Abstract
Sphingosine kinase 1 (SK1) catalyzes the conversion of sphingosine to the bioactive lipid sphingosine 1-phosphate. We have previously demonstrated that FTY720 and (S)-FTY720 vinylphosphonate are novel inhibitors of SK1 activity. Here, we show that (S)-FTY720 vinylphosphonate binds to a putative allosteric site in SK1 contingent on formation of the enzyme-sphingosine complex. We report that SK1 is an oligomeric protein (minimally a dimer) containing noncooperative catalytic sites and that the allosteric site exerts an autoinhibition of the catalytic site. A model is proposed in which (S)-FTY720 vinylphosphonate binding to and stabilization of the allosteric site might enhance the autoinhibitory effect on SK1 activity. Further evidence for the existence of allosteric site(s) in SK1 was demonstrated by data showing that two new FTY720 analogues (a conjugate of sphingosine with a fluorophore and (S)-FTY720 regioisomer) increased SK1 activity, suggesting relief of autoinhibition of SK1 activity. Comparisons with the SK1 inhibitor, SKi or siRNA knockdown of SK1 indicated that (S)-FTY720 vinylphosphonate and FTY720 behave as typical SK1 inhibitors in preventing sphingosine 1-phosphate-stimulated rearrangement of actin in MCF-7 cells. These findings are discussed in relation to the anticancer properties of SK1 inhibitors.
Keywords: Apoptosis, Cytoskeleton, Enzyme Inhibitors, Enzyme Kinetics, Sphingolipid
Introduction
Sphingosine 1-phosphate (S1P)2 is a bioactive lipid that binds to specific G protein-coupled receptors (S1P1–5). There is a wealth of evidence to demonstrate that S1P plays key roles in regulating many critical processes in cell biology and, indeed, in pathophysiological diseases such as cancer (1). The levels of S1P are defined by its synthesis (involving sphingosine kinase, which phosphorylates sphingosine; there are two isoforms termed SK1 and SK2) and removal (involving hydrolysis of S1P catalyzed by S1P lyase or dephosphorylation by S1P phosphatase). To date, emphasis has been placed on the discovery of new molecules that are capable of agonizing/antagonizing S1P1–5. In this regard, FTY720 (FingolimodTM) has been developed as a functional S1P1 antagonist by virtue of its ability to down-regulate S1P1 in T lymphocytes (2) and is now licensed (as Gilenya) for oral treatment of multiple sclerosis (3). FTY720 is a synthetic sphingosine analogue that is taken up by cells and phosphorylated to FTY720 phosphate by SK2 (4). On release from cells, (S)-FTY720 phosphate binds to four of the five S1P receptors (S1P2 being the exception) (2), to elicit polyubiquitination, endocytosis, and degradation of S1P1 in T lymphocytes (5). S1P/S1P1 is essential for effective T lymphocyte egress and, therefore, the FTY720 phosphate-induced down-regulation of S1P1 on T lymphocytes results in their retention in the lymph (6).
An alternative therapeutic approach is to use a S1P-neutralizing antibody that reduces bioavailability of S1P at its receptors. This has proven very successful as an antitumorigenic and antiangiogenic agent in a number of disease models (7) and is being tested in clinical trials for cancer and age-related macular degeneration. Thus, current target intervention is confined to inhibiting S1P binding to receptors or limiting bioavailability of S1P at these receptors.
Recently, novel intracellular targets of S1P have been identified. Prominent examples are histone deacetylase (8) and TNF receptor-associated factor-2 (9). These findings underscore the importance of limiting the intracellular action of S1P. Indeed, there is evidence for a role for SK1 in human cancers, such as increased levels of SK1 mRNA transcript and/or SK1 protein in cancers of the stomach, lung, brain, colon, kidney, non-Hodgkins lymphoma, and breast (see review, see Ref. 1). For example, high expression of SK1 and cognate S1P1 and S1P3 receptors is correlated with poor survival rates and induction of Tamoxifen resistance in estrogen receptor-positive breast cancer patients, where the average survival time is reduced from 18 years to 7.5 years and recurrence of the disease in patients receiving Tamoxifen is shortened by 8 years in the high SK1 expression group (10, 11). In addition, SK1 induces chemotherapeutic resistance in androgen-independent prostate cancer cells (a model for castrate-independent prostate cancer), indicating that this enzyme might have a role in the generic development of chemotherapeutic resistance in cancer (12). Moreover, siRNA knockdown of SK1 significantly inhibited aberrant crypt formation and cancer development in a mouse model of colon cancer, suggesting a key role for SK1 in both early and late stages of carcinogenesis (13).
