Abstract
Scavenger receptor class B, type I (SR-BI), a CD36 superfamily member, is an oligomeric high density lipoprotein (HDL) receptor that mediates negatively cooperative HDL binding and selective lipid uptake. We identified in the N-terminal transmembrane (N-TM) domain of SR-BI a conserved glycine dimerization motif, G15X2G18X3AX2G25, of which the submotif G18X3AX2G25 significantly contributes to homodimerization and lipid uptake activity. SR-BI variants were generated by mutations (single or multiple Gly → Leu substitutions) or by replacing the N-TM domain with those from other CD36 superfamily members containing (croquemort) or lacking (lysosomal integral membrane protein (LIMP) II) this glycine motif (chimeras). None of the SR-BI variants exhibited altered surface expression (based on antibody binding) or HDL binding. However, the G15L/G18L/G25L triple mutant exhibited reductions in cell surface homo-oligomerization (>10-fold) and the rate of selective lipid uptake (∼2-fold). Gly18 and Gly25 were necessary for normal lipid uptake activity of SR-BI and the SR-BI/croquemort chimera. The lipid uptake activity of the glycine motif-deficient SR-BI/LIMP II chimera was low but could be increased by introducing glycines at positions 18 and 25. The rate of lipid uptake mediated by SR-BI/LIMP II chimeras was proportional to the extent of receptor oligomerization. Thus, the glycine dimerization motif G18X3AX2G25 in the N-TM domain of SR-BI contributes substantially to the homo-oligomerization and lipid transport activity of SR-BI but does not influence the negative cooperativity of HDL binding. Oligomerization-independent binding cooperativity suggests that classic allostery is not involved and that the negative cooperativity is probably the consequence of a “lattice effect” (interligand steric interference accompanying binding to adjacent receptors).
Keywords: Cooperativity, High Density Lipoprotein (HDL), Lipoprotein, Lipoprotein Receptor, Receptor Structure-Function, CD36, SR-BI, Transmembrane Domain
Introduction
The high density lipoprotein (HDL) receptor SR-BI2 is a cell surface receptor expressed in the liver and other tissues that regulates plasma lipoprotein cholesterol levels and metabolism (Refs. 1 and 3–6; for a review, see Ref. 2). SR-BI mediates cellular uptake of cholesteryl esters and other lipids from HDL and has been proposed to play a key role in moving cholesterol from peripheral tissues to the liver and then out of the body (Refs. 7–10; for a review, see Ref. 2). SR-BI influences HDL metabolism in humans (11, 12) in whom the risk of atherosclerotic disease is inversely or directly proportional to plasma concentrations of HDL cholesterol and low density lipoprotein (LDL) cholesterol, respectively (13–15). Studies in mice have shown that SR-BI protects against atherosclerosis and coronary heart disease (Refs. 16–22; for a review, see Ref. 2).
SR-BI is a member of the CD36 superfamily of proteins in metazoans (∼25–30% sequence identities). Members of this superfamily include lysosomal integral membrane protein (LIMP) II (23) and croquemort (a Drosophila melanogaster hemocyte/macrophage receptor for apoptotic cells) (24). Most CD36 superfamily members share similar membrane topologies with a large, glycosylated extracellular loop that is anchored to the plasma membrane by N- and C-terminal transmembrane domains (N-TM and C-TM, respectively), extensions of each forming short cytoplasmic (cyto) N- and C-terminal domains (25).
SR-BI homodimers or higher order oligomers have been observed in cultured cells and tissues (26–31). Sahoo et al. (29) have concluded from fluorescence resonance energy transfer studies that in SR-BI oligomers the C-cyto, but possibly not the N-cyto, domains are in close proximity and that the C-cyto domain is not required for dimerization. They proposed that the C-TM domain or the juxtaposed portion of the extracellular domain may mediate oligomerization (31). However, there have been no reports of mutations or chemical treatments of SR-BI that disrupt oligomerization, and the sequences/structures of SR-BI responsible for oligomerization have not been identified.
The concentration dependence of HDL binding to SR-BI results in nonlinear, concave up Scatchard plots (32). Thus, SR-BI exhibits either two independent classes of binding sites (two-site model) or one class of sites exhibiting negative cooperativity (one-site plus Hill coefficient model). Dissociation kinetics experiments (33–35) indicate that HDL binding is most parsimoniously described by the model with one class of negatively cooperative binding sites (32). The mechanism underlying negative cooperativity is unclear; however, one possibility is that ligand binding to one monomer in an oligomer induces allosteric changes in neighboring monomers.
The mechanism by which SR-BI delivers the lipids of HDL to cells, called selective lipid uptake (1, 36, 37), is fundamentally different from the classic receptor-mediated endocytosis mediated by LDL receptors (38). After binding HDL, SR-BI selectively transfers the cholesteryl esters of HDL into the cells, and the lipid-depleted HDL particles subsequently dissociate from the cells and re-enter the circulation (7). In addition, SR-BI mediates the bidirectional transfer of unesterified cholesterol between cells and lipoproteins (3, 39, 40).
Strikingly, although SR-BI and CD36 share significant sequence similarities throughout their entire extracellular loops and CD36 can bind HDL tightly, CD36 cannot mediate efficient selective lipid uptake (41, 42). Analysis of the activities of chimeras of SR-BI and CD36 established that either the N-TM domain of SR-BI with the adjacent N-cyto domain (N-TM/N-cyto), its C-TM/C-cyto domains, or both could be replaced with those from CD36 without substantial loss of selective lipid uptake activity. Replacement of these domains in CD36 with those from SR-BI does not confer efficient selective uptake activity. Given that there is little sequence similarity in the N-TM and C-TM domains of SR-BI and CD36 (see “Results” for additional discussion), it appeared that there were no specific sequences in the membrane anchors of SR-BI that were essential for its normal, efficient lipid uptake (41, 42).
The apparent lack of influence on efficient SR-BI-mediated lipid uptake of specific sequences in its TM domains was somewhat surprising. There is an expanding body of evidence showing that transmembrane segments of membrane proteins can direct protein-protein interactions within the membrane (e.g. oligomerization) and influence the functions of these proteins (43–52) and that mutations within TM domains of some of these proteins are involved in diseases (44, 47). Many TM helices contain glycine (Gly) hetero- and homodimerization motifs (see “Results” for a detailed description of such motifs), and these influence the equilibrium between inactive monomers and active dimers (e.g. receptor tyrosine kinases (47, 53)) or active and inactive states (e.g. integrins (51, 52, 54)) and thus receptor activity.
In the current study, we explored the possible role of the N-TM domain of SR-BI in its oligomerization and function. We identified a glycine dimerization motif in the N-TM domain of SR-BI that significantly contributes to the homodimerization of SR-BI. Gly → Leu substitutions in this motif did not appear to alter the surface expression of SR-BI nor the binding of HDL to SR-BI, which involves negative cooperativity. However, these substitutions dramatically reduced receptor dimerization and the rate of selective lipid uptake.
EXPERIMENTAL PROCEDURES
Lipoproteins
Human HDL was isolated as eight separate density fractions using KBr gradient centrifugation from freshly harvested, EDTA-treated plasma as described previously (32, 55, 56) (see supplemental materials for additional details). For the studies reported here, HDL fraction 7 (density, 1.113 g/ml) or an equal volume mixture of either fractions 5 and 6 (mean density, 1.126 g/ml) or fractions 7 and 8 (mean density, 1.107 g/ml) were used for radiolabeling and the binding and uptake studies. The unlabeled HDL used was a mixture of fractions (density range, 1.155–1.102 g/ml). Protein concentrations were determined by the method of Lowry et al. (57) (bovine serum albumin was the standard).
HDL was labeled with 125I (125I-HDL) or with [3H]cholesteryl oleate ([3H]CE; Perkin Elmer Life Sciences catalogue number NET746L) ([3H]CE-HDL) as described previously (32, 58, 59). The specific activities were 229–361 cpm/ng of protein for 125I-HDL and 17–36 cpm/ng of protein for [3H]CE-HDL.
DNA Mutagenesis and Cloning
Site-directed mutagenesis to generate mutants was performed on murine SR-BI cDNA in the pcDNA4/TO vector (Novagen) using the QuikChange II site-directed mutagenesis kit (Stratagene). SR-BI transmembrane chimeras (SR-BI/croquemort (SR-BI/croq) and SR-BI/LIMP II (SR-BI/LIMP)) and rhodopsin (rho)- and myc-tagged variants were constructed using PCR. The rho (TETSQVAPA) and/or myc (EQKLISEEDL) tags were introduced directly at the 3′-ends of wild-type (WT) SR-BI, G15L/G18L/G25L, and SR-BI/LIMP chimera cDNAs (see supplemental Experimental Procedures for details).
Cell Culture and Transient Transfection
All cells were grown and incubated at 37 °C in a humidified 5% CO2, 95% air incubator. COS M6 cells were grown in Dulbecco's modified Eagle's medium with 50 units/ml penicillin, 50 μg/ml streptomycin, and 2 mm glutamine (medium A) supplemented with 10% fetal bovine serum (medium B) and were transiently transfected with expression vectors for wild-type, mutant, or chimeric SR-BI or with the “empty vector” plasmid (pcDNA4/TO) without a cDNA insert. For transient transfections, cells were plated in the wells of 24- or 6-well dishes on day 0, transfected on day 1, and on day 3 were used for co-immunoprecipitation, chemical cross-linking, receptor activity, or flow cytometry assays (see supplemental Experimental Procedures for details). Stably transfected ldlA-7 cells were grown in Ham's F-12 medium containing 50 units/ml penicillin, 50 μg/ml streptomycin, and 2 mm glutamine (medium C) supplemented with 5% fetal bovine serum (medium D) and 250 μg/ml G418 (medium E). On day 0, 5.0 × 104 or 3.0 × 105 cells/well were plated in the wells of 24- or 6-well dishes, respectively, in medium E. On day 2, stably transfected cells were used for chemical cross-linking, receptor activity, or flow cytometry assays. Untransfected control ldlA-7 cells were maintained in medium D.
