]
- 9. Wittmeyer J., Formosa T. (1997) Mol. Cell. Biol. 17, 4178–4190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schlesinger M. B., Formosa T. (2000) Genetics 155, 1593–1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Keller D. M., Lu H. (2002) J. Biol. Chem. 277, 50206–50213 [DOI] [PubMed] [Google Scholar]
- 12. Heo K., Kim H., Choi S. H., Choi J., Kim K., Gu J., Lieber M. R., Yang A. S., An W. (2008) Mol. Cell 30, 86–97 [DOI] [PubMed] [Google Scholar]
- 13. Belotserkovskaya R., Oh S., Bondarenko V. A., Orphanides G., Studitsky V. M., Reinberg D. (2003) Science 301, 1090–1093 [DOI] [PubMed] [Google Scholar]
- 14. Orphanides G., Wu W. H., Lane W. S., Hampsey M., Reinberg D. (1999) Nature 400, 284–288 [DOI] [PubMed] [Google Scholar]
- 15. Brewster N. K., Johnston G. C., Singer R. A. (1998) J. Biol. Chem. 273, 21972–21979 [DOI] [PubMed] [Google Scholar]
- 16. Formosa T., Eriksson P., Wittmeyer J., Ginn J., Yu Y., Stillman D. J. (2001) EMBO J. 20, 3506–3517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Brewster N. K., Johnston G. C., Singer R. A. (2001) Mol. Cell. Biol. 21, 3491–3502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ruone S., Rhoades A. R., Formosa T. (2003) J. Biol. Chem. 278, 45288–45295 [DOI] [PubMed] [Google Scholar]
- 19. VanDemark A. P., Blanksma M., Ferris E., Heroux A., Hill C. P., Formosa T. (2006) Mol. Cell 22, 363–374 [DOI] [PubMed] [Google Scholar]
- 20. Tsunaka Y., Toga J., Yamaguchi H., Tate S., Hirose S., Morikawa K. (2009) J. Biol. Chem. 284, 24610–24621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Stuwe T., Hothorn M., Lejeune E., Rybin V., Bortfeld M., Scheffzek K., Ladurner A. G. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 8884–8889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. VanDemark A. P., Xin H., McCullough L., Rawlins R., Bentley S., Heroux A., Stillman D. J., Hill C. P., Formosa T. (2008) J. Biol. Chem. 283, 5058–5068 [DOI] [PubMed] [Google Scholar]
- 23. O'Donnell A. F., Brewster N. K., Kurniawan J., Minard L. V., Johnston G. C., Singer R. A. (2004) Nucleic Acids Res. 32, 5894–5906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Philpott A., Krude T., Laskey R. A. (2000) Semin. Cell Dev. Biol. 11, 7–14 [DOI] [PubMed] [Google Scholar]
- 25. Morillo-Huesca M., Maya D., Muñoz-Centeno M. C., Singh R. K., Oreal V., Reddy G. U., Liang D., Géli V., Gunjan A., Chávez S. (2010) PLoS Genet. 6, e1000964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yu J. W., Mendrola J. M., Audhya A., Singh S., Keleti D., DeWald D. B., Murray D., Emr S. D., Lemmon M. A. (2004) Mol. Cell 13, 677–688 [DOI] [PubMed] [Google Scholar]
- 27. Moreira J. M., Holmberg S. (2000) EMBO J. 19, 6804–6813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ner S. S., Blank T., Pérez-Paralle M. L., Grigliatti T. A., Becker P. B., Travers A. A. (2001) J. Biol. Chem. 276, 37569–37576 [DOI] [PubMed] [Google Scholar]
- 29. Kasai N., Tsunaka Y., Ohki I., Hirose S., Morikawa K., Tate S. (2005) J. Biomol. NMR 32, 83–88 [DOI] [PubMed] [Google Scholar]
- 30. Masse J. E., Wong B., Yen Y. M., Allain F. H., Johnson R. C., Feigon J. (2002) J. Mol. Biol. 323, 263–284 [DOI] [PubMed] [Google Scholar]
- 31. Campos E. I., Reinberg D. (2009) Annu. Rev. Genet. 43, 559–599 [DOI] [PubMed] [Google Scholar]
- 32. Simic R., Lindstrom D. L., Tran H. G., Roinick K. L., Costa P. J., Johnson A. D., Hartzog G. A., Arndt K. M. (2003) EMBO J. 22, 1846–1856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sims R. J., 3rd, Chen C. F., Santos-Rosa H., Kouzarides T., Patel S. S., Reinberg D. (2005) J. Biol. Chem. 280, 41789–41792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Flanagan J. F., Mi L. Z., Chruszcz M., Cymborowski M., Clines K. L., Kim Y., Minor W., Rastinejad F., Khorasanizadeh S. (2005) Nature 438, 1181–1185 [DOI] [PubMed] [Google Scholar]