Previously, we reported that FTY720 inhibits purified SK1 activity and induces the proteasomal degradation of SK1 in mammalian cells (14). We also established that the (R) and (S) enantiomers of FTY720 vinylphosphonate (S > R) are inhibitors of purified SK1 and demonstrated that (S)-FTY720 vinylphosphonate induced its proteasomal degradation in cells, whereas their saturated counterpart, (R)- or (S)-FTY720 phosphonate, and (S)-FTY720 phosphate do not significantly inhibit the enzyme (14). To elucidate the mechanisms by which specific FTY720 analogues function as SK1 inhibitors, we have now studied their behavior with respect to enzyme inhibition kinetics, proteasomal degradation of SK1, and rearrangement of actin in MCF-7 breast cancer cells. These studies revealed that (S)-FTY720 vinylphosphonate appears to inhibit SK1 via a putative allosteric site interaction. FTY720 and (S)-FTY720 vinylphosphonate induced the proteasomal degradation of SK1 and abrogated the S1P-stimulated rearrangement of actin into lamellipodia/membrane ruffles required for cell migration. These findings indicate that FTY720 analogues are SK1 inhibitors that exhibit novel properties that are favorable for anticancer activity.
EXPERIMENTAL PROCEDURES
Materials
All general biochemicals and anti-actin antibody were from Sigma. High glucose Dulbecco's modified Eagle's medium (DMEM), Minimum Essential Medium (MEM), penicillin-streptomycin (10,000 units/ml penicillin and 10,000 μg/ml streptomycin) and Lipofectamine 2000TM were from Invitrogen. MCF-7 Neo and MCF-7 HER218 cells were gifts from R. Schiff (Baylor College of Medicine, Houston, TX). Anti-ERK2 antibody was from BD Transduction Laboratories (Oxford, UK), and anti-SK1 antibody was a gift from A. Huwiler (University of Bern, Switzerland). Anti-FLAG M2 antibody was from Stratagene. Anti-myc antibody was from Santa Cruz Biotechnology (Santa Cruz, CA). Sphingosine and S1P were from Avanti Polar Lipids (Alabaster, AL). MG132 and purified SK1 were from Enzo Life Sciences (Exeter, UK). SKi (2-(p-hydroxyanilino)-4-(p-chlorophenyl)thiazole) was from Merck. FTY720 was from Cayman Chemicals (Tallinn, Estonia). 4′,6-Diamidino-2-phenylindole (DAPI) was from Vector Laboratories. The G113A SK1 mutant was a kind gift from S. Pitson (SA Pathology, Adelaide, Australia).
Cell Culture
MCF-7 Neo and HER218 breast cancer cells were grown in a monolayer culture in high glucose DMEM with 10% European FCS and 100 units/ml penicillin, 100 μg/ml streptomycin, 0.4% Geneticin (absent for parental cells), and 15 μg/ml insulin at 37 °C with 5% CO2. HEK 293 cells were cultured in MEM supplemented with 10% European FCS, 100 units/ml penicillin, 100 μg/ml streptomycin, and 1% nonessential amino acids at 37 °C in 5% CO2 and in the presence of DMEM (instead of MEM) and 0.8% Geneticin for HEK 293 cells stably overexpressing GFP-tagged SK1. HEK 293 cells were transfected with Lipofectamine 2000TM reagent and myc- or FLAG-tagged SK1 or G113A SK1 plasmid constructs and grown for 48 h.