Determination of Cell Surface Receptor Expression by Flow Cytometry
The levels of cell surface expression of WT and mutant SR-BIs on intact cells were determined by flow cytometry using the SR-BI-specific polyclonal rabbit antibody KKB-1 that recognizes epitopes on the extracellular loop of SR-BI (40) (see supplemental Experimental Procedures for details). Cell surface receptor expression was used to normalize the results for receptor activity assays (125I-HDL binding and [3H]CE-HDL and 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate (DiI)-HDL lipid uptake). To do this, the same populations of cells for the surface expression and activity assays were simultaneously plated, and then surface expression and receptor activity were measured on the same day. The activities of SR-BI mutants were normalized by multiplying by the ratio of wild-type to mutant surface expression determined using the KKB-1 antibody. In all cases, the differences in cell surface expression between the wild-type and mutant receptors were less than 10% of the wild-type surface levels. This normalization, which did not qualitatively influence any of the findings, was based on the assumption that the KKB-1 antibody binds with equal affinity and to the same extent to the wild-type and variant receptors (mutants/chimeras). Several observations suggest that this is likely to be the case (see supplemental Experimental Procedures) and that the normalization was appropriate. If the variants in the N-TM domain did alter KKB-1 binding to the exoplasmic domain of the receptor, then it is possible that these variants might have caused changes in the absolute amounts of HDL binding. The conclusion that there was a significant decrease in lipid uptake relative to HDL binding in variants with defective N-TM glycine dimerization motifs is independent of any potential alterations in KKB-1 binding to the variants.
Generation of Stably Transfected Cell Lines
Stable cell lines expressing wild-type murine SR-BI and the triple glycine (G15L/G18L/G25L) mutant were generated in ldlA-7 cells, which are LDL receptor-deficient CHO cells (60) that express very low levels of endogenous SR-BI (1). We have previously described ldlA[SR-BI] cells that express wild-type murine SR-BI under the control of a cytomegalovirus promoter (1). We periodically used flow cytometry and the KKB-1 antibody to remove from the population cells that spontaneously lose expression of the transgene. A stable cell line expressing the triple glycine mutant (G15L/G18L/G25L), called ldlA[G15L/G18L/G25L], was generated by co-transfection of ldlA-7 cells with an expression vector encoding the triple mutant and the pBK-CMV plasmid (G418 resistance marker, Stratagene; 0.1 μg of DNA) using FuGENE 6 transfection reagent (Roche Applied Science) according to the manufacturer's instructions and selection in medium E as described previously (40). Surface expression of the mutant SR-BI was detected by flow cytometry as described above. Individual cells with high expression levels were sorted and grown from single cells into individual clones in medium E as described previously (61). One clone, ldlA[G15L/G18L/G25L], exhibiting surface expression of the triple mutant form of SR-BI that was similar to ldlA[SR-BI] was used for the experiments shown. Both ldlA[SR-BI] and ldlA[G15L/G18L/G25L] cells were maintained in stock culture in medium E. For additional details, see supplemental Experimental Procedures.
Chemical Cross-linking
Cell surface chemical cross-linking was performed with bis(sulfosuccinimidyl)suberate (BS3; Thermo Scientific), a water-soluble, membrane-impermeable cross-linker that reacts with primary amines. For cross-linking of rho-tagged SR-BI/LIMP chimeras in transiently transfected COS cells, on day 0, 6.0 × 105 cells/well were plated in 6-well dishes in medium B without antibiotics. On day 1, cells were transfected with a 1:10 mixture of rho-tagged receptor plasmid DNA:empty vector pcDNA4/TO (4 μg of total DNA/well). The receptor-encoding plasmid was diluted with the empty vector to reduce total and cell surface receptor expression. For stably transfected ldlA[SR-BI] and ldlA[G15L/G18L/G25L] cells, on day 1, 3.0 × 105 cells/well were plated into the wells of 6-well dishes in medium E. On day 3, transiently or stably transfected cells were washed twice with PBS-inh (ice-cold PBS supplemented with Complete Mini protease inhibitor mixture and PhosSTOP phosphatase inhibitor mixture (Roche Applied Science)) and then incubated for 1 h at 4 °C in 0.5 ml of PBS-inh containing the indicated concentrations of BS3 (0–1 mm). The reaction was stopped by adding 0.02 ml of 1 m Tris-HCl, pH 7.5 for 15 min. Cells were then quickly washed with ice-cold PBS and lysed with 0.25 ml of 1% sodium dodecyl sulfate (SDS) in PBS-inh (cell lysate). Lysate protein content was determined with the BCA protein assay kit (Thermo Scientific). Cell lysates (1–10 μg of total protein) were analyzed by 8% SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting on PVDF membranes with either anti-rho 1D4 mouse monoclonal antibody in the case of transiently transfected cells or with polyclonal rabbit anti-C terminus SR-BI antibody 495 (1, 10) in the case of stably transfected cells. SR-BI-containing proteins were visualized using horseradish peroxidase (HRP)-conjugated anti-mouse or anti-rabbit IgG (Invitrogen), respectively, and the ECL chemiluminescence detection system (GE Healthcare) and either x-ray film or an Eastman Kodak Co. Image Station for quantitation of band intensities. The same filters were also subjected to immunoblotting analysis using a polyclonal rabbit anti-ϵ-COP antibody (62) as a loading control. ϵ-COP is a cytoplasmic protein that is a component of the multiprotein COPI complex (62). Under the conditions used here, treatment of the intact cells with BS3 (0–1 mm) did not induce cross-linking of ϵ-COP to itself, other COPI components, or any other proteins (supplemental Fig. S1). This suggests that the cross-linking mediated by BS3 was restricted to cell surface proteins exposed to the extracellular space.
Co-immunoprecipitation
COS M6 cells in 6-well dishes were grown and transfected (4 μg of total plasmid DNA/well) as described above using either the empty vector pcDNA4/TO or wild-type SR-BI or triple glycine mutant G15L/G18L/G25L expression vectors encoding receptors in which the C termini bear either rho or myc epitope tag extensions (see above). Cells were transfected with either (i) only one of the four pure individual expression vectors (SR-BI-rho, SR-BI-myc, G15L/G18L/G25L-rho, or G15L/G18L/G25L-myc), (ii) 1:1 mixtures of vectors encoding the wild-type SR-BI rho- and myc-tagged receptors or 1:1 mixtures of the vectors encoding the G15L/G18L/G25L rho- and myc-tagged receptors, or (iii) the same mixtures of vectors as in condition ii but diluted with the empty vector either 1:20 (SR-BI-tags) or 1:10 (G15L/G18L/G25L-tags) to reduce total and cell surface receptor expression. Transfection with each of the four individual expression plasmids expressed alone (condition i) resulted in virtually identical levels of surface expression of the encoded proteins measured on day 3 using flow cytometry and the KKB-1 antibody (∼100% expression). Similar levels of total receptor surface expression were observed for the undiluted binary mixtures of vectors (condition ii), whereas the empty vector-diluted mixed samples (condition iii) for both the wild-type and triple mutant tagged receptors resulted in reduced surface expression (∼75%).
The co-immunoprecipitation assay was performed on day 3. Cells were washed twice with ice-cold Tris-buffered saline (TBS; 25 mm Tris-HCl, 0.15 m NaCl, pH 7.2) and lysed with 300 μl of mammalian protein extraction reagent (M-PER; Thermo Scientific) supplemented with the protease and phosphatase inhibitor mixtures described above. Aliquots of total cell lysates (10 μg of total protein) were separated by 8% SDS-PAGE; analyzed by immunoblotting on PVDF membranes with rabbit polyclonal anti-SR-BI KKB-1 antiserum, mouse monoclonal anti-myc (Invitrogen), and mouse monoclonal anti-rho 1D4 (ATCC) antibodies together with HRP-conjugated rabbit and mouse IgG (Invitrogen); and visualized by ECL chemiluminescence and either x-ray film or a Kodak Image Station for quantitation of band intensities. The expression levels of the wild-type SR-BI and G15L/G18L/G25L mutant tagged with myc were essentially identical (see “Results”). The same was observed for the rho-tagged wild-type and G15L/G18L/G25L mutant receptors. Co-immunoprecipitation was performed using the Pierce Mammalian c-Myc Tag IP/Co-IP kit (Thermo Scientific). Co-immunoprecipitation samples in a total volume of 150 μl were prepared by mixing lysates (150 μg of total cellular protein), 2.5 mg/ml BSA, and 7.0 μl of anti-c-myc-agarose slurry (containing 3.5 μg of anti-c-myc antibody). These samples were rotated at room temperature for 2 h. The beads were then washed three times with TBS containing 0.05% Tween 20 and heated at 100 °C for 5 min, and then myc-tagged proteins were eluted with 25 μl of 2× non-reducing sample buffer (80 mm Tris-HCl, pH 6.8, 4% SDS, 8% glycerol, 0.02% bromphenol blue) at 100 °C for 5 min. Samples (10 μl) were supplemented with 2 mm β-mercaptoethanol, size-fractionated by 8% SDS-PAGE, and transferred to PVDF membranes, and then the rho- and myc-tagged proteins in the membranes were visualized using anti-rho 1D4 or anti-myc mouse monoclonal antibody, HRP-conjugated mouse IgG, and chemiluminescence detection. Quantitation of the relative amounts of signal was determined using a Kodak Image Station chemiluminescence instrument.
HDL Receptor Activity Assays
125I-HDL binding and uptake of [3H]CE from [3H]CE-HDL or uptake of the fluorescent lipid DiI from DiI-HDL were determined as described previously (1, 32, 40) using the HDL concentrations indicated in the figure legends (see supplemental Experimental Procedures for details). The amounts of cell-associated [3H]cholesteryl oleate (uptake) are expressed as the equivalent amount of [3H]CE-HDL protein (ng of lipoprotein protein/mg of cell protein) to permit direct comparison of the relative amounts of 125I-HDL binding and [3H]CE uptake (63). The values presented represent SR-BI-specific binding or specific uptake and were calculated as the differences between the average of replicates of total cell-associated binding or uptake minus the average value for control wells representing nonspecific uptake. For all experiments, total cell-associated binding or uptake values are based on quadruplicate determinations and nonspecific values from duplicate determinations made in the presence of a 40-fold excess of the corresponding unlabeled lipoprotein.
125I-HDL binding and [3H]CE uptake in transiently transfected COS cells were measured at a single HDL concentration of 10 μg protein/ml without (quadruplicate determinations) or with (duplicate determinations) a 40-fold excess of unlabeled HDL. Receptor-specific activity values for transfected COS cells were defined as the differences between specific binding or uptake values for the receptor-expressing cells calculated as described above and control cells transfected with the empty vector pcDNA4/TO (COS[Control]). Specific activities of SR-BI mutants were expressed as the percentage of wild-type SR-BI activity measured in the same experiment. These values were corrected by the relative levels of surface expression determined using flow cytometry and the KKB-1 anti-SR-BI antibody as described above. All binding and uptake data represent the mean ± S.D. of at least three independent experiments.