- 35. Krogan N. J., Kim M., Ahn S. H., Zhong G., Kobor M. S., Cagney G., Emili A., Shilatifard A., Buratowski S., Greenblatt J. F. (2002) Mol. Cell. Biol. 22, 6979–6992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kelley D. E., Stokes D. G., Perry R. P. (1999) Chromosoma 108, 10–25 [DOI] [PubMed] [Google Scholar]
- 37. Shi X., Finkelstein A., Wolf A. J., Wade P. A., Burton Z. F., Jaehning J. A. (1996) Mol. Cell. Biol. 16, 669–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rondón A. G., Gallardo M., García-Rubio M., Aguilera A. (2004) EMBO Rep. 5, 47–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Xiao T., Kao C. F., Krogan N. J., Sun Z. W., Greenblatt J. F., Osley M. A., Strahl B. D. (2005) Mol. Cell. Biol. 25, 637–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pavri R., Zhu B., Li G., Trojer P., Mandal S., Shilatifard A., Reinberg D. (2006) Cell 125, 703–717 [DOI] [PubMed] [Google Scholar]
- 41. Squazzo S. L., Costa P. J., Lindstrom D. L., Kumer K. E., Simic R., Jennings J. L., Link A. J., Arndt K. M., Hartzog G. A. (2002) EMBO J. 21, 1764–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhou W., Zhu P., Wang J., Pascual G., Ohgi K. A., Lozach J., Glass C. K., Rosenfeld M. G. (2008) Mol. Cell 29, 69–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Celeste A., Difilippantonio S., Difilippantonio M. J., Fernandez-Capetillo O., Pilch D. R., Sedelnikova O. A., Eckhaus M., Ried T., Bonner W. M., Nussenzweig A. (2003) Cell 114, 371–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Furuta T., Takemura H., Liao Z. Y., Aune G. J., Redon C., Sedelnikova O. A., Pilch D. R., Rogakou E. P., Celeste A., Chen H. T., Nussenzweig A., Aladjem M. I., Bonner W. M., Pommier Y. (2003) J. Biol. Chem. 278, 20303–20312 [DOI] [PubMed] [Google Scholar]
- 45. Rogakou E. P., Boon C., Redon C., Bonner W. M. (1999) J. Cell Biol. 146, 905–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Huang J. Y., Chen W. H., Chang Y. L., Wang H. T., Chuang W. T., Lee S. C. (2006) Nucleic Acids Res. 34, 2398–2407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Keller D. M., Zeng X., Wang Y., Zhang Q. H., Kapoor M., Shu H., Goodman R., Lozano G., Zhao Y., Lu H. (2001) Mol. Cell 7, 283–292 [DOI] [PubMed] [Google Scholar]
- 48. Reinberg D., Sims R. J., 3rd (2006) J. Biol. Chem. 281, 23297–23301 [DOI] [PubMed] [Google Scholar]
- 49. Formosa T. (2008) Mol. Biosyst. 4, 1085–1093 [DOI] [PubMed] [Google Scholar]
- 50. Ransom M., Williams S. K., Dechassa M. L., Das C., Linger J., Adkins M., Liu C., Bartholomew B., Tyler J. K. (2009) J. Biol. Chem. 284, 23461–23471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kireeva M. L., Walter W., Tchernajenko V., Bondarenko V., Kashlev M., Studitsky V. M. (2002) Mol. Cell 9, 541–552 [DOI] [PubMed] [Google Scholar]
- 52. Chang C. H., Luse D. S. (1997) J. Biol. Chem. 272, 23427–23434 [DOI] [PubMed] [Google Scholar]
- 53. Jackson V. (1990) Biochemistry 29, 719–731 [DOI] [PubMed] [Google Scholar]
- 54. Formosa T., Ruone S., Adams M. D., Olsen A. E., Eriksson P., Yu Y., Rhoades A. R., Kaufman P. D., Stillman D. J. (2002) Genetics 162, 1557–1571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Mason P. B., Struhl K. (2003) Mol. Cell. Biol. 23, 8323–8333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Rhoades A. R., Ruone S., Formosa T. (2004) Mol. Cell. Biol. 24, 3907–3917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Biswas D., Yu Y., Prall M., Formosa T., Stillman D. J. (2005) Mol. Cell. Biol. 25, 5812–5822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Andrews A. J., Chen X., Zevin A., Stargell L. A., Luger K. (2010) Mol. Cell 37, 834–842 [DOI] [PMC free article] [PubMed] [Google Scholar]