Abstract
Chromosomal translocations are a major source of genetic abnormalities causally linked to certain malignancies. Synovial sarcoma is an aggressive soft tissue tumor characterized by a chromosomal translocation between chromosome 18 and X, generating oncoproteins such as SYT-SSX1 and SYT-SSX2. The molecular mechanism underlying the oncogenic potential of SYT-SSX1/2 is not clear. Here we show that SYT-SSX1 leads to up-regulation of NCOA3, a protein critical for the formation of various cancers. The increase of NCOA3 is essential for SYT-SSX1-mediated synovial sarcoma formation. SYT-SSX1 does so by increasing the sumoylation of NCOA3 through interaction with a SUMO E3 ligase, PIASy, as well as the sumoylation of NEMO. NEMO has also been shown to physically interact with NCOA3. Increased sumoylation of NCOA3 leads to its increased steady state level and nuclear localization. Our findings represent the first example that an oncoprotein directly regulates substrate modification by a SUMO E3 ligase, and leads to overexpression of a protein essential for tumor formation. Such a mechanistic finding provides an opportunity to design specific therapeutic interventions to treat synovial sarcoma.
Keywords: Nuclear Translocation, Oncogene, Post-translational Modification, Protein Turnover, Protein-Protein Interactions, Sumoylation, Transcription Coactivators
Introduction
Synovial sarcoma is an aggressive soft tissue tumor that accounts for about 7–10% of all human sarcomas (1). It has a characteristic translocation between chromosomes 18 and X. In almost all the cases, the synovial sarcoma translocated (SYT)4 gene on chromosome 18 is fused to one of the SSX-family genes on the X chromosome, mainly SSX1 and SSX2, yielding a fusion protein SYT-SSX1 or SYT-SSX2 (2–9). The reciprocal fusion transcript of SSX-SYT is rarely found in the tumor, implicating SYT-SSX as the etiological culprit (7, 10–14). Consistent with this, SYT-SSX1 has been shown to be an oncoprotein as overexpression of SYT-SSX1 in rat fibroblasts leads to anchorage-independent growth in culture and tumor formation in immunocompromised mice (15). Furthermore, expression of SYT-SSX2 within skeletal muscle-specific Myf5 lineage leads to synovial-sarcoma-like tumors with 100% penetrance in mice while expression in more differentiated tissues leads to myopathy or even embryonic lethality with ubiquitous expression (16).
The molecular mechanisms responsible for the oncogenic potential of the SYT-SSX1/2 remain unclear. SYT is a nuclear protein with no particular homology to any protein of known functions. SYT protein does not appear to have any recognizable DNA binding motif, although it can regulate transcription through binding to a variety of transcriptional regulators, including p300, CoAA, β-catenin, as well as chromatin remodeling complex SWI/SNF2 (15, 17–21). The SSX protein has also been shown to interact with proteins involved in transcription such as core histones (22). We show here that SYT and SYT-SSX can interact with SUMO E3 ligase PIASy, and consequently leads to enhanced sumoylation of its substrates, NCOA3 and NEMO. Increased sumoylation of NCOA3 results in its increased protein level, which is critical for SYT-SSX1-mediated tumor formation.
EXPERIMENTAL PROCEDURES
Culture of Mammalian Cells
Rat1 3Y1, SYO-1, and 293T were cultured in Dulbecco's modified Eagle's medium, supplemented with 10% heat inactivated fetal calf serum, 2 mm glutamine, penicillin, and streptomycin at 37 °C with 5% CO2. Anchorage-independent growth in 1.5% methylcellulose was done as described (23).
Nucleic Acids
Antisense phosphothioate oligonucleotide against SYT-SSX was used as described (23). For conditional siRNA expression for Ncoa3, pSico vector was used with the following sequence: 5′-gccaagaagcagcagtaat-3′. A U6/H1 double promoter-driven siRNA expression (24) for Ncoa3 was also used with the same sequence. Chemical small interference RNA for Nemo was from Qiagen with the product code Rn_RGD:735223_1 FlexiTube siRNA (SI02021187). Two sets of chemical siRNA for Piasy from IDT. Inc. were used with Piasy-1 for the following sequences: reverse 5′-rGrUrCrArGrCrUrGrUrCrGrCrArCrCrArGrGrUrArCrArArArGrCrC-3′; forward 5′-/5Phos/rCrUrUrUrGrUrArCrCrUrGrGrUrGrCrGrArCrArGrCrUrGAC-3′; and Piasy-2 with the catalogue number of HSC.RNAI.N015897.12.1. Calcium phosphate was used to transfect DNA into 293T cells, while Dharmafect was used to transfect siRNA/DNA. PCR primers for Ncoa3 quantification: forward 5′-TCACTTGGGACAGATGACCA-3′; reverse 5′-AGAACACCATGCCACACAGA-3′. PCR primers for Gapdh quantification: forward 5′-CATGGGTGTGAACCATGAGA-3′; reverse 5′-CAGTGATGGCATGGACTGTG-3′. RNA was isolated using Qiagen RNaeasy kit, and cDNA was generated using SuperscriptII reverse transcriptase (Invitrogen).
Biochemical and Cell Biological Reagents and Procedures
Anti-NCOA3, PIASy, SYT, NEMO antibodies were from Santa Cruz Biotechnology Inc. Anti-Flag M2 antibody was from Sigma (A1205). For immunoprecipitation and immunoblotting, cells were collected by scraping or trypsin and lysed in lysis buffer (20 mm KCl, 150 mm NaCl, 1% IGEPAL, 50 mm TrisHCl (pH 7.5), 50 mm NaF, 50 mm β-glycerolphosphate, 1 mm EGTA, 1 mm DTT, and 1× protease inhibitor mixture (Roche) and 10% glycerol). Immunoprecipitations were performed with appropriate antibody and protein A- or G-Sepharose (Upstate Biotechnology). Beads were washed three times in lysis buffer and immunoprecipitated proteins were separated on SDS-PAGE followed by Western blotting with primary and horseradish peroxidase-conjugated secondary antibody (Bio-Rad). Immunoreactive proteins were visualized by enhanced chemiluminescence (SuperSignal West Femto, Pierce Biotechnology).
TUNEL and in Vivo Tumor Formation Assay
TUNEL assay on cells grown in methylcellulose was performed as described (23). Tumor formation assay in immunocompromised mice was done also as described (23) with the measurement of tumor volume as the (1/2)L1(L2)2 where L1 is the long axis and L2 is the short axis of the tumor mass measured with a caliper in millimeter.
In Vitro Ubiquitination Assay
20 or 40 μg of Hela Fraction II (Enzo Life Sciences) was used as a source for E1, E2s, and E3s in 30 μl reaction with 20 mm Tris, pH 7.5, 5 mm MgCl2, 0.5 mm DTT, and 2 mm ATP, 0.13 mm MG-132, 1.8 μm ubiquitin aldehyde, 0.67 mm NEM. The immnoprecipitated protein bound to the protein G beads after washing with the reaction buffer was used as a substrate at 30 °C for 60′. The reaction was again extensively washed followed by SDS-PAGE and immunoblotting analysis.