Cellular uptake of DiI from DiI-HDL was measured by flow cytometry in transiently transfected COS cells as described previously (40). Briefly, COS cells were grown in 6-well dishes and transiently transfected on day 1 with plasmids (4 μg of total DNA) encoding the indicated receptors as described above. On day 3, the transfected cells were washed twice with medium A and incubated in medium A containing 0.5% (w/v) fatty acid-free BSA and 10 μg protein/ml DiI-HDL for 2 h at 37 °C. Cells were then washed twice with medium A and incubated with anti-SR-BI KKB-1 antiserum followed by FITC-conjugated goat anti-rabbit IgG as described above. Cells transfected with the empty vector only were used to determine background staining and to set the receptor-specific surface expression gate in which mean values of DiI fluorescence intensity were measured. These values, defined as DiI uptake activity, are expressed as the percentage of wild-type SR-BI activity (100%) measured in the same experiment. Values represent mean ± S.D. of at least three independent experiments, whereas the values from each experiment are the means of duplicate determinations.
Data Analysis
Data were analyzed using the program Prism 5 (GraphPad Software, Inc., San Diego, CA). Non-linear regression analysis used the standard equations for a one-site binding model, a two-site binding model, and a one-site binding model plus a Hill slope (Hill coefficient). For each experiment, all three models were fit, and standard statistical pairwise comparisons of the models were made using the software with an extra sum-of-squares F test. In all cases, either the two-site or one-site plus Hill slope models were preferred over the simple one-site model. Statistical analyses comparing HDL binding and lipid uptake at a single substrate concentration for wild-type SR-BI and its mutants/chimeras were performed using the unpaired two-tailed t test at 95% confidence intervals. Statistical analysis for the comparison between multiple SR-BI mutants/chimeras was performed with one-way analysis of variance and Tukey post-test at 95% confidence intervals.
RESULTS
Sequence Alignment of SR-BI and CD36 Reveals Existence of Conserved Glycine Dimerization Motifs in Their N-terminal Transmembrane Domains
The activity of SR-BI is not substantially altered when its N-TM domain is substituted by that of CD36 (41, 42). We compared the sequences of the 24–28-residue-long N-TM domains from eight mammalian SR-BIs with sequences of eight CD36s (Fig. 1). Sixteen positions in SR-BI and ∼26 positions in CD36 are highly conserved (shaded in Fig. 1 if 7 or 8 of 8 residues are identical). However, only four residues (Gly18, Ala22, Gly25, and Ile36; SR-BI numbering) are invariant for both superfamily members (rectangles on the consensus sequences in the center of Fig. 1).
FIGURE 1.
Conserved glycine dimerization motifs in N-TM domains of mammalian SR-BIs and CD36s. Sequences of the N-cyto, N-TM, and part of the extracellular (exo) domains of mammalian SR-BI (top) and CD36 (bottom) were aligned manually. Residues conserved in ≥7 of 8 sequences are shaded. Consensus sequences for SR-BI and CD36 and glycine motifs are indicated (center) with the residues conserved in all sequences boxed. Symbols are as follows: “+,” similarity; “α,” aliphatic residue; and “.” or “x,” any residue. The glycine heptad is indicated by “AbcDefgAbcD” where glycine or alanines appear in the A and D positions.
These strictly conserved glycine and alanine residues compose sequence motifs (GxxGxxxAxxG or GxxxGxxxAxxGG) that have been associated with homo- and heterodimerization of transmembrane helices (43, 44). These include the glycophorin A dimerization motif (GX3G), the glycine zipper (GX3GX3G), and the glycine heptad (a–g) repeat with Gly in the a and d positions (e.g. GX2GX3GX2G) (64–67). Substitution of Gly by Leu in these glycine-based dimerization motifs introduces a larger apolar residue that is expected to interfere with helical TM domain associations and thus interfere with dimerization without perturbing the helical backbone. These Gly-containing dimerization motifs in SR-BI and CD36 may be capable of driving inter- or intramolecular transmembrane helix association and of influencing the activity of SR-BI.
Cell Surface Cross-linking of SR-BI and Its G15L/G18L/G25L Triple Mutant
To investigate the influence of the glycine dimerization motif on the oligomerization of SR-BI, we generated a mutant cDNA expression vector that encodes murine SR-BI in which the glycines at positions 15, 18, and 25 were converted to leucines (“G15L/G18L/G25L”). A stable cell line expressing this triple glycine mutant, ldlA[G15L/G18L/G25L], was generated and compared with ldlA[SR-BI] cells expressing similar cell surface levels of wild-type SR-BI. Untransfected ldlA-7 cells are LDL receptor-deficient cells originally isolated from mutagen-treated CHO cells (60).
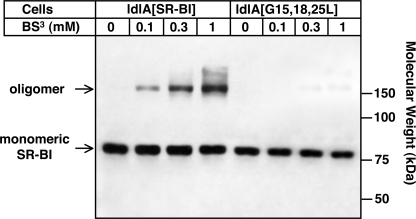
We examined the oligomerization of WT SR-BI and the G15L/G18L/G25L triple mutant by treating intact ldlA[SR-BI] and ldlA[G15L/G18L/G25L] cells for 60 min at 4 °C with varying concentrations (0–1 mm) of a water-soluble, membrane-impermeable, chemical cross-linker, BS3, that reacts with the exposed primary amines of cell surface proteins (68, 69). The cells were then lysed, and the lysates were subjected to SDS-PAGE and immunoblotting with a polyclonal antibody raised against the C terminus of SR-BI. Fig. 2 shows that in the absence of the cross-linker both wild-type and mutant SR-BI proteins appeared as single bands of ∼82 kDa, corresponding to fully glycosylated SR-BI monomers (70, 71). In ldlA[SR-BI] cells, treatment with increasing concentrations of BS3 led to the appearance of high molecular mass (≥150-kDa) bands of increasing intensity that represent SR-BI-containing homo- or hetero-oligomers. These observations are consistent with previous studies in cultured cells and tissues (28–31). Quantitative analysis of the immunoblots using a chemiluminescence image scanner showed that at 1 mm BS3, the highest concentration used, the oligomeric forms of WT SR-BI represented ∼50% of the total SR-BI protein. Thus, at least 50% (and probably significantly more) of the SR-BI was potentially cross-linkable. The precise molecular weights of the oligomers cannot be determined by immunoblotting because cross-linking can distort the log-linear relationship of electrophoretic mobility and protein mass in SDS-PAGE (72).
FIGURE 2.
Cell surface cross-linking of SR-BI and its G15L/G18L/G25L triple mutant. Stably transfected ldlA[SR-BI] and ldlA[G15L/G18L/G25L] cells expressing the wild-type and triple glycine → leucine substitution mutant receptors, respectively, were treated for 60 min at 4 °C with the indicated concentrations of the water-soluble, membrane-impermeable chemical cross-linker BS3. The cells were then lysed, and the lysates were subjected to SDS-PAGE and immunoblotting with an anti-C terminus SR-BI polyclonal antibody as described under “Experimental Procedures.” The intensities of the bands corresponding to the monomeric (∼82-kDa) and oligomeric (>150-kDa) species (see text) were determined using a Kodak Image Station.
In contrast to wild-type SR-BI, very little of the triple glycine mutant in ldlA[G15L/G18L/G25L] cells was converted to oligomers by cross-linking (<5% oligomers at 1 mm BS3) (Fig. 2). These results suggest that the conserved glycines in the N-TM domain of SR-BI significantly contribute to the association of SR-BI with itself and/or other molecules in the plasma membrane because the triple mutation led to a dramatic (>10-fold) reduction of cross-linked SR-BI.
Co-immunoprecipitation Analysis of Receptor Homo-oligomerization
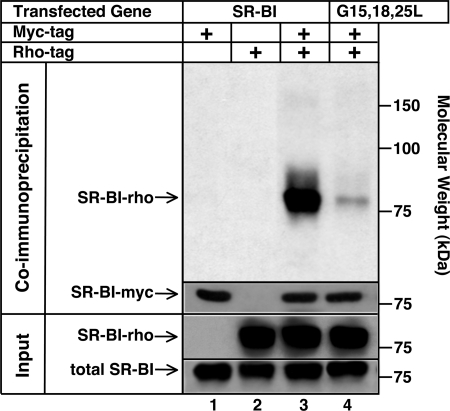
To determine if the SR-BI oligomers detected by cross-linking are composed, at least in part, of homo-oligomers, we performed co-immunoprecipitation studies using receptors fused to myc or rho epitope tags at their C termini (see “Experimental Procedures”). COS cells were transiently transfected with myc- or rho-tagged wild-type SR-BI-expressing plasmids individually or simultaneously. Parallel transfections were performed with the vectors encoding the two tagged G15L/G18L/G25L mutants. Total protein expression levels of the wild-type SR-BI and the G15L/G18L/G25L mutant were assessed by immunoblotting of detergent-solubilized whole cell lysates (“Input”; 10 μg of protein) using monoclonal anti-rho antibody (“SR-BI-rho”), anti-myc antibody (“SR-BI-myc”; not shown), and the rabbit polyclonal anti-murine SR-BI antibody KKB-1 (“total SR-BI”) (e.g. see Fig. 3, lower “Input” panels). Cell surface expression was quantified by flow cytometry using the KKB-1 antibody that recognizes the extracellular loop of SR-BI (40). The transfection conditions used resulted in each of the four plasmids producing essentially the same level of surface receptor protein expression. Each of the four epitope-tagged receptors also bound similar amounts of HDL and mediated similar levels of selective lipid uptake, indicating that the epitope tags did not dramatically alter receptor activity.
FIGURE 3.
Co-immunoprecipitation of rho and myc epitope-tagged receptors. COS cells were transiently transfected with plasmids encoding myc- or rho-tagged wild-type SR-BI either individually or with a 1:1 mixture of both plasmid cDNAs. Parallel transfections were performed with the vectors encoding the two tagged G15L/G18L/G25L mutants. Recombinant protein expression levels in detergent-solubilized whole cell lysates (Input; 10 μg of lysate protein) were assessed by immunoblotting using KKB-1, a rabbit anti-murine SR-BI polyclonal antibody that recognizes the extracellular loop of SR-BI (total SR-BI) and anti-rho antibodies (SR-BI-rho) (lower Input panels). Cell surface expression of receptors was quantified by flow cytometry of intact cells stained with KKB-1 and secondary FITC-labeled anti-rabbit IgG antibody. For co-immunoprecipitations, whole cell lysates were precipitated with agarose beads covalently linked to anti-myc antibody. Bead-bound proteins were eluted, size-fractionated by SDS-PAGE, and transferred to PVDF membranes, and then the myc- (SR-BI-myc) and rho-tagged proteins were visualized using anti-myc or anti-rho antibody and chemiluminescence detection (upper Co-immunoprecipitation panels). Band intensities (see text) were determined using a Kodak Image Station.