Western Blotting
Cells were harvested in sample buffer (125 mm Tris, pH 6.7, 0.5 mm Na4P2O7, 1.25 mm EDTA, 0.5% w/v SDS containing 1.25% v/v glycerol, 0.06% w/v bromphenol blue, and 50 mm dithiothreitol) and were subjected to SDS-PAGE and Western blotting as described previously (15). Resolved proteins were Western blotted with anti-myc, anti-FLAG, anti-SK1, anti-ERK2, or anti-actin antibodies.
Immunoprecipitation
Forty-eight hours after transfection with the indicated plasmid constructs, the medium was removed, and HEK 293 cells were lysed in ice-cold immunoprecipitation buffer (500 μl) containing 20 mm Tris, 137 mm NaCl, 2.7 mm KCl, 1 mm MgCl2, 1 mm CaCl2, 1% (v/v) Nonidet P-40, 10% (v/v) glycerol, 1 mg/ml bovine serum albumin (BSA), 0.5 mm Na4P2O7, 0.2 mm phenylmethanesulfonyl fluoride (PMSF), leupeptin, and aprotinin (all protease inhibitors were at 10 μg/ml, pH 8.0) for 20 min at 4 °C. The material was harvested and transferred to microcentrifuge tubes, which were further mixed for 60 min at 4 °C, and then centrifuged at 14,000 × g for 10 min at 4 °C to remove cellular debris. After preclearing with protein G-Sepharose beads, equal amounts of supernatant from each sample were taken for immunoprecipitation with protein G-Sepharose beads and anti-myc or anti-FLAG antibodies. After 2 h or overnight agitation at 4 °C, the supernatant was removed by centrifugation at 14,000 × g for 15 s at 4 °C. Immunoprecipitates were washed twice with 1 ml of buffer A containing 10 mm HEPES, pH 7.0, 100 mm NaCl, and 0.5% (v/v) Nonidet P-40 and once in 1 ml of buffer A without Nonidet P-40. Immunoprecipitates were collected by centrifugation at 14,000 × g for 15 s at 4 °C and combined with boiling sample buffer for SDS-PAGE.
SK1 Activity Assay
SK1 activity was assayed as described previously (16). Briefly, sphingosine was solubilized in Triton X-100 (final concentration 0.063% w/v) and combined with buffer 1 containing 20 mm Tris, pH 7.4, 1 mm EDTA, 1 mm Na3VO4, 40 mm β-glycerophosphate, 1 mm NaF, 0.007% (v/v) β-mercaptoethanol, 20% (v/v) glycerol, 10 μg/ml aprotinin, 10 μg/ml soybean trypsin inhibitor, 1 mm PMSF, and 0.5 mm 4-deoxypyridoxine. Modulation of SK1 activity was determined by incubating 15 ng of purified SK1 or 15 μg of HEK 293 cell lysates containing stably overexpressed or transiently expressed recombinant SK1 for 15–20 min at 30 °C, in the presence of 0.5–20 μm sphingosine, 250 μm [γ-32P]ATP (4.4 × 104 cpm/nmol, in 10 mm MgCl2), and varying concentrations of inhibitors dissolved in dimethyl sulfoxide or control (5% dimethyl sulfoxide). Reactions were terminated by the addition of 500 μ1 of n-butyl alcohol and mixed with 1 ml of 2 m KCl. The organic phase containing [32P]S1P was then extracted by washing twice with 1 ml of 2 m KCl before quantification by Cerenkov counting. The kinetic parameters were calculated using the graph plotting and curve fitting programs Biograph (University of Strathclyde, Glasgow, UK) and Prism 4.03 (GraphPad). Substrate kinetics were analyzed according to the Michaelis-Menten equation, and the inhibition constants (Kicand Kiu) were determined using Dixon and Cornish-Bowden plots (17).