Tissue Microarray, Immunohistochemical Staining, and Image Analysis
The method for the construction of tissue microarray (TMA) has been described before (25). The TMA used in the study contained two cores from each of 11 cases of diffuse neurofibromas and 15 cases of synovial sarcomas. A 5-μm paraffin section of the TMA was stained with the anti-NCOA3 antibody. Pretreatment was performed with 0.001 m EDTA, pH 8.0 (Invitrogen, Cat. 00-5501) in a steamer at 95 °C for 25 min. The section was incubated with the primary Ab (1:150) for 45 min, followed by incubation with the secondary antibody (Dakocytomation Envision System Labeled Polymer HRP anti-rabbit, DakoCytomation Cat. 4003) for 30 min. DAB (diaminobenzidine) was added for 10 min followed by hematoxylin counter stain.
The stained slide was scanned and analyzed through the Translational Pathology Core Laboratory, Department of Pathology and Laboratory Medicine, David Geffen School of Medicine at UCLA. The slide was analyzed using the Ariol SL-50 automated slide scanner (Applied Imaging, San Jose) to quantitate the amount of nuclear staining for each core. Thresholds for each image were applied using the Ariol analytical software based on RGB algorithm, shape, and size. All analyses were performed with the MultiStain script. Accuracy of thresholding was verified by a pathologist prior to analysis.
For assessing nuclear staining, positive DAB staining was calculated by applying two color thresholds with one recognizing blue background (hematoxylin stained) cells, and another recognizing brown positive cells and blue non-positive cells (total cell number). Individual cells were discriminated by incorporating the shape and size thresholds, providing, together with the color thresholds, actual cell counts. Percent of positivity was determined by dividing the cell number detected by the brown threshold by the total cell number, detected by the sum of the brown and blue thresholds. Total tissue area analyzed was also included in the final analysis (μm2).
RESULTS
NCOA3 Is Functionally Linked with SYT-SSX1
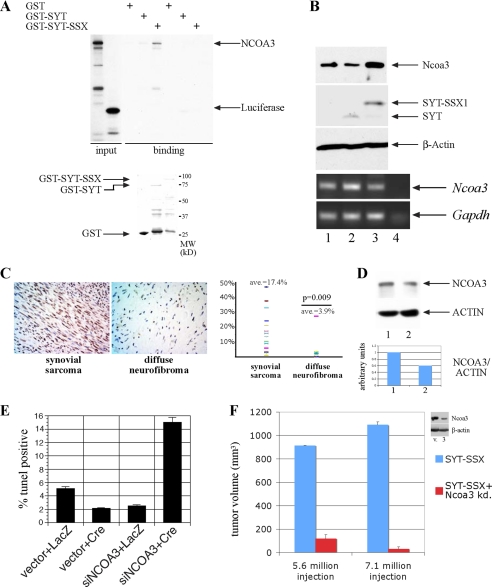
Because SYT can interact with acetyltransferase p300 (17), we examined whether SYT-SSX1 through acquisition of a novel domain interacts with acetyltransferases other than p300. In an in vitro binding assay between glutathione transferase-SYT or -SYT-SSX1 fusion protein and a panel of known acetyltransferases, we found that NCOA3 specifically interacted with SYT-SSX1 but only weakly interacted with SYT alone (Fig. 1A). NCOA3, also known as AIB1 (amplified in breast cancer), is a member of the nuclear receptor coactivators family, and is overexpressed in various cancers (26–30). Overexpression of Ncoa3 can promote tumor formation in a transgenic animal model (31). In addition, Ncoa3 is required for both prostate and breast cancer formation in mouse models (32, 33). In light of the role of NCOA3 in tumor formation, we examined its expression in rat1 3Y1 cells where SYT-SSX1 can cause tumor formation. As seen in Fig. 1B, Ncoa3 was more significantly expressed in cells stably overexpressing SYT-SSX1 than in vector control cells or those expressing SYT only. The high expression of Ncoa3 was not due to a higher transcript level (Fig. 1B, lower panel), thus unlikely a result of genomic amplification as seen in breast cancer. Consistent with this finding, we studied the expression of NCOA3 by immunohistochemistry using a TMA and found high levels of expression of NCOA3 in 13 cases of human synovial sarcoma, while 11 cases of diffuse neurofibroma, a benign soft tissue tumor, expressed lower levels of NCOA3 (Fig. 1C). To test whether SYT-SSX regulates NCOA3 expression in human sarcoma cells, we introduced into a human synovial sarcoma cell line, SYO-1 (34), an antisense oligonucleotide directed specifically against the junction of SYT/SSX fusion, with a scrambled oligonucleotide as the control. As shown in Fig. 1D, in response to the antisense oligonucleotide, the level of NCOA3 was decreased by half as compared with the cells treated with the control oligonucleotide, supporting the notion that SYT-SSX (in the SYO-1 cells, SYT-SSX2) is necessary for maintaining the level of NCOA3 in human synovial sarcoma cells. Taken together, these results are consistent with the scenario that SYT-SSX1 or SYT-SSX2 can directly interact with NCOA3 to modulate its protein level.
FIGURE 1.
SYT-SSX regulates NCOA3 protein. A, NCOA3 specifically interacts with SYT-SSX1. NCOA3 and luciferase as a control protein were radioactively labeled with [35S]methionine through in vitro translation, and were bound to GST, GST-SYT, and GST-SYT-SSX1. The bound proteins were washed and analyzed by SDS-PAGE. The input recombinant GST proteins used in these experiments were visualized through Coomassie staining shown in the lower panel. B, Ncoa3 protein and transcript level in rat1 3Y1 cells with stable expression of vector (1), SYT (2), and SYT-SSX1 (3) expression, were detected by immunoblot using antibody against NCOA3 and RT-PCR. SYT and SYT-SSX1 expression were detected with anti-Flag antibody (15). Lane 4 is a negative control without input. C, NCOA3 is significantly more expressed in synovial sarcomas compared with that in benign neurofibromas. Arbitrary staining intensity was measured by scoring methods outlined in methods section. The plot indicates the percentage of positive staining cells in the two tumor populations. Student's t test was applied with p value of 0.009. D, NCOA3 is decreased in SYO-1 cells treated with antisense oligonucleotide for SYT-SSX1/2. Lane 1: control oligonucleotide; lane 2: antisense oligonucleotide. The relative intensity of the NCOA3 and ACTIN level were quantitated by PhotoshopTM histogram analysis. E, rat1 3Y1cells with reduced Ncoa3 are more apoptotic in anchorage-independent growth. Cells infected with a conditional siRNA for Ncoa3 or a control construct was treated with Adeno-LacZ or Adeno-Cre to activate the siRNA expression for 2 days before cells were seeded into methylcellulose. After 48 h, cells were collected from methylcellulose, and the percentage of TUNEL-positive cells was quantitated through FACS analysis. F, decreased Ncoa3 expression results abrogation of tumor formation in vivo. SYT-SSX1-expressing rat fibroblast and those with reduced Ncoa3 expression (clone 3) were injected into both flanks of two nude mice, tumor formation was assessed 4 weeks later with measurement of tumor volume.