The co-immunoprecipitations were performed as follows (see “Experimental Procedures” for details). Cells lysates were precipitated with agarose beads covalently linked to anti-myc antibody. Bead-bound proteins were eluted and analyzed by SDS-PAGE/immunoblotting with anti-rho or anti-myc antibody and chemiluminescence detection (Fig. 3, upper “Co-immunoprecipitation” panels). Quantitation of the relative amounts of signal was determined using a Kodak Image Station.
Fig. 3, upper Co-immunoprecipitation panels, show that wild-type SR-BI-myc (lane 1), but not wild-type SR-BI-rho (lane 2), was precipitated by the anti-myc beads from cells transfected with the plasmids individually. The anti-myc beads were able to co-precipitate ∼10% of the total wild-type SR-BI-rho expressed in cells cotransfected with vectors encoding both the myc- and rho-tagged wild type receptor (lane 3). This confirms previous reports of SR-BI homo-oligomerization detected by co-immunoprecipitation of co-expressed epitope-tagged SR-BIs (28, 29). In contrast, very little rho-tagged G15L/G18L/G25L mutant (∼0.5% of the total G15L/G18L/G25L-rho present in the cells) co-immunoprecipitated with the co-expressed myc-tagged mutant (lane 4). There was 20-fold less co-immunoprecipitation of the triple glycine mutant than the wild-type receptor. Comparable results were obtained when the amounts of the cDNA expression vectors used for transfections were reduced 10–20-fold by dilution with noncoding “empty” vector control DNA (supplemental Fig. S2). Thus, the homo-oligomerization of wild-type SR-BI and dramatically reduced homo-oligomerization of the G15L/G18L/G25L mutant are unlikely to be artifactual consequences of excessive overexpression induced by transfection with undiluted vectors. Based on the cell surface cross-linking results (Fig. 2 above) and these co-immunoprecipitation results, we conclude that at least 10% (and probably much more (>50%)) of the wild-type SR-BI on the surfaces of transfected cells is present as homo-oligomers (possibly dimers) and that one or more of the conserved glycines at positions 15, 18, and 25 in the N-TM domain (Fig. 1) play a key role in mediating oligomerization. This is probably a consequence of these glycines stabilizing intermolecular transmembrane helix associations via classic glycine dimerization motifs.
Effects of Triple G15L/G18L/G25L Substitutions on Receptor-mediated HDL Binding and Lipid Uptake
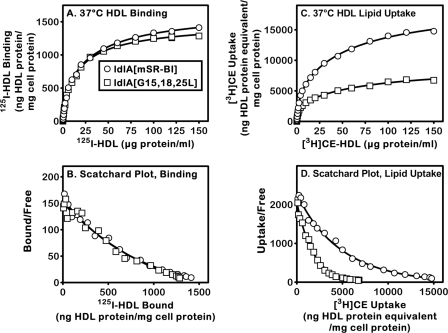
To determine if the triple G15L/G18L/G25L substitutions influenced receptor activity as well as receptor homo-oligomerization, we measured in ldlA[SR-BI] and ldlA[G15L/G18L/G25L] cells at 37 °C the lipoprotein concentration dependence of 125I-HDL binding and uptake of [3H]CE from [3H]CE-HDL. Fig. 4 shows the results from one of three independent experiments whose results were concordant. The data were corrected for small differences (<10%) in surface expression of the wild-type and mutant receptors measured using flow cytometry and a polyclonal anti-SR-BI antibody. The corrections assume that the substitutions did not alter antibody binding (see “Experimental Procedures”). Fig. 4, A and C, show the HDL concentration dependences of binding and uptake, respectively, whereas B and D show the same data in the form of Scatchard plots. The data are best fit by models (described in more detail under “Discussion”) in which SR-BI exhibits either two independent classes of binding sites (two-site model) or one class of sites exhibiting negative cooperativity (one-site plus Hill coefficient model) (32). As reported previously (32), it is likely that the nonlinear, concave up Scatchard plots (Fig. 4, B and D) are due to a single class of binding sites exhibiting negative cooperativity.
FIGURE 4.
Concentration dependences (A and C) and corresponding Scatchard analyses (B and D) of specific SR-BI-mediated 125I-HDL binding (A and B) and uptake of [3H]cholesteryl ester from [3H]CE-HDL (C and D) at 37 °C. ldlA[mSR-BI] and ldlA[G15L/G18L/G25L] cells were plated on day 0 at a density of 50,000 cells/well in 24-well dishes. On day 2, the cells were washed and incubated for 2 h at 37 °C with the indicated amounts of 125I-HDL or [3H]CE-HDL in the absence (quadruplicate determinations) or presence (duplicate determinations) of a 40-fold excess of unlabeled HDL. 125I-HDL cell surface binding (A and B) and cell-associated [3H]CE uptake (C and D) were determined as described under “Experimental Procedures.” Averaged nonspecific background measured in the presence of a 40-fold excess of unlabeled HDL was subtracted from the average of quadruplicate determinations of the total values measured in the absence of unlabeled HDL to calculate the specific values. The specific data from individual representative experiments are shown (the derived binding and uptake parameters are presented in Table 1).
Fig. 4, A and B, show that the triple G15L/G18L/G25L substitutions did not substantially alter the concentration dependence of 125I-HDL binding to SR-BI. Table 1 (top) shows the parameters determined by least squares fitting of these 125I-HDL binding data using either the two-site model (apparent dissociation constants (KD1 and KD2) and apparent maximum binding for each site (Bm1 and Bm2) or one-site plus Hill coefficient model (apparent dissociation constant (KHD), apparent maximum binding (BHm), and Hill coefficient). The values of these parameters were essentially identical for the wild-type and triple glycine mutant receptors. In two independent experiments, there were no statistically significant differences in the values for the wild-type and mutant receptors for six of these seven parameters (the BHm values were statistically different). Similar results (nearly identical binding parameters for the wild-type and mutant receptors) were observed in a preliminary experiment measuring 125I-HDL binding at 4 °C (data not shown).
TABLE 1.
Effects of G15L/G18L/G25L triple substitution on specific, SR-BI-mediated 125I-HDL binding and uptake of [3H]cholesteryl ester from [3H]CE-HDL at 37 °C
KD1 and KD2, apparent dissociation constants; Bm1 and Bm2, apparent maximum binding values; KHD, apparent dissociation constant for the one class of site with negative cooperativity model; BHm, apparent maximum binding value; Km1 and Km2, Michaelis constants; Vm1 and Vm2, maximum lipid uptake rates; KHm, Michaelis constant for the one class of site with negative cooperativity model; VHm, maximum lipid uptake rate for the one site with negative cooperativity model; Hill coef., Hill coefficient.
| 125I-HDL binding, 37 °C | Two-site modela |
One-site plus Hill coefficient modela |
|||||
|---|---|---|---|---|---|---|---|
| KD1b | KD2 | Bm1 | Bm2 | KHD | Hill coef. | BHm | |
| μg/ml | μg/ml | ng/mg cell protein | ng/mg cell protein | μg/ml | ng/mg cell protein | ||
| SR-BI | 3.8 ± 1.3 | 59 ± 15 | 548 ± 170 | 1,508 ± 129 | 21 ± 2 | 0.80 ± 0.02 | 1,704 ± 42 |
| G15L/G18L/G25L | 3.4 ± 1.4 | 43 ± 5 | 414 ± 164 | 1,450 ± 143 | 17 ± 1 | 0.82 ± 0.03 | 1,523 ± 36 |
| [3H]CE uptake from [3H]CE-HDL, 37 °C | Two-site modela |
One-site plus Hill coefficient modela |
|||||
|---|---|---|---|---|---|---|---|
| Km1b | Km2 | Vm1 | Vm2 | KHm | Hill coef. | VHm | |
| μg/ml | μg/ml | ng/mg cell protein | ng/mg cell protein | μg/ml | ng/mg cell protein | ||
| SR-BI | 3.3 ± 0.5 | 71 ± 13 | 5,326 ± 586 | 12,543 ± 385 | 45 ± 6 | 0.65 ± 0.02 | 19,824 ± 722 |
| G15L/G18L/G25L | 1.9 ± 0.2 | 51 ± 8 | 3,079 ± 227 | 5,722 ± 153 | 26 ± 3 | 0.61 ± 0.02 | 9,762 ± 303 |
a Binding and uptake parameters were obtained by fitting the data with either a two-site binding model or one-site binding model with Hill slope (negative cooperativity). The parameters ± S.E. were determined from one representative experiment in which each experimental value was the average of four determinations (see “Experimental Procedures”).
b For experiments performed at 37 °C the binding values represent apparent dissociation constants and maximal binding as the measurements were not made under equilibrium conditions.
Although the triple G15L/G18L/G25L substitutions did not appreciably affect 125I-HDL binding at 37 °C, they had a profound effect on the uptake of [3H]CE from [3H]CE-HDL (Fig. 4, C and D). The most striking difference apparent in Fig. 4C is that the amount of lipid uptake was decreased by ∼50%, whereas the shapes of the curves appear to be similar. This impression is confirmed by the least squares analysis of the data using the two models (Table 1, bottom). The Michaelis-Menten parameters for the lipid transfer from HDL to the cells are Km1, Km2, Vm1, and Vm2 for the two-site model and KHm, VHm, and Hill coefficient for the one-site with Hill coefficient model. In both models, the Vm values for G15L/G18L/G25L were about half that of the wild-type SR-BI. The Km values were also lower for the mutant than for the wild-type receptors. These results are consistent with a model in which the triple G15L/G18L/G25L substitutions do not alter HDL binding (Michaelis-Menten rate constants k1 and k−1 unchanged) but do decrease by about half the rate at which the receptor transfers HDL cholesteryl ester into cells (Michaelis-Menten rate constant k2 decreased by ∼50%). Under these conditions, the ratio of the Km values for the mutant (Kmm) and wild-type (KmWT) receptors ((Kmm/KmWT) = (k−1 + k2m)/(k−1 + k2WT)) is expected to be less than 1 and similar to the ratio of k2m/k2WT when k2 ≫ k−1. Therefore, a reduced Km1 or KHm in the triple mutant is consistent with the mutations decreasing lipid uptake (k2) without changing HDL binding.