Fluorescence Microscopy
Cells were plated onto autoclaved 13-mm glass coverslips and grown to 60% confluence before serum starvation for 48 h prior to stimulation. Cells were fixed with 3.7% formaldehyde in PBS for 10 min and permeabilized with 0.1% Triton X-100 in PBS for 1 min before incubation in blocking solution (5% European FCS and 1% BSA in PBS) for 1 h at room temperature. Coverslips were then incubated with phalloidin red (1:100 dilution in blocking solution) for 1 h at room temperature. Coverslips were washed with PBS and mounted on glass slides using Vectashield® hard set mounting medium with DAPI. Actin rearrangement was assessed using phalloidin red staining. Fluorescence was visualized using a Nikon (Surrey, UK) E600 epifluorescence microscope.
Densitometry
Densitometric quantification of Western blots was performed using ScanImage program (Scion Corporation, Frederick, MD). Statistical analysis was performed using an unpaired Student's t test.
RESULTS
Synthesis of New FTY720 Analogues
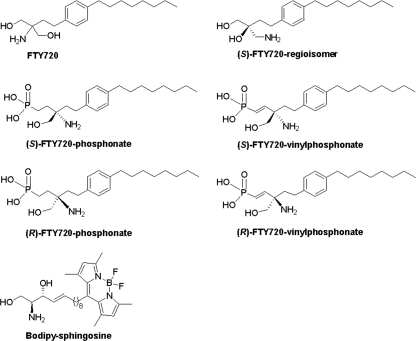
The structural formulas of the FTY720 analogues are shown in Fig. 1, and the synthetic schemes employed to prepare (S)-FTY720 regioisomer and Bodipy-sphingosine (Bdp-So) are outlined in supplemental Fig. 1 and supplemental Experimental Procedures.
FIGURE 1.
Structures of FTY720 analogues.
Kinetic Analysis of Inhibition of SK1 Activity by FTY720 Analogues
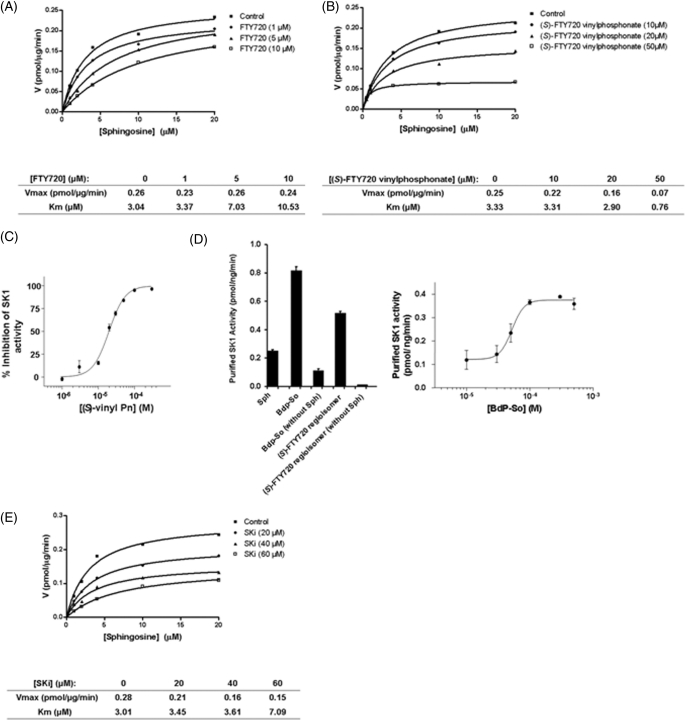
We have previously demonstrated that SKi (which was identified by French et al.) (18), FTY720, and (S)-and (R)-FTY720 vinylphosphonate inhibited purified SK1 activity, whereas (S)- and (R)-FTY720 phosphonate are inactive (12, 14). Using HEK 293 cells stably overexpressing SK1 (30-fold increase in SK1 activity versus stable vector-transfected cells), we have now investigated the kinetic mechanism by which SKi, FTY720, and (S)-FTY720 vinylphosphonate inhibit SK1 activity (Table 1). We found that FTY720 is a competitive inhibitor (with sphingosine) with a Kic = 2 ± 0.5 μm (Table 1, Fig. 2A, and supplemental Fig. 2A). (S)-FTY720 vinylphosphonate is an uncompetitive inhibitor (with sphingosine) with a Kiu = 14.5 ± 4.4 μm (Table 1, Fig. 2B, and supplemental Fig. 2B). This kinetic behavior indicates that (S)-FTY720 vinylphosphonate binds to a putative allosteric site in SK1 contingent on formation of the SK1-sphingosine complex. Indeed, when (S)-FTY720 vinylphosphonate concentration was plotted against inhibition of catalysis at a fixed concentration of sphingosine (10 μm), then a Hill coefficient of 1.9 ± 0.2 and an IC50 = 20 ± 1.1 μm were obtained (Fig. 2C). A Hill coefficient of 2.2 ± 0.3 was obtained at 20 μm sphingosine (data not shown). The