NCOA3 Is Required for SYT-SSX1-mediated Tumor Formation
To determine whether increased Ncoa3 protein level is essential for the oncogenic activity of SYT-SSX1, we attempted siRNA knockdown of Ncoa3 in rat1 3Y1 cells expressing SYT-SSX1. We generated a conditionally expressing shRNA for Ncoa3 through Cre-mediated excision of the stop sequence in front of the shRNA expression cassette (35). As shown in Fig. 1E, compared with the control cells, the cells with shRNA for Ncoa3 exhibited a higher percentage of TUNEL positivity when they were suspended in methylcellulose, suggesting that these cells are more prone to anoikis, a parameter that is closely correlated with transformation. We also generated several individual clones with reduced Ncoa3 levels by the constitutive expression of the shRNA for Ncoa3 (clone 3 as in Fig. 1F, inset), and directly measured the tumor forming ability of the stable clone with reduced Ncoa3 level in immunocompromised mice. Cells with normal or reduced Ncoa3 were injected into the bilateral flanks of the mice, with 2 mice in each group. As shown in Fig. 1F, in a total of 8 injection sites of two inoculum sizes, SYT-SSX1-transformed rat1 fibroblasts with reduced Ncoa3 nearly completely lost its ability to form tumors compared with the vector control cells which formed robust tumors. These experiments establish a direct connection between SYT-SSX1 protein and the overexpression of Ncoa3, and demonstrate that Ncoa3 is required for the tumorigenic activity of SYT-SSX1.
Molecular Mechanisms of Increased NCOA3 Expression Induced by SYT-SSX1
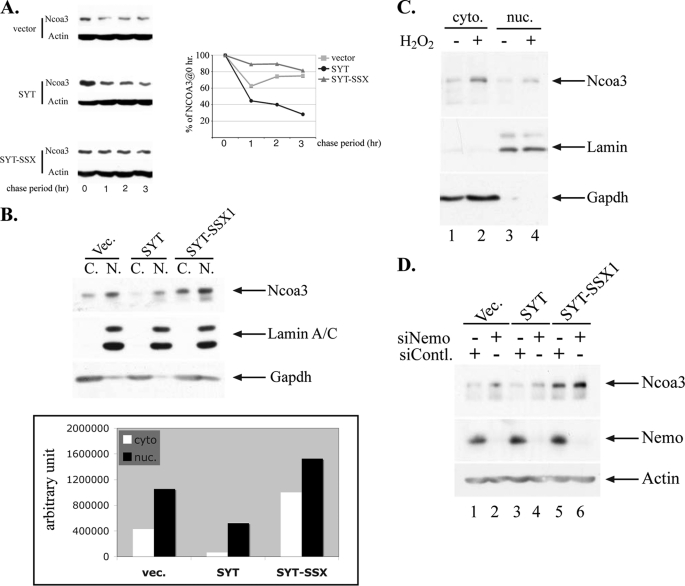
Expression of SYT-SSX1 is necessary and sufficient for an increased protein level of NCOA3, which is critical for SYT-SSX1-mediated tumor formation. To determine whether SYT-SSX1 can directly stabilize NCOA3 protein to increase its level, we measured the protein stability of NCOA3. Rat1 3Y1 cells stably expressing vector, SYT and SYT-SSX1 were treated with cyclohexamide to block the de novo protein synthesis, and the endogenous Ncoa3 levels measured at various time intervals after cyclohexamide addition. As can be seen in Fig. 2A, Ncoa3 stability was increased in SYT-SSX1-expressing cells compared with those expressing the vector or SYT alone. This is consistent with the finding that there is no increased transcript level of Ncoa3 in SYT-SSX1-expressing cells (Fig. 1B). As cellular localization is frequently correlated with protein stability (36), we also examined the subcellular distribution of Ncoa3. As shown in Fig. 2B, both nuclear and cytoplasmic Ncoa3 was increased in cells overexpressing SYT-SSX1.
FIGURE 2.
Molecular mechanisms of increased NCOA3 by SYT-SSX1. A, NCOA3 is more stable in SYT-SSX1-expressing cells. Rat1 3Y1 cells stably expressing vector, SYT and SYT-SSX1 (as in Fig. 1B) were treated with 10 μg/ml cycloheximide (CHX), and were collected at 0, 1, 2, 3 h post CHX addition. Ncoa3 was analyzed by immunoblot, and the graph depicts the quantitation of Ncoa3 level at those time points. B, Ncoa3 protein was more abundant in both cytoplasm (C.) and nucleus (N.) of SYT-SSX1-expressing cells than that in control and SYT-expressing cells. Rat fibroblast stably expressing various genes (as in Fig. 1B) were fractionated, and the cytosolic and nuclear fraction were loaded with equal volume and analyzed by SDS-PAGE. Gapdh and nuclear Lamin A/C were used as a marker for cytosolic and nuclear proteins. In the lower panel, relative intensity of the Ncoa3 level was quantitated by PhotoshopTM histogram analysis. C, Ncoa3 increases in response to oxidative stress. Rat1 3Y1 cells were treated with 2 mm H2O2 for 45 min, cells were collected followed by fractionation. The cytosolic (cyto.) and nuclear (nuc.) fraction were loaded with equal volume and analyzed by SDS-PAGE. The success of the fractionation was confirmed as in B. D, Nemo reduction leads to increased NCOA3 expression. Small interference RNA for Nemo or a control was transiently transfected for 48 h, and the lysate was analyzed by SDS-PAGE followed by immunoblot analysis of the Ncoa3 and Nemo level. Vec., vector.
It has been shown that sumoylation can regulate protein stability by antagonizing ubiquitin-mediated degradation (37, 38) as well as cytoplasmic-nuclear trafficking of a variety of molecules (39). NEMO, a structural component of the NF-kb kinase complex, is sumoylated specifically by the SUMO E3 ligase PIASy in response to DNA damage or oxidative stress, resulting in its nuclear translocation (40–42). As NEMO has also been shown to interact with NCOA3 (43), we examined whether NCOA3 followed the pattern of movement of NEMO in response to oxidative stress. We treated rat1 3Y1 cells with H2O2 and observed a rapid increase of NCOA3 protein level with a clear increase in both the cytoplasmic and nuclear compartment (Fig. 2C). These results support the notion that subcellular localization and protein level of NCOA3, similar to NEMO, can be regulated by oxidative stress.
We further hypothesized that SYT-SSX1 can modulate sumoylation of NEMO or NCOA3 thus regulate the subcellular localization as well as protein stability as sumoylation can directly compete with ubiquitination to negate proteasome-mediated protein degradation. Since NCOA3 can form a complex with NEMO, we examined whether NCOA3 is dependent upon NEMO for its nuclear localization and increased level. We measured Ncoa3 levels in cells treated with siRNA for Nemo or a control siRNA. As can be seen in Fig. 2D, siRNA-mediated knockdown of Nemo in the three cell lines led to an increase of Ncoa3 as demonstrated by immunoblot analysis. One mechanistic explanation for this phenomenon is that Nemo and Ncoa3 compete with each other for SUMO-E3 ligase-mediated sumoylation and thus reducing Nemo leads to increased sumoylation and protein level of Ncoa3.