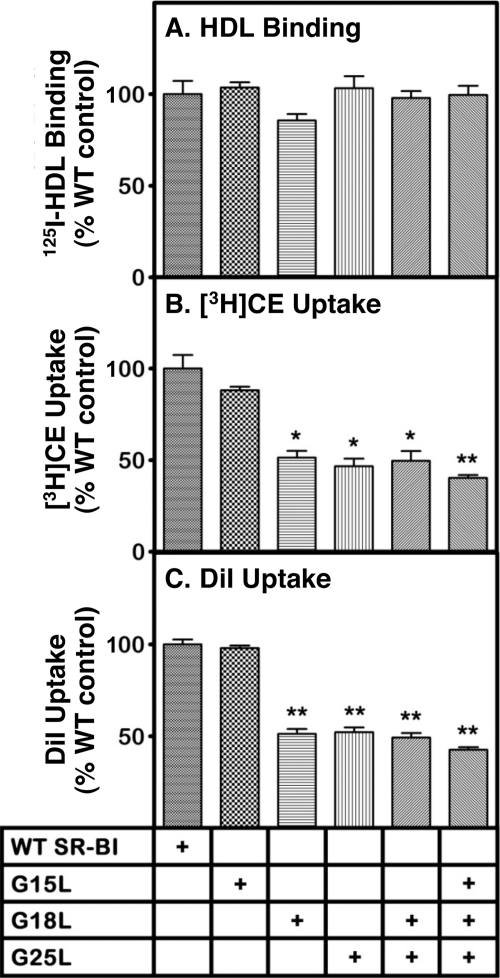
SR-BI-mediated HDL Binding and Lipid Uptake Activities of Individual Glycine Mutants
To analyze the role of each of the conserved glycines (positions 15, 18, and 25) individually on SR-BI activity, we generated mutant cDNA expression vectors that encode SR-BI in which glycines at these positions were substituted individually by leucine (G15L, G18L, or G25L) and a G18L/G25L double mutant. These vectors, together with vectors encoding wild-type SR-BI and G15L/G18L/G25L, were expressed transiently in COS cells, and receptor activity was assessed. We measured at 37 °C the following receptor activities at a single HDL concentration (10 μg of protein/ml) in three to four independent experiments: (i) 125I-HDL binding (Fig. 5A), (ii) uptake of [3H]CE from [3H]CE-HDL (Fig. 5B), and (iii) uptake of the fluorescent lipid DiI from DiI-HDL (Fig. 5C), which is an independent method to assess SR-BI-mediated selective lipid uptake (1, 73).
FIGURE 5.
Effects of individual Gly to Leu substitutions on SR-BI-mediated 125I-HDL binding and lipid uptake activities. COS cells were transiently transfected with cDNA expression vectors encoding wild-type SR-BI with single substitutions (G15L, G18L, and G25L) or with the double substitution (G18L/G25L) and triple substitution (G15L/G18L/G25L). Receptor activities for 125I-HDL binding (A), uptake of [3H]CE from [3H]CE-HDL (B), and uptake of the fluorescent lipid DiI from DiI-HDL (C) were measured at 37 °C at a single HDL concentration (10 μg of protein/ml). The data represent mean ± S.D. from three to four independent experiments normalized to 100% for wild-type SR-BI. Statistical analyses comparing wild-type SR-BI and the individual mutants were performed using the unpaired two-tailed t test at 95% confidence intervals (*, p < 0.01; **, p < 0.001). Values marked by * or ** were not significantly different from each other but were each significantly different from those for SR-BI and the G15L variant based on multiple comparisons using one-way analysis of variance with Tukey post testing. The 100% control values were as follows: 125I-HDL binding, 519 ± 100 ng of HDL protein/mg of cell protein; uptake of [3H]CE, 5380 ± 1054 ng of HDL protein equivalents/mg of cell protein; and uptake of DiI, 358 ± 18 relative fluorescence units.
None of the mutations significantly altered 125I-HDL binding (Fig. 5A). The single G15L substitution did not significantly influence [3H]CE or DiI uptake (Fig. 5, B and C). Thus, a glycine or alanine at position 15 is not essential for normal receptor lipid uptake activity under these conditions. In contrast, the single G18L and G25L mutants as well as the double G18L/G25L mutant exhibited significant (∼50%) reductions in [3H]CE and DiI uptake activities compared with wild-type SR-BI. The uptake values for these single and double mutants were not significantly different from those of the triple glycine mutant (Fig. 5, B and C). We conclude, therefore, that individually both Gly18 and Gly25 play important roles in determining the ability of SR-BI to mediate efficient lipid transport to cells from HDL.
Testing the Generality of the Role of N-TM Domain Glycine Dimerization Motifs with SR-BI Chimeras
Because the sequence context in which Gly18 and Gly25 are embedded may play a critical role in determining their influence on SR-BI activity, we examined the sequences of N-TM domains from other CD36 superfamily members with the goal of finding N-TM domain sequences that might be used to replace the endogenous N-TM domain in murine SR-BI. In particular, we looked for sequences that contained either two glycines with spacing identical to Gly18 and Gly25 in SR-BI (GX6G) or contained no glycines or only one glycine and thus might serve as a potential negative control.
SR-BI/Croquemort Chimera
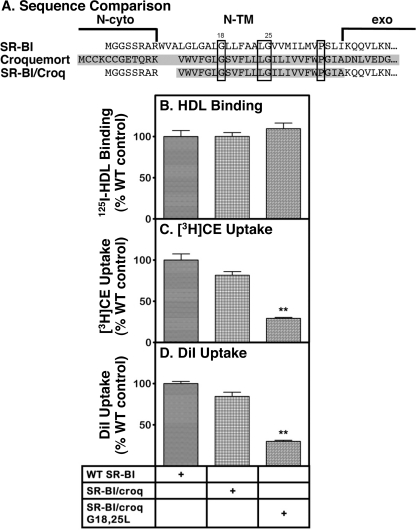
Croquemort is a CD36 paralogue from D. melanogaster that is a receptor for apoptotic cells (24). Fig. 6A shows the sequence alignment of the N-terminal region of murine SR-BI (top, no shading) with that of croquemort (middle, shaded). Boxes indicate those residues in the N-TM domain that are shared with the consensus sequence of the N-TM domain of SR-BI (Fig. 1) and constitute the following motif: GX5LGX7P. The putative length of the N-TM domain of croquemort (defined by flanking charged residues) is 3 residues shorter than that proposed for SR-BI. We generated cDNA expression vectors encoding an N-TM SR-BI/croq chimera (Fig. 6A, bottom; residues from croquemort are shaded) without or with a G18L/G25L double mutation (SR-BI numbering system) and determined their activities in transiently transfected COS cells. The activities of the SR-BI/croq chimera (125I-HDL binding, [3H]CE-HDL uptake, and DiI-HDL uptake; Fig. 6, B–D, respectively) were not significantly different from those of the wild-type SR-BI. Thus, the N-TM domain of D. melanogaster croquemort can functionally substitute for that of SR-BI. Strikingly, the double substitution of leucines for Gly18 and Gly25 in the chimera (SR-BI/croq-G18L/G25L) had qualitatively the same effect as the same double substitutions in wild-type SR-BI (Fig. 5). There was no significant change in 125I-HDL binding (Fig. 6B), but there was a substantial (≥2-fold) and statistically significant decrease in [3H]CE and DiI uptake activities (Fig. 6, C and D). We also explored the potential function of the conserved proline (Pro33) in SR-BI and croquemort sequences. Substitution of alanine for proline at position 33 of wild-type SR-BI resulted in small, insignificant reductions in 125I-HDL binding (89% of wild type) and in uptake of [3H]CE from [3H]CE-HDL (92% of wild type). Taken together, these results provide additional support for the necessary roles of Gly18 and Gly25, but not Pro33, in maintaining normal levels of lipid uptake activity mediated by SR-BI.
FIGURE 6.
Effects of replacing N-TM domain of SR-BI with that from native and mutated croquemort on receptor-mediated 125I-HDL binding and lipid uptake activities. A, sequences of the N-cyto, N-TM, and part of the extracellular (exo) domains of murine SR-BI (top), croquemort (middle; shaded), and an SR-BI/croq chimera (bottom; croquemort sequence is shaded) were aligned manually. Residues shared with the SR-BI consensus sequence are boxed. The putative length of the N-TM domain of croquemort (defined by flanking charged groups) is 3 residues shorter than that proposed for SR-BI. B–D, COS cells were transiently transfected with cDNA expression vectors encoding wild-type SR-BI and the chimeras SR-BI/croq and SR-BI/croq-G18L/G25L in which the glycines at positions 18 and 25 (SR-BI numbering system) were replaced with leucines. Receptor activities for 125I-HDL binding (B), uptake of [3H]CE from [3H]CE-HDL (C), and uptake of the fluorescent lipid DiI from DiI-HDL (D) were measured at 37 °C at a single HDL concentration (10 μg of protein/ml). The data represent mean ± S.D. from three to four independent experiments normalized to 100% for wild-type SR-BI. Statistical analyses comparing wild-type SR-BI and the individual mutants were performed using the unpaired two-tailed t test at 95% confidence intervals (**, p < 0.001). Values marked by ** were significantly different from those for SR-BI and the SR-BI/croq based on multiple comparisons using one-way analysis of variance with Tukey post-testing. The 100% control values were as follows: 125I-HDL binding, 542 ± 90 ng of HDL protein/mg of cell protein; uptake of [3H]CE, 5566 ± 1063 ng of HDL protein equivalents/mg of cell protein; and uptake of DiI, 288 ± 15 relative fluorescence units.
SR-BI/LIMP II Chimera
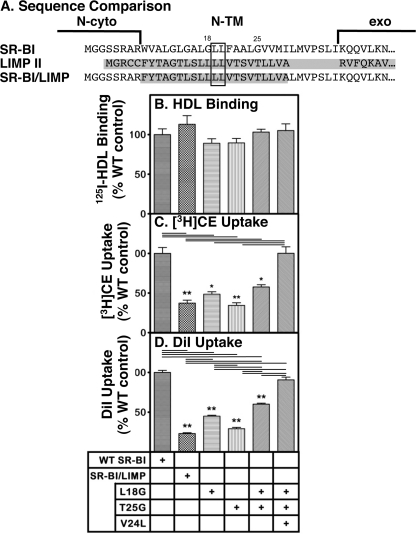
The N-TM domain of murine LIMP II, another member of the CD36 superfamily (23), has a short N-TM domain (∼21–23 residues). This domain contains only a single glycine residue that does not align with Gly18 or Gly25 of the SR-BI consensus without introducing arbitrary gaps (Fig. 7A; LIMP II sequence is shaded). There is a double leucine in the N-TM of LIMP II that can align with a double leucine in the SR-BI consensus sequence (Fig. 7A, boxed residues).
FIGURE 7.