percent inhibition of stably overexpressed SK1 in HEK 293 cells by 50 μm (S)-FTY720 vinylphosphonate (using 10 μm sphingosine) showed good agreement with our previous findings using purified SK1 (14), and in each case, inhibition of SK1 activity was ∼80%. Additional evidence that SK1 contains an allosteric site(s) was demonstrated by data showing that micromolar concentrations of Bdp-So (a conjugate of sphingosine with the Bodipy fluorophore) and (S)-FTY720 regioisomer, both of which are also relatively weak substrates of SK1, activated purified SK1 activity (Fig. 2D). Bdp-So stimulated SK1 activity in a concentration-dependent manner at a fixed concentration of sphingosine (10 μm) (Fig. 2D). Indeed, SK1 phosphorylated sphingosine in the presence of Bdp-So with a Hill coefficient of 4.5 ± 2.7 (Fig. 2D). SKi is a mixed inhibitor of SK1 with a Kic = 17 ± 3.5 μm and a Kiu = 48.3 ± 11.5 μm and thus was biased toward competitive inhibition at low micromolar concentrations of SKi (Table 1, Fig. 2E, and supplemental Fig. 2C).
TABLE 1.
Kinetic constants
Ki values are means ± S.D. for n = 3 experiments.
| Inhibitor | Inhibition mechanism | Inhibition constant(s) |
|---|---|---|
| μm | ||
| FTY720 | Competitive | Kic = 2.0 ± 0.5 |
| (S)-FTY720 vinylphosphonate | Uncompetitive | Kiu = 14.5 ± 4.4 |
| SKi | Mixed | Kic = 17.0 ± 3.5; Kiu = 48.3 ± 11.5 |
FIGURE 2.
Inhibitor kinetic analysis of SK1 in HEK 293 cells. A and B, V versus S nonlinear regression analysis of stably expressed recombinant SK1 in HEK 293 cells for FTY720 (A) and (S)-FTY720 vinylphosphonate (B). C, effect of varied concentrations of (S)-FTY720 vinylphosphonate on stably expressed recombinant SK1 activity in HEK293 cells assayed with 10 μm sphingosine. D, effect of Bdp-So (50 μm final concentration and varied concentrations, right) and (S)-FTY720 regioisomer (50 μm final concentration) on purified SK1 activity assayed with and without 10 μm sphingosine (left). E, V versus S nonlinear regression analysis for the effect of SKi on recombinant SK1 stably expressed in HEK 293 cells. Results are representative of three independent experiments.
Putative Allosteric Site(s) in SK1
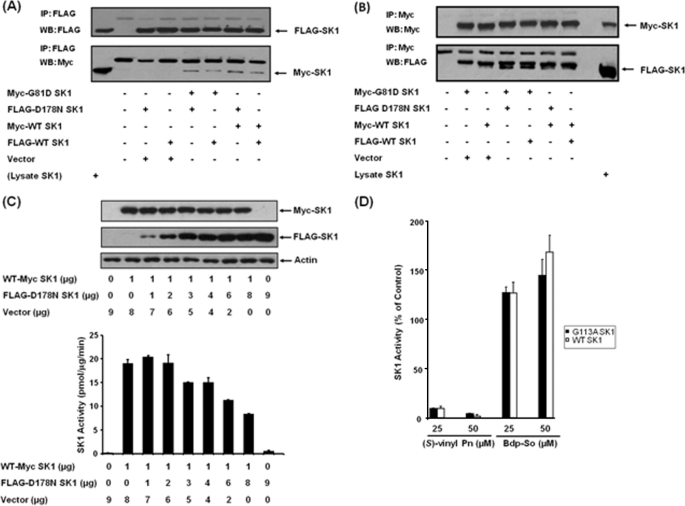
The data presented in Fig. 3 demonstrate that SK1 is an oligomeric protein. This conclusion is based on results from experiments in which wild-type myc- and FLAG-tagged SK1 are transiently co-expressed in HEK 293 cells and where myc- and FLAG-tagged SK1 (molecular mass = 42 kDa) were co-immunoprecipitated with anti-FLAG antibody (Fig. 3A). Identical results were obtained using anti-myc antibody (Fig. 3B). The ability of SK1 to form oligomers confirms data previously published by Kihara et al. (19).