SYT/SYT-SSX1 Binds PIASy and Regulates Its Sumoylation Activity
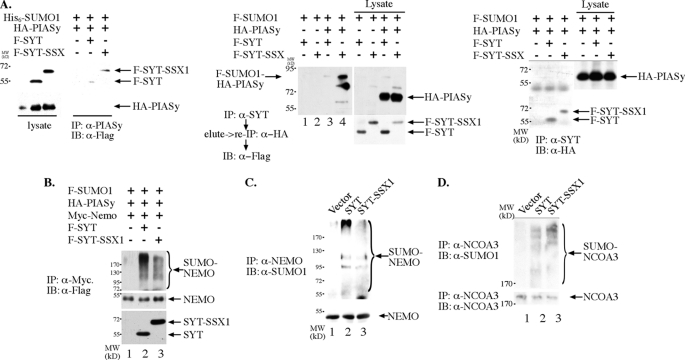
Because PIASy can directly sumoylate NEMO in response to DNA damage or oxidative stress (40–42), we examined whether SYT and SYT-SSX1 can directly interact with PIASy to influence the sumoylation of its substrates. As shown in the left panel of Fig. 3A, both SYT and SYT-SSX1 can interact with PIASy by their presence in the immunoprecipitates of PIASy. Their interaction is only detectable by co-expression of SUMO1 protein (data not shown), suggesting that sumoylation of PIASy might promote its interaction with SYT/SYT-SSX1. Consistent with that, when SYT or SYT-SSX1 was immunoprecipitated, SYT-SSX1 in particular as well as SYT was associated with the sumoylated form of the PIASy (the middle panel of Fig. 3A), but not with the unmodified form of PIASy (the right panel of Fig. 3A). PIASy is known to autosumoylate itself to increase its activity (44, 45). Thus the above results suggest that SYT and SYT-SSX1 can directly associate with the active form of PIASy. To examine whether SYT or SYT-SSX1 can regulate the SUMO E3 ligase activity of PIASy, we reconstituted the SUMO modification of NEMO in 293T cells with the expression of PIASy and epitope-tagged SUMO-1. As shown in Fig. 3B, expression of SYT as well as SYT-SSX1 dramatically increased the level of SUMO-1 in the immunoprecipitated NEMO. This increase may not have been due to an increased association of NEMO with sumoylated proteins in 293T cells since NEMO protein was immunoprecipitated from the heat-denatured lysate after centrifugation. Thus we conclude that wild type as well as SSX-fused SYT upon overexpression is capable of increasing sumoylation by PIASy, in this case, sumoylation of NEMO. We then verified that in rat1 3Y1 cells expressing either SYT or SYT-SSX1, Nemo protein was more sumoylated than that in the control cells (Fig. 3C). These results suggest that SYT and SYT-SSX1, through direct physical interaction with PIASy, can enhance its ability to sumoylate one of its substrates, NEMO.
FIGURE 3.
SYT and SYT-SSX can regulate sumoylation by SUMO E3 PIASy. A, SYT and SYT-SSX1 interact with PIASy. Epitope-tagged SYT, SYT-SSX1, and PIASy were co-transfected into 293T cells. PIASy was immunoprecipitated with anti-PIASy antibody, separated by SDS-PAGE, and analyzed with anti-Flag antibody to reveal the interacting SYT and SYT-SSX1 protein. For the middle panel, SYT and SYT-SSX1 were immunoprecipitated with anti-SYT antibody. The bound proteins were released with 1% SDS buffer with heating at 70 °C for 5 min followed by dilution to 0.1% SDS and immunoprecipitation with anti-HA antibody to captures the HA-PIASy protein. The immunoprecipitates were analyzed with SDS-PAGE followed by immunoblot with anti-Flag antibody to reveal the sumoylated PIASy. On the right panel, SYT and SYT-SSX1 were immunoprecipitated with anti-SYT antibody, and the immunoprecipitates separated by SDS-PAGE followed by immunoblot with anti-HA antibody but failed to detect any interacting PIASy. B, SYT and SYT-SSX1 can stimulate NEMO sumoylation by PIASy. Epitope-tagged SYT, SYT-SSX1, NEMO, PIASy, and SUMO-1were co-transfected into 293T cells, the lysate was boiled in 95 °C for 5 min followed by centrifugation. The cleared supernatant was immunoprecipitated with anti-Myc antibody, separated on SDS-PAGE and analyzed with anti-Flag antibody to reveal the extent of NEMO sumoylation. C, Nemo protein was more sumoylated in SYT- and SYT-SSX-expressing rat1 fibroblast cells. The Nemo protein was immunoprecipitated, separated by SDS-PAGE, and immunoblotted with anti-SUMO1 antibody to reveal the extent of Nemo sumoylation in vector (1), SYT (2), and SYT-SSX (3) cells. D, Ncoa3 sumoylation in rat1 3Y1 cells. Ncoa3 was immunoprecipitated from rat1 3Y1 cells stably expressing vector, SYT, and SYT-SSX1 and immunoblotted with anti-SUMO1 antibody to reveal the extent of Ncoa3 sumoylation. The amount of immunoprecipitated Ncoa3 was visualized through anti-NCOA3 immunoblot shown in the lower panel.