Effects of replacing N-TM domain of SR-BI with that from native and mutated LIMP II on receptor-mediated 125I-HDL binding and lipid uptake activities. A, sequences of the N-cyto, N-TM, and part of the extracellular (exo) domains of murine SR-BI (top), murine LIMP II (middle; shaded), and an SR-BI/LIMP chimera (bottom; LIMP II sequence is shaded) were aligned manually. Residues shared with the SR-BI consensus sequence are boxed. Because the putative length of the N-TM domain of LIMP II (defined by flanking charged residues) is 7 residues shorter than that proposed for SR-BI, 7 residues of the N-TM of SR-BI (LMVPSLI) were incorporated into the N-TM domain of the chimera. B–D, COS cells were transiently transfected with cDNA expression vectors encoding wild-type SR-BI and the chimeras SR-BI/LIMP and SR-BI/LIMP in which the following substitutions, singly or combined as indicated, were made: L18G, T25G, and V24L (SR-BI numbering system). Receptor activities for 125I-HDL binding (B), uptake of [3H]CE from [3H]CE-HDL (C), and uptake of the fluorescent lipid DiI from DiI-HDL (D) were measured at 37 °C at a single HDL concentration (10 μg of protein/ml). The data represent mean ± S.D. from three to four independent experiments normalized to 100% for wild-type SR-BI. Statistical analyses comparing wild-type SR-BI and the individual mutants were performed using the unpaired two-tailed t test at 95% confidence intervals (*, p < 0.01; **, p < 0.001). Pairs of values that were significantly different based on multiple comparisons using one-way analysis of variance with Tukey post-testing are designated by the horizontal lines at the top of C and D. The 100% control values were as follows: 125I-HDL binding, 510 ± 66 ng of HDL protein/mg of cell protein; uptake of [3H]CE, 5047 ± 360 ng of HDL protein equivalents/mg of cell protein; and uptake of DiI, 326 ± 21 relative fluorescence units.
We constructed a cDNA vector encoding an SR-BI/LIMP N-TM chimera in which 7 residues from the N-TM domain of murine SR-BI (LMVPSLI) were added so that the length of the chimeric protein would be the same as that of the wild-type SR-BI (Fig. 7A, bottom; LIMP II sequence is shaded). When transiently expressed in COS cells, the SR-BI/LIMP chimera exhibited 125I-HDL binding not statistically different from SR-BI (Fig. 7B); however, it exhibited substantially (<2-fold) and significantly lower [3H]CE and DiI uptake activities (Fig. 7, C and D; horizontal lines at the top of the panels indicate significant differences based on Tukey's multiple comparison test). The N-TM domain of LIMP II could not fully functionally substitute for that of SR-BI. The activity of the SR-BI/LIMP chimera was reminiscent of that of SR-BI with single or multiple Gly18/Gly25 to leucine substitutions (Fig. 5, B and C). We then asked if normal lipid uptake activity could be conferred on the SR-BI/LIMP chimera by substituting a glycine for either the leucine at position 18 (L18G; SR-BI numbering), the threonine at position 25 (T25G), or both (L18G/T25G). Fig. 7C shows that the single substitution L18G or T25G into SR-BI/LIMP did not increase its relatively low [3H]CE uptake activity from [3H]CE-HDL. In contrast, the [3H]CE uptake activity of the SR-BI/LIMP-L18G/T25G double mutant chimera was significantly greater than that of the unmodified SR-BI/LIMP chimera and was not statistically different from wild-type SR-BI based on Tukey's multiple comparison test. (The activities of wild-type SR-BI and SR-BI/LIMP-L18G/T25G were significantly different when they were compared independently of the other mutants by t test.) Similar results were obtained for the DiI uptake assay (Fig. 7D). Thus, introduction of the GX6G motif, but not Gly18 or Gly25 individually, into the N-TM of the SR-BI/LIMP chimera increased lipid transport activity.
These data suggested that the GX6G motif alone was not sufficient to confer activity identical to that of wild-type SR-BI. We noted that in the sequences of the N-TM domains of murine SR-BI (Fig. 7A, top, and the consensus SR-BI sequence in Fig. 1) and of SR-BI/croq (Fig. 6A, bottom), both of which exhibit full lipid uptake activity, the Gly25 is preceded by leucine (Leu24). Interestingly, statistical analysis of amino acid patterns in transmembrane helices by Senes et al. (67) showed that the GxxxG motif was frequently found in association with large aliphatic residues (Ile, Val, and Leu) at neighboring positions. Therefore, we generated a cDNA vector encoding a triple mutant of the SR-BI/LIMP chimera, SR-BI/LIMP-L18G/T25G/V24L. Fig. 7 shows that the 125I-HDL binding activity of this receptor was not significantly different from those of wild-type SR-BI or the other SR-BI/LIMP chimeras (B) and that the [3H]CE (C) and DiI (D) uptake activities of the SR-BI/LIMP-L18G/T25G/V24L chimera were virtually identical to those of wild-type SR-BI (C and D). Thus, it appears that the sequence context of the GX6G motif (i.e. GX5LG) in the N-TM domain can influence significantly the lipid uptake activity of the chimeric receptor.
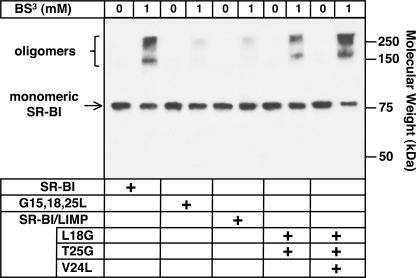
To determine if the lipid uptake activities of wild-type SR-BI and the SR-BI/LIMP chimeras were correlated with the oligomerization state of the receptors, we treated transiently transfected COS cells expressing these receptors, all having C-terminal rho epitope tags, with the membrane-impermeable cross-linker BS3 (1 mm) and assayed for cross-linking using SDS-PAGE and immunoblotting with an anti-rho antibody as described above. Fig. 8 shows that the extent of cross-linking (intensity of oligomer bands) varied with the following rank order: wild-type SR-BI ≈ SR-BI/LIMP-L18G/T25G/V24L (65 and 75% cross-linking, respectively) > SR-BI/LIMP-L18G/T25G (∼50% cross-linking) ≫ SR-BI/LIMP and SR-BI[G15L/G18L/G25L] (∼3% cross-linking). Indeed, the rank order of receptor activities (Fig. 7) matched that of their susceptibility to cross-linking, suggesting that receptor oligomerization may directly influence receptor activity.
FIGURE 8.
Effects of replacing N-TM domain of SR-BI with that from native and mutated LIMP II on cross-linking of cell surface receptors. COS cells were transiently transfected with a 10:1 mixture of empty vector plasmid and a vector encoding one of the following rho-tagged receptors: wild-type SR-BI, the G15L/G18L/G25L triple SR-BI mutant, and the chimeras SR-BI/LIMP and SR-BI/LIMP with the double L18G/T25G or triple L18G/T25G/V24L substitutions (SR-BI numbering system). Cells were treated for 60 min at 4 °C with 0 or 1 mm BS3 cross-linker. The cells were then lysed, and the lysates were subjected to SDS-PAGE and immunoblotting with the anti-rho mouse monoclonal antibody 1D4 as described under “Experimental Procedures.” The intensities of the bands corresponding to the monomeric (∼82-kDa) and oligomeric (>150-kDa) species (see text) were determined using a Kodak Image Station.
DISCUSSION
In the current study, we identified a conserved glycine dimerization motif, G18X3AX2G25, in the N-TM domain of SR-BI that plays a major role in mediating receptor homo-oligomerization. The N-TM domain of SR-BI might contribute to homo-oligomerization by interacting directly with the N-TM domains on other SR-BI molecules (intermolecular contacts), with its C-TM domain (intra- or intermolecular interactions), or with both. Future studies will be required to determine the precise mechanism of oligomerization and the stoichiometry of SR-BI oligomers (dimers or higher order oligomers).
We studied a variety of SR-BI mutants (single or multiple Gly to Leu substitutions in the dimerization motif) and SR-BI N-TM chimeras in which the N-TM domain of SR-BI was replaced by wild-type and mutated N-TM domains from other CD36 superfamily members. Strikingly, the glycine dimerization motif-defective SR-BI mutants and chimeras exhibited HDL binding activities that were virtually indistinguishable from that of wild-type SR-BI. (Cell surface receptor expression was determined by flow cytometry using a polyclonal anti-SR-BI antibody whose affinities for wild-type and variant receptors was assumed to be equal (see “Experimental Procedures”).) For example, in a G15L/G18L/G25L triple mutant of SR-BI, oligomerization was almost completely disrupted, yet the affinity of HDL binding and its characteristic negative cooperativity were identical to those of wild-type SR-BI. Although disruption of the dimerization motif was not associated with changes in HDL binding, it did significantly reduce the lipid uptake rate by ∼50%. The rates of lipid uptake mediated by wild-type SR-BI and various SR-BI N-TM chimeras with and without dimerization motifs were directly proportional to the extents of receptor oligomerization.
The detailed mechanism by which SR-BI mediates cellular uptake of cholesteryl esters from HDL remains to be established. There may be direct participation of the N-TM domain in the movement of lipids from HDL into the cells. Thus, Gly → Leu mutations in this domain might reduce the rate of lipid transport by directly interfering with lipid movements. Alternatively, the principal effect of the N-TM domain glycines on lipid transport rates may be their direct participation in oligomerization rather than their direct involvement in the transport process. Indeed, the correlation of the extent of SR-BI oligomerization and lipid uptake rate is consistent with this possibility.
HDL binding to wild-type SR-BI in cultured cells is complex, exhibiting negative cooperativity (32). Two general mechanisms might account for this negative cooperativity: classic allostery or a lattice (ensemble) effect. Classic homotropic allostery (74–78), or cooperativity, occurs when initial ligand binding to one site induces changes in the structure of a protein that alters the binding properties of an identical ligand at another site(s). Typically, such receptors when bound to ligands are multimeric, and the binding sites can be identical in the absence of bound ligand (34, 77–80). If the allosteric changes lower the affinity of the unoccupied binding site(s), negative cooperativity ensues. In contrast, the lattice model of negative cooperativity does not depend on ligand-induced receptor conformational changes. Instead, if identical binding site are near one another in a “lattice,” either as adjacent monomers or oligomers, the initial binding of a relatively large ligand to one site could, without altering the structure or intrinsic binding properties of neighboring sites, sterically interfere with the access of a second ligand to one of the neighboring binding sites (81–84). This initial binding effectively increases the KD of the neighboring unbound sites, resulting in negative cooperativity (85–87). SR-BI forms clusters on the surfaces of ldlA[mSR-BI] and other cells (70, 88), and it forms oligomers (Refs. 26–31 and this study). Thus, its lipoprotein binding sites can be in the close apposition required for lattice-based negative cooperativity. Because N-TM mutations that dramatically reduced homo-oligomerization had virtually no influence on the concentration dependence of HDL binding, classic allostery depending on conformational changes in oligomeric receptors seems unlikely to be responsible for the observed negative cooperativity. We cannot formally rule out the possibility that ligand-induced allosteric changes within independent monomers, each having multiple equivalent, allosterically coupled binding sites, is the source of the negative cooperativity of HDL binding, although this does not seem likely. Rather, we conclude that a lattice effect appears to be the most attractive model to explain the negative cooperativity of HDL binding to SR-BI.