FIGURE 3.
SK1 is an oligomer. HEK 293 cells were transiently transfected with myc-tagged G81D SK1, FLAG-tagged D178N SK1, myc-tagged WT SK1, and/or FLAG-tagged WT SK1 plasmid constructs. A and B, Western blots (WB) of anti-FLAG immunoprecipitates (IP) (A) or anti-myc immunoprecipitates (B) probed with respective anti-myc and anti-FLAG antibodies. Lysate SK1 represents lysates of cells overexpressing both myc- and FLAG-tagged WT SK1. C, Western blot and SK1 activity of HEK 293 cell lysates showing the effect of overexpressing FLAG-tagged D178N SK1 on myc-tagged WT SK1 activity (using micrograms of plasmid constructs shown). FLAG-tagged D178N SK1-myc-tagged G81D SK1 oligomers were catalytically deficient. Results are representative of three experiments. SK1 activity was also measured in immunoprecipitates: anti-myc immunoprecipitates: myc-tagged G81D SK1-transfected cells, 0.23 pmol/min; myc-tagged G81D SK1/FLAG-tagged D178N SK1-transfected cells, 1.65 pmol/min; myc-tagged WT SK1-transfected cells, 33 pmol/min. Anti-FLAG immunoprecipitates: FLAG-tagged D178N SK1-transfected cells, 2.8 pmol/min; FLAG-tagged D178N SK1/myc-tagged G81D SK-transfected cells, 2 pmol/min; FLAG-tagged WT SK1-transfected cells, 20.55 pmol/min. Results are representative of two or three experiments. D, effect of (S)-FTY720 vinylphosphonate or Bdp-So (both at 25 and 50 μm and assayed using 10 μm sphingosine) on WT and G113A SK1 (both transiently over-expressed in HEK293 cells) activity. Results are expressed as % of SK1 activity in the absence of (S)-FTY720 vinylphosphonate or Bdp-So (control = 100% SK1 activity, n = 3 experiments).
To test whether (S)-FTY720 vinylphosphonate conforms to a model in which inhibitors can stabilize an allosteric site/domain that exerts an autoinhibitory effect on SK1 activity, we created hybrid SK1 oligomers containing wild-type SK1 and kinase-deficient SK1 mutant monomers. The assumption here is that each monomer of the wild enzyme contains one catalytic site and one allosteric site. Thus, co-expression of WT SK1 with D178N SK1 mutant (mutated in the sphingosine binding site) (20) or G81D SK1 mutant (mutated in the ATP binding site) (21) will result in formation of hybrid SK1 oligomers containing more allosteric sites compared with catalytic sites. Therefore, if the allosteric site exerts an autoinhibition, then we expect the catalytically deficient SK1 mutants to reduce catalytic activity of WT SK1 in the oligomer. First, we demonstrated that transiently overexpressed FLAG-tagged D178N SK1 mutant and the myc-tagged G81D SK1 mutants can indeed form oligomers with myc-tagged and FLAG-tagged WT SK1, respectively (Fig. 3, A and B). We also confirmed that the FLAG-tagged D178N SK1 mutant exhibited <10% of the activity of the wild-type enzyme (see Fig. 3C). To address directly whether mutant SK1 inhibits the catalytic activity of WT SK1, we transiently co-expressed WT myc-tagged SK1, whose concentration was fixed, with varying amounts of FLAG-tagged D178N SK1 mutant. Under these conditions, the FLAG-tagged D178N SK1 mutant inhibited WT myc-tagged SK1 activity by ∼50% (Fig. 3C).