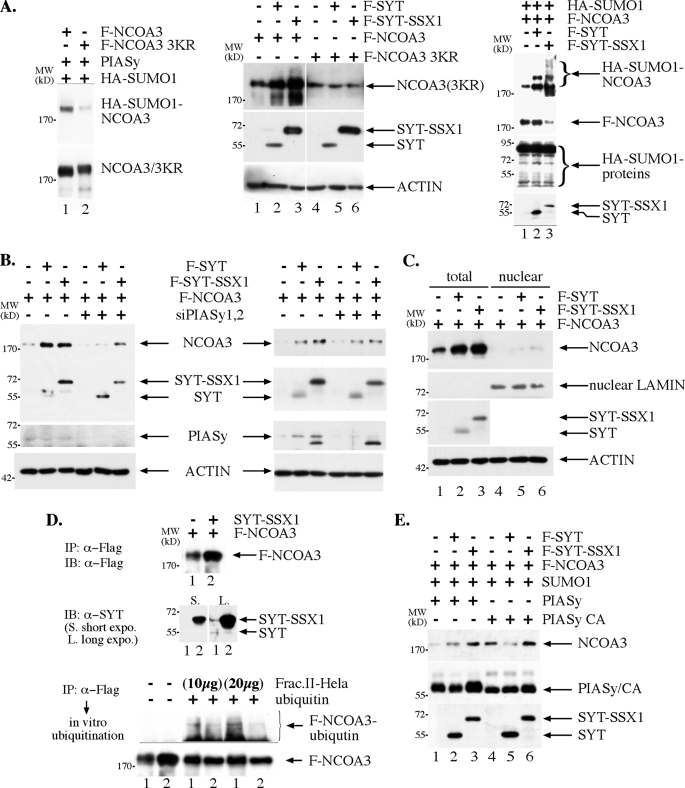
It is known that NCOA3 itself can also be modified through sumoylation (46), and sumoylation can compete with ubiquitination, if such modification occurs on the same lysine residue (37, 38). Indeed, in rat1 3Y1 fibroblast cells, expression of SYT and SYT-SSX1 results in increased sumoylation of Ncoa3 (Fig. 3D). We thus tested whether SYT and SYT-SSX1 can regulate the expression of NCOA3 in a reconstitution system such as human embryonic kidney cells (293T cells). As shown in the middle panel of Fig. 4A, expression of both SYT and SYT-SSX1 increased the protein level of NCOA3. We also generate a mutant NCOA3 where its consensus sumoylation sites were changed from lysines to arginines as shown in Fig. 4A, left panel. In contrast to wild-type NCOA3, the expression level of this mutant is not increased by the co-expression of SYT or SYT-SSX1 (Fig. 4A, middle panel), suggesting that SYT or SYT-SSX1 increases NCOA3 through enhancing the latter sumoylation. Consistent with this finding, while general protein sumoylation is not affected in cells expressing SYT and SYT-SSX1, sumoylation of NCOA3 was significantly increased in cells co-expressing either SYT or SYT-SSX as shown in right panel of Fig. 4A. As NEMO and NCOA3 can form a complex (43) and likely compete for substrate sumoylation (Fig. 2D), it suggests that PIASy is a SUMO E3 ligase for NCOA3 as well. In support of this notion, PIASy knockdown through two independent siRNA (Fig. 4B) strongly reduced the impact of SYT or SYT-SSX1 on the steady state level of NCOA3, suggesting that SYT/SYT-SSX1 through interaction with PIASy enhances the sumoylation of NCOA3, another substrate of PIASy. Although SYT-SSX seems to be able to increase NCOA3 level in cells with a knockdown of PIASy compared with those in vector control and SYT-expressing cells (Fig. 4B), it is possible that residual PIASy as a result of incomplete elimination of PIASy by siRNA is potent enough to increase the NCOA3 through sumoylation. Alternatively, it is possible that protein-protein interaction between SYT-SSX1 and NCOA3 affords NCOA3 increased stability thus a higher level compared with that in the control and SYT-expressing cells. Interestingly, the results shown in Fig. 4C suggest that in 293T cells, SYT and SYT-SSX1-mediated increase of NCOA3 sumoylation does not lead to its enhanced nuclear localization as in rat1 3Y1 cells, rather the majority of NCOA3 stays in the cytoplasm with an increased level from sumoylation-mediated protection against degradation. To directly test whether the increased NCOA3 in SYT-SSX1-expressing cells is a result of resistance to ubiquitin-mediated degradation due to increased sumoylation (Fig. 4A, right panel), we purified NCOA3 through immunoprecipitation from 293T cells where it was expressed alone or together with SYT-SSX1. These two versions of NCOA3 were then subjected to in vitro ubiquitination with Fraction II of Hela cells as a source of ubiquitin E1, E2s and E3s. As can be seen in Fig. 4D, NCOA3 isolated from co-expression of SYT-SSX1 was less ubiquitinated in comparison to when it was expressed alone although the input level of NCOA3 was higher when it was isolated from co-expression with SYT-SSX1 (Fig. 4D, upper panel). This direct biochemical evidence supports the notion that SYT-SSX1 can increase the steady level of NCOA3 through latter's resistance to ubiquitin-mediated degradation upon enhanced sumoylation.
FIGURE 4.
NCOA3 sumoylation leads to a higher steady state level. A, epitope-tagged NCOA3, SYT, SYT-SSX1, were co-transfected into 293T cells, and the amount of NCOA3 was examined with anti-Flag antibody in an immunoblot following SDS-PAGE analysis. Left panel: mutant NCOA3 protein with the consensus sumoylation sites (47) changed to arginine was significantly less sumoylated than the wild-type protein. The two proteins were immunoprecipitated by anti-Flag antibody followed by SDS-PAGE and immunoblot analysis. The PVDF membrane was first probed with anti-HA epitope antibody to reveal the extent of sumoylation shown in the top part. The blot was then stripped and re-probed with anti-Flag antibody to indicate approximate equal amount of protein between the wild type and the mutant F-NCOA3(3KR) shown in the lower part. Middle panel: expression of wild-type NCOA3 was increased by SYT and SYT-SSX1, while that of mutant was not. The exposure for the NCOA3 3KR was longer than that for the wild-type protein. Right panel: NCOA3 sumoylation in 293T was increased with co-expression of SYT and SYT-SSX1. For the right panel, Flag-NCOA3 was immunoprecipitated from 100% lysate (1), 40% lysate (2), and 30% lysate (3) with anti-Flag antibody followed by anti-HA immunoblot to reveal the sumoylation of NCOA3. The blot was stripped and reprobed with anti-Flag antibody to assess the amount of immunoprecipitated Flag-NCOA3. Separately, an equal amount of lysate from the three cell populations was analyzed in an immunoblot with anti-HA antibody to assess the general protein sumoylation in the control, SYT and SYT-SSX1-expressing cells. B, PIASy is required for the SYT and SYT-SSX1-mediated increase of NCOA3 in 293T cells. Two sets of siRNA against PIASy were cotransfected with expression plasmids for NCOA3, SYT, and SYT-SSX1, the steady state level of NCOA3 was examined as in A. C, SYT- and SYT-SSX1-mediated increase of NCOA3 is not dependent upon nuclear localization of NCOA3 in 293T cells. Expression plasmids were transfected as in A, and cellular fractionation was conducted as in Fig. 2B. D, SYT-SSX1 expression causes the NCOA3 to be less ubiquitinated in vitro. Flag-NCOA3 was immunoprecipitated with anti-Flag antibody from 293T cells alone (lane 1) or with co-expression of SYT-SSX1 (lane 2), and used as a substrate for in vitro ubiquitination assay. Upon incubation at 30 °C for 60 min with 20 or 40 μg Hela lysate, the NCOA3 protein was again washed, separated by SDS-PAGE, and immunoblotted with anti-Flag antibody to reveal the NCOA3 ubiquitination. The bottom part is a short exposure of the blot to reveal the input F-NCOA3 protein. E, a decreased PIASy activity contributes to a lower NCOA3 level in 293T cells. Expression plasmids were transfected, and NCOA3 analyzed as in A.