In conclusion, our data show that the N-TM domain of SR-BI is not simply a lipid anchor for the receptor but rather plays an important role in SR-BI homo-oligomerization and lipid uptake. Future experiments will be required to elucidate the precise mechanism by which the glycine dimerization motif in the N-TM of SR-BI influences the rate of lipid transfer and how receptor oligomerization influences this process.
Supplementary Material
Acknowledgments
We thank Elina Godes for assistance with the illustrations and Thomas Nieland for helpful discussions and suggestions.
This work was supported, in whole or in part, by National Institutes of Health Grants HL52212 and HL066105 (to M. K.). This work was also supported by a Merck/Massachusetts Institute of Technology fellowship (to L. G.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Experimental Procedures and Figs. S1 and S2.
- SR-BI
- scavenger receptor class B, type I
- [3H]CE
- [3H]cholesteryl oleate
- DiI
- 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate
- BS3
- bis(sulfosuccinimidyl)suberate
- rho
- rhodopsin
- TM
- transmembrane
- N-TM
- N-terminal transmembrane
- C-TM
- C-terminal transmembrane
- cyto
- cytoplasmic
- croq
- croquemort
- LIMP
- lysosomal integral membrane protein.
REFERENCES
- 1. Acton S., Rigotti A., Landschulz K. T., Xu S., Hobbs H. H., Krieger M. (1996) Science 271, 518–520 [DOI] [PubMed] [Google Scholar]
- 2. Rigotti A., Miettinen H. E., Krieger M. (2003) Endocr. Rev. 24, 357–387 [DOI] [PubMed] [Google Scholar]
- 3. Ji Y., Jian B., Wang N., Sun Y., Moya M. L., Phillips M. C., Rothblat G. H., Swaney J. B., Tall A. R. (1997) J. Biol. Chem. 272, 20982–20985 [DOI] [PubMed] [Google Scholar]
- 4. Hatzopoulos A. K., Rigotti A., Rosenberg R. D., Krieger M. (1998) J. Lipid Res. 39, 495–508 [PubMed] [Google Scholar]
- 5. Yuhanna I. S., Zhu Y., Cox B. E., Hahner L. D., Osborne-Lawrence S., Lu P., Marcel Y. L., Anderson R. G., Mendelsohn M. E., Hobbs H. H., Shaul P. W. (2001) Nat. Med. 7, 853–857 [DOI] [PubMed] [Google Scholar]
- 6. Uittenbogaard A., Shaul P. W., Yuhanna I. S., Blair A., Smart E. J. (2000) J. Biol. Chem. 275, 11278–11283 [DOI] [PubMed] [Google Scholar]
- 7. Krieger M. (1999) Annu. Rev. Biochem. 68, 523–558 [DOI] [PubMed] [Google Scholar]
- 8. Rothblat G. H., Phillips M. C. (2010) Curr. Opin. Lipidol. 21, 229–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Glomset J. A. (1968) J. Lipid Res. 9, 155–167 [PubMed] [Google Scholar]
- 10. Rigotti A., Trigatti B. L., Penman M., Rayburn H., Herz J., Krieger M. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 12610–12615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Teslovich T. M., Musunuru K., Smith A. V., Edmondson A. C., Stylianou I. M., Koseki M., Pirruccello J. P., Ripatti S., Chasman D. I., Willer C. J., Johansen C. T., Fouchier S. W., Isaacs A., Peloso G. M., Barbalic M., Ricketts S. L., Bis J. C., Aulchenko Y. S., Thorleifsson G., Feitosa M. F., Chambers J., Orho-Melander M., Melander O., Johnson T., Li X., Guo X., Li M., Shin Cho Y., Jin Go M., Jin Kim Y., Lee J. Y., Park T., Kim K., Sim X., Twee-Hee Ong R., Croteau-Chonka D. C., Lange L. A., Smith J. D., Song K., Hua Zhao J., Yuan X., Luan J., Lamina C., Ziegler A., Zhang W., Zee R. Y., Wright A. F., Witteman J. C., Wilson J. F., Willemsen G., Wichmann H. E., Whitfield J. B., Waterworth D. M., Wareham N. J., Waeber G., Vollenweider P., Voight B. F., Vitart V., Uitterlinden A. G., Uda M., Tuomilehto J., Thompson J. R., Tanaka T., Surakka I., Stringham H. M., Spector T. D., Soranzo N., Smit J. H., Sinisalo J., Silander K., Sijbrands E. J., Scuteri A., Scott J., Schlessinger D., Sanna S., Salomaa V., Saharinen J., Sabatti C., Ruokonen A., Rudan I., Rose L. M., Roberts R., Rieder M., Psaty B. M., Pramstaller P. P., Pichler I., Perola M., Penninx B. W., Pedersen N. L., Pattaro C., Parker A. N., Pare G., Oostra B. A., O'Donnell C. J., Nieminen M. S., Nickerson D. A., Montgomery G. W., Meitinger T., McPherson R., McCarthy M. I., McArdle W., Masson D., Martin N. G., Marroni F., Mangino M., Magnusson P. K., Lucas G., Luben R., Loos R. J., Lokki M. L., Lettre G., Langenberg C., Launer L. J., Lakatta E. G., Laaksonen R., Kyvik K. O., Kronenberg F., König I. R., Khaw K. T., Kaprio J., Kaplan L. M., Johansson A., Jarvelin M. R., Janssens A. C., Ingelsson E., Igl W., Kees Hovingh G., Hottenga J. J., Hofman A., Hicks A. A., Hengstenberg C., Heid I. M., Hayward C., Havulinna A. S., Hastie N. D., Harris T. B., Haritunians T., Hall A. S., Gyllensten U., Guiducci C., Groop L. C., Gonzalez E., Gieger C., Freimer N. B., Ferrucci L., Erdmann J., Elliott P., Ejebe K. G., Döring A., Dominiczak A. F., Demissie S., Deloukas P., de Geus E. J., de Faire U., Crawford G., Collins F. S., Chen Y. D., Caulfield M. J., Campbell H., Burtt N. P., Bonnycastle L. L., Boomsma D. I., Boekholdt S. M., Bergman R. N., Barroso I., Bandinelli S., Ballantyne C. M., Assimes T. L., Quertermous T., Altshuler D., Seielstad M., Wong T. Y., Tai E. S., Feranil A. B., Kuzawa C. W., Adair L. S., Taylor H. A., Jr., Borecki I. B., Gabriel S. B., Wilson J. G., Holm H., Thorsteinsdottir U., Gudnason V., Krauss R. M., Mohlke K. L., Ordovas J. M., Munroe P. B., Kooner J. S., Tall A. R., Hegele R. A., Kastelein J. J., Schadt E. E., Rotter J. I., Boerwinkle E., Strachan D. P., Mooser V., Stefansson K., Reilly M. P., Samani N. J., Schunkert H., Cupples L. A., Sandhu M. S., Ridker P. M., Rader D. J., van Duijn C. M., Peltonen L., Abecasis G. R., Boehnke M., Kathiresan S. (2010) Nature 466, 707–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Vergeer M., Korporaal S. J., Franssen R., Meurs I., Out R., Hovingh G. K., Hoekstra M., Sierts J. A., Dallinga-Thie G. M., Motazacker M. M., Holleboom A. G., Van Berkel T. J., Kastelein J. J., Van Eck M., Kuivenhoven J. A. (2011) N. Engl. J. Med. 364, 136–145 [DOI] [PubMed] [Google Scholar]
- 13. Gordon T., Castelli W. P., Hjortland M. C., Kannel W. B., Dawber T. R. (1977) Am. J. Med. 62, 707–714 [DOI] [PubMed] [Google Scholar]
- 14. Miller G. J., Miller N. E. (1975) Lancet 1, 16–19 [DOI] [PubMed] [Google Scholar]
- 15. Gordon D. J., Probstfield J. L., Garrison R. J., Neaton J. D., Castelli W. P., Knoke J. D., Jacobs D. R., Jr., Bangdiwala S., Tyroler H. A. (1989) Circulation 79, 8–15 [DOI] [PubMed] [Google Scholar]
- 16. Kozarsky K. F., Donahee M. H., Glick J. M., Krieger M., Rader D. J. (2000) Arterioscler. Thromb. Vasc. Biol. 20, 721–727 [DOI] [PubMed] [Google Scholar]
- 17. Arai T., Wang N., Bezouevski M., Welch C., Tall A. R. (1999) J. Biol. Chem. 274, 2366–2371 [DOI] [PubMed] [Google Scholar]
- 18. Ueda Y., Gong E., Royer L., Cooper P. N., Francone O. L., Rubin E. M. (2000) J. Biol. Chem. 275, 20368–20373 [DOI] [PubMed] [Google Scholar]
- 19. Trigatti B., Rayburn H., Viñals M., Braun A., Miettinen H., Penman M., Hertz M., Schrenzel M., Amigo L., Rigotti A., Krieger M. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 9322–9327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Huszar D., Varban M. L., Rinninger F., Feeley R., Arai T., Fairchild-Huntress V., Donovan M. J., Tall A. R. (2000) Arterioscler. Thromb. Vasc. Biol. 20, 1068–1073 [DOI] [PubMed] [Google Scholar]
- 21. Braun A., Trigatti B. L., Post M. J., Sato K., Simons M., Edelberg J. M., Rosenberg R. D., Schrenzel M., Krieger M. (2002) Circ. Res. 90, 270–276 [DOI] [PubMed] [Google Scholar]
- 22. Braun A., Zhang S., Miettinen H. E., Ebrahim S., Holm T. M., Vasile E., Post M. J., Yoerger D. M., Picard M. H., Krieger J. L., Andrews N. C., Simons M., Krieger M. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 7283–7288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Vega M. A., Seguí-Real B., García J. A., Calés C., Rodríguez F., Vanderkerckhove J., Sandoval I. V. (1991) J. Biol. Chem. 266, 16818–16824 [PubMed] [Google Scholar]
- 24. Franc N. C., Dimarcq J. L., Lagueux M., Hoffmann J., Ezekowitz R. A. (1996) Immunity 4, 431–443 [DOI] [PubMed] [Google Scholar]
- 25. Greenwalt D. E., Lipsky R. H., Ockenhouse C. F., Ikeda H., Tandon N. N., Jamieson G. A. (1992) Blood 80, 1105–1115 [PubMed] [Google Scholar]
- 26. Landschulz K. T., Pathak R. K., Rigotti A., Krieger M., Hobbs H. H. (1996) J. Clin. Investig. 98, 984–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Williams D. L., de La Llera-Moya M., Thuahnai S. T., Lund-Katz S., Connelly M. A., Azhar S., Anantharamaiah G. M., Phillips M. C. (2000) J. Biol. Chem. 275, 18897–18904 [DOI] [PubMed] [Google Scholar]
- 28. Reaven E., Cortez Y., Leers-Sucheta S., Nomoto A., Azhar S. (2004) J. Lipid Res. 45, 513–528 [DOI] [PubMed] [Google Scholar]
- 29. Sahoo D., Darlington Y. F., Pop D., Williams D. L., Connelly M. A. (2007) Biochim. Biophys. Acta 1771, 807–817 [DOI] [PubMed] [Google Scholar]
- 30. Azhar S., Nomoto A., Reaven E. (2002) J. Lipid Res. 43, 861–871 [PubMed] [Google Scholar]
- 31. Sahoo D., Peng Y., Smith J. R., Darlington Y. F., Connelly M. A. (2007) Biochim. Biophys. Acta 1771, 818–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nieland T. J., Xu S., Penman M., Krieger M. (2011) Biochemistry 50, 1818–1830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. de Meyts P., Roth J., Neville D. M., Jr., Gavin J. R., 3rd, Lesniak M. A. (1973) Biochem. Biophys. Res. Commun. 55, 154–161 [DOI] [PubMed] [Google Scholar]
- 34. DeMeyts P., Bainco A. R., Roth J. (1976) J. Biol. Chem. 251, 1877–1888 [PubMed] [Google Scholar]
- 35. Limbird L. E. (2005) Cell Surface Receptors: a Short Course on Theory and Methods, pp. 113–114, Springer, New York [Google Scholar]
- 36. Glass C., Pittman R. C., Weinstein D. B., Steinberg D. (1983) Proc. Natl. Acad. Sci. U.S.A. 80, 5435–5439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Stein Y., Dabach Y., Hollander G., Halperin G., Stein O. (1983) Biochim. Biophys. Acta 752, 98–105 [DOI] [PubMed] [Google Scholar]
- 38. Goldstein J. L., Brown M. S. (2009) Arterioscler. Thromb. Vasc. Biol. 29, 431–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Liu T., Krieger M., Kan H. Y., Zannis V. I. (2002) J. Biol. Chem. 277, 21576–21584 [DOI] [PubMed] [Google Scholar]
- 40. Gu X., Kozarsky K., Krieger M. (2000) J. Biol. Chem. 275, 29993–30001 [DOI] [PubMed] [Google Scholar]
- 41. Connelly M. A., Klein S. M., Azhar S., Abumrad N. A., Williams D. L. (1999) J. Biol. Chem. 274, 41–47 [DOI] [PubMed] [Google Scholar]
- 42. Gu X., Trigatti B., Xu S., Acton S., Babitt J., Krieger M. (1998) J. Biol. Chem. 273, 26338–26348 [DOI] [PubMed] [Google Scholar]
- 43. Mackenzie K. R. (2006) Chem. Rev. 106, 1931–1977 [DOI] [PubMed] [Google Scholar]
- 44. Moore D. T., Berger B. W., DeGrado W. F. (2008) Structure 16, 991–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Senes A., Engel D. E., DeGrado W. F. (2004) Curr. Opin. Struct. Biol. 14, 465–479 [DOI] [PubMed] [Google Scholar]
- 46. Ebie A. Z., Fleming K. G. (2007) J. Mol. Biol. 366, 517–524 [DOI] [PubMed] [Google Scholar]
- 47. Li E., Hristova K. (2006) Biochemistry 45, 6241–6251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Constantinescu S. N., Keren T., Socolovsky M., Nam H., Henis Y. I., Lodish H. F. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 4379–4384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Schnell J. R., Chou J. J. (2008) Nature 451, 591–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Stouffer A. L., Acharya R., Salom D., Levine A. S., Di Costanzo L., Soto C. S., Tereshko V., Nanda V., Stayrook S., DeGrado W. F. (2008) Nature 451, 596–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lau T. L., Kim C., Ginsberg M. H., Ulmer T. S. (2009) EMBO J. 28, 1351–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Luo B. H., Carman C. V., Takagi J., Springer T. A. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 3679–3684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Mendrola J. M., Berger M. B., King M. C., Lemmon M. A. (2002) J. Biol. Chem. 277, 4704–4712 [DOI] [PubMed] [Google Scholar]
- 54. Li W., Metcalf D. G., Gorelik R., Li R., Mitra N., Nanda V., Law P. B., Lear J. D., Degrado W. F., Bennett J. S. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 1424–1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Chung B. H., Wilkinson T., Geer J. C., Segrest J. P. (1980) J. Lipid Res. 21, 284–291 [PubMed] [Google Scholar]
- 56. Patsch J. R., Patsch W. (1986) Methods Enzymol. 129, 3–26 [DOI] [PubMed] [Google Scholar]
- 57. Lowry O. H., Rosebrough N. J., Farr A. L., Randall R. J. (1951) J. Biol. Chem. 193, 265–275 [PubMed] [Google Scholar]
- 58. Goldstein J. L., Basu S. K., Brown M. S. (1983) Methods Enzymol. 98, 241–260 [DOI] [PubMed] [Google Scholar]
- 59. Gwynne J. T., Mahaffee D. D. (1989) J. Biol. Chem. 264, 8141–8150 [PubMed] [Google Scholar]
- 60. Kingsley D. M., Krieger M. (1984) Proc. Natl. Acad. Sci. U.S.A. 81, 5454–5458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Gu X., Lawrence R., Krieger M. (2000) J. Biol. Chem. 275, 9120–9130 [DOI] [PubMed] [Google Scholar]
- 62. Guo Q., Penman M., Trigatti B. L., Krieger M. (1996) J. Biol. Chem. 271, 11191–11196 [DOI] [PubMed] [Google Scholar]
- 63. Pittman R. C., Knecht T. P., Rosenbaum M. S., Taylor C. A., Jr. (1987) J. Biol. Chem. 262, 2443–2450 [PubMed] [Google Scholar]
- 64. Lemmon M. A., Flanagan J. M., Hunt J. F., Adair B. D., Bormann B. J., Dempsey C. E., Engelman D. M. (1992) J. Biol. Chem. 267, 7683–7689 [PubMed] [Google Scholar]
- 65. Riek R. P., Finch A. A., Begg G. E., Graham R. M. (2008) Mol. Pharmacol. 73, 1092–1104 [DOI] [PubMed] [Google Scholar]
- 66. Lear J. D., Stouffer A. L., Gratkowski H., Nanda V., Degrado W. F. (2004) Biophys. J. 87, 3421–3429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Senes A., Gerstein M., Engelman D. M. (2000) J. Mol. Biol. 296, 921–936 [DOI] [PubMed] [Google Scholar]
- 68. Archibald K., Perry M. J., Molnár E., Henley J. M. (1998) Neuropharmacology 37, 1345–1353 [DOI] [PubMed] [Google Scholar]
- 69. Salhany J. M., Sloan R. L. (1989) Biochem. Biophys. Res. Commun. 159, 1337–1344 [DOI] [PubMed] [Google Scholar]
- 70. Babitt J., Trigatti B., Rigotti A., Smart E. J., Anderson R. G., Xu S., Krieger M. (1997) J. Biol. Chem. 272, 13242–13249 [DOI] [PubMed] [Google Scholar]
- 71. Viñals M., Xu S., Vasile E., Krieger M. (2003) J. Biol. Chem. 278, 5325–5332 [DOI] [PubMed] [Google Scholar]
- 72. Goetz H., Kuschel M., Wulff T., Sauber C., Miller C., Fisher S., Woodward C. (2004) J. Biochem. Biophys. Methods 60, 281–293 [DOI] [PubMed] [Google Scholar]
- 73. Schouten D., Kleinherenbrink-Stins M. F., Brouwer A., Knook D. L., Kamps J. A., Kuiper J., van Berkel T. J. (1990) Arteriosclerosis 10, 1127–1135 [DOI] [PubMed] [Google Scholar]
- 74. Monod J., Wyman J., Changeux J. P. (1965) J. Mol. Biol. 12, 88–118 [DOI] [PubMed] [Google Scholar]
- 75. Levitzki A., Koshland D. E., Jr. (1969) Proc. Natl. Acad. Sci. U.S.A. 62, 1121–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Koshland D. E., Jr. (1996) Curr. Opin. Struct. Biol. 6, 757–761 [DOI] [PubMed] [Google Scholar]
- 77. De Meyts P., Gauguin L., Svendsen A. M., Sarhan M., Knudsen L., Nøhr J., Kiselyov V. V. (2009) Ann. N.Y. Acad. Sci. 1160, 45–53 [DOI] [PubMed] [Google Scholar]
- 78. De Meyts P. (2008) Trends Biochem. Sci. 33, 376–384 [DOI] [PubMed] [Google Scholar]
- 79. Macdonald J. L., Pike L. J. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 112–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Alvarado D., Klein D. E., Lemmon M. A. (2010) Cell 142, 568–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. McGhee J. D., von Hippel P. H. (1974) J. Mol. Biol. 86, 469–489 [DOI] [PubMed] [Google Scholar]
- 82. Sackett D. L., Saroff H. A. (1996) FEBS Lett. 397, 1–6 [DOI] [PubMed] [Google Scholar]
- 83. Stankowski S. (1983) Biochim. Biophys. Acta 735, 341–351 [Google Scholar]
- 84. Stankowski S. (1983) Biochim. Biophys. Acta 735, 352–360 [Google Scholar]
- 85. Chappell D. A., Fry G. L., Waknitz M. A., Berns J. J. (1991) J. Biol. Chem. 266, 19296–19302 [PubMed] [Google Scholar]
- 86. Hardy M. R., Townsend R. R., Parkhurst S. M., Lee Y. C. (1985) Biochemistry 24, 22–28 [DOI] [PubMed] [Google Scholar]
- 87. Ashkenas J., Penman M., Vasile E., Acton S., Freeman M., Krieger M. (1993) J. Lipid Res. 34, 983–1000 [PubMed] [Google Scholar]
- 88. Peng Y., Akmentin W., Connelly M. A., Lund-Katz S., Phillips M. C., Williams D. L. (2004) Mol. Biol. Cell 15, 384–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.