SYT and SYT-SSX1 Are Functionally Different
The results in the reconstituted 293T cells demonstrate that both SYT and SYT-SSX1 can similarly enhance the expression of NCOA3. Yet in rat1 3Y1 fibroblast cells, SYT expression actually leads to a decrease of NCOA3 protein level (Fig. 1B). It is known that NCOA3 protein level is subject to regulation through both ubiquitin-dependent and ubiquitin-independent mechanisms by REGγ and aPKC as well as several ubiquitin E3 ligases, such as Fbw7α, E6AP, and CHIP (47–52). It is possible that SYT is less potent than SYT-SSX1 in increasing sumoylation of NCOA3, thus at certain cellular level of PIASy activity, SYT is no longer able to protect NCOA3 protein through sumoylation. We therefore compared SYT and SYT-SSX1 activity in regulating NCOA3 levels in response to different levels of PIASy expression. As shown in Fig. 4E, when SYT and SYT-SSX1 were coexpressed with the active form of PIASy together with SUMO-1, they similarly increased NCOA3 expression. However, in response to expression of a catalytically inactive PIASy, SYT expression decreased NCOA3 level while SYT-SSX1 continued to increase the expression of NCOA3 in 293T cells. These results suggest that SYT-SSX1 is more potent than SYT in inducing NCOA3 sumoylation when limited amount of active PIASy is present. The higher potency of SYT-SSX1 in inducing NCOA3 sumoylation may derive from its interaction with NCOA3 to forge a stronger enzyme and substrate interaction. SYT, on the other hand, will decrease the expression of NCOA3 in response to decreased PIASy sumoylation activity through a mechanism not yet understood. These data also suggest that in rat1 3Y1 fibroblast cells, there may be a lower PIASy activity toward the NCOA3 protein. As a result, SYT expression leads to a lower protein level of NCOA3, similar to cells with expression of catalytically inactive PIASy. Thus, there is a fundamental difference between SYT and its oncogenic descendant SYT-SSX1 in that the latter is more potent in increasing sumoylation of NCOA3, leading to a higher steady state protein level of NCOA3 (Fig. 5).
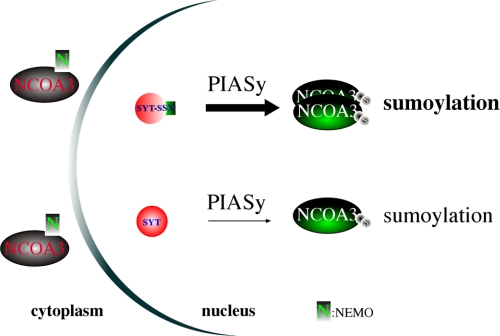
FIGURE 5.
A schematic diagram illustrating differential sumoylation of NCOA3 by SYT-SSX1 results in its overexpression.
DISCUSSION
We report here that NCOA3 protein is critical for tumor formation induced by the synovial sarcoma oncoprotein SYT-SSX1. SYT-SSX1 increases the steady state level of NCOA3 through direct protein-protein interaction with a SUMO E3 ligase PIASy that increases sumoylation of NCOA3 as well as NEMO, an NCOA3-interacting protein.
It is clear that increased steady state level of NCOA3 is directly linked with increased sumoylation of NCOA3 as SYT-SSX1/SYT cannot enhance the protein level of NCOA3 3KR mutant protein with consensus sumoylation sites changed (Fig. 4A). Moreover, in vitro biochemical evidence also supports the notion that sumoylation suppresses ubiquitination (Fig. 4D), thus likely reduces ubiquitin-mediated degradation, hence increases protein abundance. Because there are several ubiquitin-E3 ligases that can lead to degradation of NCOA3, including Fbw7α, E6AP, and CHIP (47, 51–52), it is not clear at the moment which one of them, or all of them, is sensitive to SUMO-induced resistance. It is also possible that sumoylation alters NCOA3 sensitivity toward ubiquitin-independent REGγ-mediated degradation of NCOA3 (49). Future experiments will be required to clarify these issues. We found that the expression of NCOA3 3KR mutant driven by the same promoter for the wild-type protein will result a lower level than that of the wild-type protein (Fig. 4A, middle panel). Such a decreased expression level, although appears counter-intuitive because the ubiquitination sites are mutated thus protein should be more stable and higher in level, is still compatible with our proposed molecular mechanism (Fig. 5). One possibility is that these lysines are also ubiquitination sites that are important for the transactivation potency by NCOA3, a phospho-dependent ubiquitin clock for this transactivator (47). Without ubiquitination, mutant NCOA3 cannot serve as an efficient transcriptional coactivator to generate transcript for this protein, thus low in protein level, yet the mutant 3KR protein is no longer sensitive to sumoylation-enhanced protein stability. It is also possible that separate, additional lysine residues on NCOA3 are responsible for ubiquitin-mediated degradation via ubiquitin-dependent and -independent mechanisms (47–52). As such, the sumoylation on the consensus sumoylation sites may alter the conformation of the NCOA3 to reduce degradation, and without sumoylation the protein is more labile for degradation. Alternatively, these lysines are necessary ubiquitination sites without which there is no robust protein degradation. Yet without modification by either ubiquitination or sumoylation, the NCOA3 protein will adopt an unstable conformation resulting in lower protein level, and is not responsive to SYT- and SYT-SSX1-induced stability. In all these potential scenarios, sumoylation on these sites does act as a direct modulation against ubiquitination to confer increased stability of NCOA3.
We also found that NCOA3 behaves differently in the 293T reconstitution system than in the rat1 fibroblast cells. In the latter, there is preferential nuclear localization of NCOA3 while NCOA3 largely resides in the cytoplasm in 293T cells (Fig. 4C). A possible explanation is that the highly expressed NCOA3 in the transient expression system of 293T cells overwhelmed the nuclear transport machinery, while stable expression of SYT, SYT-SSX1, and Ncoa3 at moderate levels in rat1 fibroblast cells results in their enhanced nuclear localization in a sumoylation-dependent manner. Consistent with this, Ncoa3 expression is enhanced when Nemo is reduced as these two proteins may compete for PIASy-mediated sumoylation.
Our study demonstrates that at the molecular level SYT and SYT-SSX are very different proteins even though they possess similar biochemical activity in their interaction with and regulation of PIASy. In the context of 293T reconstitution system where SYT, SYT-SSX1 and NCOA3 are all driven by ectopic expression, SYT and SYT-SSX1 behave similarly in that they can both increase the sumoylation and thus level of NCOA3 (Fig. 4, A–E). Yet in rat1 3Y1 fibroblast cells with stable expression of SYT and SYT-SSX1, only SYT-SSX1 leads to enhanced expression of Ncoa3 while SYT actually results in a reduction of Ncoa3 (Figs. 1B and 2B). We suggest that this reduction might result from an insufficient level of PIASy activity in rat1 fibroblast cells as we could mimic this scenario in the 293T cells by expression of a catalytic inactive PIASy (Fig. 4E). SYT as a normal cellular protein participates in a variety of protein-protein interactions and cellular activities, is thus less potent in interacting with and enhancing PIASy activity while SYT-SSX seems to be locked in a state of protein-protein interaction with PIASy to regulate NCOA3. In this sense, SYT-SSX behaves as a dominant oncoprotein, similar to the dominant active mutant Ras, Bcr-Abl, and others, and stays in a constitutively “ON“ state to promote downstream signaling ultimately resulting in cellular transformation.
The role of sumoylation in tumor formation has been well documented. A variety of oncoproteins and tumor suppressors have been found to be modified by sumoylation. For example, the PML-RARα fusion that is causal for acute promyelocytic leukemia requires intact sumoylation for its ability to transform cells (53), while arsenic trioxide-induced hypersumoylation likely causes its degradation, resulting in a therapeutic efficacy (54–56). The activities of p19Arf, p53, and Mdm2 have all been linked with sumoylation although how sumoylation regulates their abilities to cause cellular transformation is not clear (57). Reduced sumoylation by a hypomorphic allele of a SUMO E3 ligase RanBP2 on Topoisomerase 2 and enhanced chromosomal abnormalities have been causally linked to enhanced tumor formation in a mouse model (58). Our results show, for the first time, that regulation of SUMO E3 ligase by a human oncoprotein results in differential substrate modification and causes an increase of a protein critical for cellular transformation. The mechanistic understanding of SYT-SSX1 ability to up-regulate NCOA3 makes it possible to design target-specific therapies for synovial sarcoma, a cancer currently without an effective treatment beyond surgical resection.
Acknowledgments
We thank Dr. Shigeki Miyamoto for sharing the plasmids for NEMO, wild type, and mutant PIASy and SUMO-1; Dr. Tyler Jacks for pSico plasmid. We thank Dr. Dingcheng Gao for initial in vitro identification of NCOA3 binding to SYT-SSX1. We also thank Ngan B. Doan for performing the immunohistochemical staining and Clara Magyar and Tim Wen for help on tissue microarray analysis.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 CA093848 (to Y. S.). This work was also supported by Grants from the American Cancer Society (RSG-07-092-01-TBE), Dept. of Defense Prostate Cancer Research Program (PC061456), a developmental research award from UCLA SPORE in prostate cancer (PI: R. Reiter) and a challenge award from the Prostate Cancer Foundation (PI: O. Witte) (to J. H.).
- SYT
- synovial sarcoma translocated
- TMA
- tissue microarray.
REFERENCES
- 1. Ladanyi M. (2001) Oncogene 20, 5755–5762 [DOI] [PubMed] [Google Scholar]
- 2. Clark J., Rocques P. J., Crew A. J., Gill S., Shipley J., Chan A. M., Gusterson B. A., Cooper C. S. (1994) Nat. Genet. 7, 502–508 [DOI] [PubMed] [Google Scholar]
- 3. Crew A. J., Clark J., Fisher C., Gill S., Grimer R., Chand A., Shipley J., Gusterson B. A., Cooper C. S. (1995) EMBO J. 14, 2333–2340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. de Leeuw B., Balemans M., Olde Weghuis D., Geurts van Kessel A. (1995) Hum. Mol. Genet. 4, 1097–1099 [DOI] [PubMed] [Google Scholar]
- 5. dos Santos N. R., de Bruijn D. R., van Kessel A. G. (2001) Genes. Chromosomes Cancer. 30, 1–14 [DOI] [PubMed] [Google Scholar]
- 6. Limon J., Dal Cin P., Sandberg A. A. (1986) Cancer. Genet. Cytogenet. 23, 87–91 [DOI] [PubMed] [Google Scholar]
- 7. Panagopoulos I., Mertens F., Isaksson M., Limon J., Gustafson P., Skytting B., Akerman M., Sciot R., Dal Cin P., Samson I., Iliszko M., Ryoe J., Dêbiec-Rychter M., Szadowska A., Brosjö O., Larsson O., Rydholm A., Mandahl N. (2001) Genes Chromosomes Cancer 31, 362–372 [DOI] [PubMed] [Google Scholar]
- 8. Skytting B., Nilsson G., Brodin B., Xie Y., Lundeberg J., Uhlén M., Larsson O. (1999) J. Natl. Cancer. Inst. 91, 974–975 [DOI] [PubMed] [Google Scholar]
- 9. Smith S., Reeves B. R., Wong L., Fisher C. (1987) Cancer. Genet. Cytogenet. 26, 179–180 [DOI] [PubMed] [Google Scholar]
- 10. Guillou L., Coindre J., Gallagher G., Terrier P., Gebhard S., de Saint Aubain Somerhausen N., Michels J., Jundt G., Vince D. R., Collin F., Trassard M., Le Doussal V., Benhattar J. (2001) Hum. Pathol. 32, 105–112 [DOI] [PubMed] [Google Scholar]
- 11. Hiraga H., Nojima T., Abe S., Sawa H., Yamashiro K., Yamawaki S., Kaneda K., Nagashima K. (1998) Diagn. Mol. Pathol. 7, 102–110 [DOI] [PubMed] [Google Scholar]
- 12. Ladanyi M., Bridge J. A. (2000) Hum. Pathol. 31, 532–538 [DOI] [PubMed] [Google Scholar]
- 13. Poteat H. T., Corson J. M., Fletcher J. A. (1995) Cancer. Genet. Cytogenet. 84, 76–81 [DOI] [PubMed] [Google Scholar]
- 14. Willeke F., Mechtersheimer G., Schwarzbach M., Weitz J., Zimmer D., Lehnert T., Herfarth C., von Knebel Doeberitz M., Ridder R. (1998) Eur. J. Cancer. 34, 2087–2093 [DOI] [PubMed] [Google Scholar]
- 15. Nagai M., Tanaka S., Tsuda M., Endo S., Kato H., Sonobe H., Minami A., Hiraga H., Nishihara H., Sawa H., Nagashima K. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 3843–3848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Haldar M., Hancock J. D., Coffin C. M., Lessnick S. L., Capecchi M. R. (2007) Cancer Cell 11, 375–388 [DOI] [PubMed] [Google Scholar]
- 17. Eid J. E., Kung A. L., Scully R., Livingston D. M. (2000) Cell 102, 839–848 [DOI] [PubMed] [Google Scholar]
- 18. Iwasaki T., Koibuchi N., Chin W. W. (2005) Endocrinology 146, 3892–3899 [DOI] [PubMed] [Google Scholar]
- 19. Perani M., Antonson P., Hamoudi R., Ingram C. J., Cooper C. S., Garrett M. D., Goodwin G. H. (2005) J. Biol. Chem. 280, 42863–42876 [DOI] [PubMed] [Google Scholar]
- 20. Thaete C., Brett D., Monaghan P., Whitehouse S., Rennie G., Rayner E., Cooper C. S., Goodwin G. (1999) Hum. Mol. Genet. 8, 585–591 [DOI] [PubMed] [Google Scholar]
- 21. Pretto D., Barco R., Rivera J., Neel N., Gustavson M. D., Eid J. E. (2006) Oncogene. 25, 3661–3669 [DOI] [PubMed] [Google Scholar]
- 22. Kato H., Tjernberg A., Zhang W., Krutchinsky A. N., An W., Takeuchi T., Ohtsuki Y., Sugano S., de Bruijn D. R., Chait B. T., Roeder R. G. (2002) J. Biol. Chem. 277, 5498–5505 [DOI] [PubMed] [Google Scholar]
- 23. Sun Y., Gao D., Liu Y., Huang J., Lessnick S., Tanaka S. (2006) Oncogene 25, 1042–1052 [DOI] [PubMed] [Google Scholar]
- 24. Zheng L., Liu J., Batalov S., Zhou D., Orth A., Ding S., Schultz P. G. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 135–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Palapattu G. S., Wu C., Silvers C. R., Martin H. B., Williams K., Salamone L., Bushnell T., Huang L. S., Yang Q., Huang J. (2009) Prostate 69, 787–798 [DOI] [PubMed] [Google Scholar]
- 26. Anzick S. L., Kononen J., Walker R. L., Azorsa D. O., Tanner M. M., Guan X. Y., Sauter G., Kallioniemi O. P., Trent J. M., Meltzer P. S. (1997) Science 277, 965–968 [DOI] [PubMed] [Google Scholar]
- 27. Glaeser M., Floetotto T., Hanstein B., Beckmann M. W., Niederacher D. (2001) Horm. Metab. Res. 33, 121–126 [DOI] [PubMed] [Google Scholar]
- 28. Gnanapragasam V. J., Leung H. Y., Pulimood A. S., Neal D. E., Robson C. N. (2001) Br. J. Cancer. 85, 1928–1936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. List H. J., Reiter R., Singh B., Wellstein A., Riegel A. T. (2001) Breast. Cancer. Res. Treat. 68, 21–28 [DOI] [PubMed] [Google Scholar]
- 30. Sakakura C., Hagiwara A., Yasuoka R., Fujita Y., Nakanishi M., Masuda K., Kimura A., Nakamura Y., Inazawa J., Abe T., Yamagishi H. (2000) Int. J. Cancer. 89, 217–223 [PubMed] [Google Scholar]
- 31. Torres-Arzayus M. I., Font de Mora J., Yuan J., Vazquez F., Bronson R., Rue M., Sellers W. R., Brown M. (2004) Cancer. Cell. 6, 263–274 [DOI] [PubMed] [Google Scholar]
- 32. Chung A. C., Zhou S., Liao L., Tien J. C., Greenberg N. M., Xu J. (2007) Cancer Res. 67, 5965–5975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kuang S. Q., Liao L., Zhang H., Lee A. V., O'Malley B. W., Xu J. (2004) Cancer Res. 64, 1875–1885 [DOI] [PubMed] [Google Scholar]
- 34. Kawai A., Naito N., Yoshida A., Morimoto Y., Ouchida M., Shimizu K., Beppu Y. (2004) Cancer Lett. 204, 105–113 [DOI] [PubMed] [Google Scholar]
- 35. Ventura A., Meissner A., Dillon C. P., McManus M., Sharp P. A., Van Parijs L., Jaenisch R., Jacks T. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 10380–10385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Freedman D. A., Levine A. J. (1998) Mol. Cell. Biol. 18, 7288–7293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bae S. H., Jeong J. W., Park J. A., Kim S. H., Bae M. K., Choi S. J., Kim K. W. (2004) Biochem. Biophys. Res. Commun. 324, 394–400 [DOI] [PubMed] [Google Scholar]
- 38. Desterro J. M., Rodriguez M. S., Hay R. T. (1998) Mol. Cell 2, 233–239 [DOI] [PubMed] [Google Scholar]
- 39. Pichler A., Melchior F. (2002) Traffic. 3, 381–387 [DOI] [PubMed] [Google Scholar]
- 40. Huang T. T., Wuerzberger-Davis S. M., Wu Z. H., Miyamoto S. (2003) Cell 115, 565–576 [DOI] [PubMed] [Google Scholar]
- 41. Wuerzberger-Davis S. M., Nakamura Y., Seufzer B. J., Miyamoto S. (2007) Oncogene. 26, 641–651 [DOI] [PubMed] [Google Scholar]
- 42. Mabb A. M., Wuerzberger-Davis S. M., Miyamoto S. (2006) Nat. Cell. Biol. 8, 986–993 [DOI] [PubMed] [Google Scholar]
- 43. Wu R. C., Qin J., Hashimoto Y., Wong J., Xu J., Tsai S. Y., Tsai M. J., O'Malley B. W. (2002) Mol. Cell. Biol. 22, 3549–3561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Schmidt D., Müller S. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 2872–2877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gocke C. B., Yu H., Kang J. (2005) J. Biol. Chem. 280, 5004–5012 [DOI] [PubMed] [Google Scholar]
- 46. Wu H., Sun L., Zhang Y., Chen Y., Shi B., Li R., Wang Y., Liang J., Fan D., Wu G., Wang D., Li S., Shang Y. (2006) J. Biol. Chem. 281, 21848–21856 [DOI] [PubMed] [Google Scholar]
- 47. Wu R. C., Feng Q., Lonard D. M., O'Malley B. W. (2007) Cell. 129, 1125–1140 [DOI] [PubMed] [Google Scholar]
- 48. Yi P., Feng Q., Amazit L., Lonard D. M., Tsai S. Y., Tsai M. J., O'Malley B. W. (2008) Mol. Cell. 29, 465–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Li X., Amazit L., Long W., Lonard D. M., Monaco J. J., O'Malley B. W. (2007) Mol. Cell. 26, 831–842 [DOI] [PubMed] [Google Scholar]
- 50. Lonard D. M., Nawaz Z., Smith C. L., O'Malley B. W. (2000) Mol. Cell. 5, 939–948 [DOI] [PubMed] [Google Scholar]
- 51. Mani A., Oh A. S., Bowden E. T., Lahusen T., Lorick K. L., Weissman A. M., Schlegel R., Wellstein A., Riegel A. T. (2006) Cancer. Res. 66, 8680–8686 [DOI] [PubMed] [Google Scholar]
- 52. Kajiro M., Hirota R., Nakajima Y., Kawanowa K., So-ma K., Ito I., Yamaguchi Y., Ohie S. H., Kobayashi Y., Seino Y., Kawano M., Kawabe Y., Takei H., Hayashi S., Kurosumi M., Murayama A., Kimura K., Yanagisawa J. (2009) Nat. Cell. Biol. 11, 312–319 [DOI] [PubMed] [Google Scholar]
- 53. Zhu J., Zhou J., Peres L., Riaucoux F., Honoré N., Kogan S., de Thé H. (2005) Cancer. Cell 7, 143–153 [DOI] [PubMed] [Google Scholar]
- 54. Zhang X. W., Yan X. J., Zhou Z. R., Yang F. F., Wu Z. Y., Sun H. B., Liang W. X., Song A. X., Lallemand-Breitenbach V., Jeanne M., Zhang Q. Y., Yang H. Y., Huang Q. H., Zhou G. B., Tong J. H., Zhang Y., Wu J. H., Hu H. Y., de The H., Chen S. J., Chen Z. (2010) Science 328, 240–243 [DOI] [PubMed] [Google Scholar]
- 55. Tatham M. H., Geoffroy M. C., Shen L., Plechanovova A., Hattersley N., Jaffray E. G., Palvimo J. J., Hay R. T. (2008) Nat. Cell. Biol. 10, 538–546 [DOI] [PubMed] [Google Scholar]
- 56. Lallemand-Breitenbach V., Jeanne M., Benhenda S., Nasr R., Lei M., Peres L., Zhou J., Zhu J., Raught B., de Thé H. (2008) Nat. Cell. Biol. 10, 547–555 [DOI] [PubMed] [Google Scholar]
- 57. Kim K. I., Baek S. H. (2006) Mol. Cells 22, 247–253 [PubMed] [Google Scholar]
- 58. Dawlaty M. M., Malureanu L., Jeganathan K. B., Kao E., Sustmann C., Tahk S., Shuai K., Grosschedl R., van Deursen J. M. (2008) Cell 133, 103–115 [DOI] [PMC free article] [PubMed] [Google Scholar]