We also evaluated the nature of the inhibition kinetics with (S)-FTY720 vinylphosphonate or FTY720 in HEK 293 cell lysates containing transiently overexpressed WT FLAG/myc-tagged SK1. In common with stably overexpressed WT SK1, (S)-FTY720 vinylphosphonate was an uncompetitive inhibitor (with sphingosine) with a Kiu = 7 μm, whereas FTY720 was a competitive inhibitor with a Kic = 7 μm (data not shown).
Finally, we explored the possibility that Gly113 in SK1 might be part of the putative allosteric site as mutation to alanine in SK1 results in constitutive activation of the lipid kinase (22). This is a result of an increase in Vmax, without alteration in substrate binding to the enzyme. Therefore, we tested the sensitivity of the G113A mutant SK1 (transiently overexpressed in HEK 293 cells) to (S)-FTY720 vinylphosphonate and Bdp-So. In this regard, (S)-FTY720 vinylphosphonate inhibited and Bdp-So activated the G113A SK1 mutant to an extent similar to that with WT SK1 (Fig. 3D).
Effects of FTY720, (S)-FTY720 Vinylphosphonate, and SKi on SK1 Expression in MCF-7 Cells
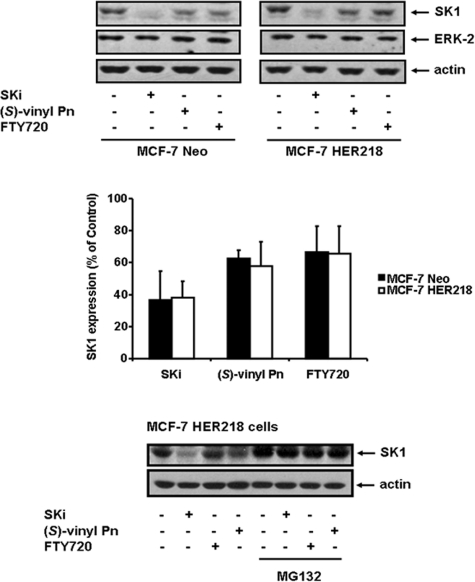
Previously, we demonstrated an entirely novel mechanism of functional interaction of SK1 inhibitors with SK1 that offers additional opportunities for drug discovery (12, 14). These studies involved polyubiquitination, which directs proteasomal degradation of proteins (12), and employed the proteasomal inhibitor, MG132 (12). We showed that a number of SK1 inhibitors, including (S)-FTY720 vinylphosphonate, induce MG132-sensitive degradation of SK1 (treatment of cells for 6–48 h), thereby removing SK1 from MCF-7 breast cancer and LNCaP prostate cancer cells (12, 14). Indeed SK1 is polyubiquitinated, and SKi enhanced the rate at which polyubiquitinated SK1 is degraded by the proteasome (12). Moreover, SKi induces a ubiquitin-proteasomal degradation of SK1 via a ceramide-dependent mechanism in LNCaP cells, a consequence of an initial inhibition of SK1 catalytic activity and perturbation of the ceramide-sphingosine-S1P rheostat (12). The creation of an SK1-null cancer cell or one with severely reduced SK1 expression results in the onset of apoptosis, as indicated by increased poly(ADP-ribose) polymerase cleavage (12). Therefore, we used MCF-7 Neo (which expresses a vector-encoded neomycin resistance gene) (10) to establish the effects of SKi, FTY720, and (S)-FTY720 vinylphosphonate on the proteasomal degradation of SK1; we determined whether this might be influenced by an oncogenic background of HER2. As shown in Fig. 4, treatment of MCF-7 Neo cells with SKi, FTY720 or (S)-FTY720 vinylphosphonate for 24 h induced the degradation of SK1 (molecular mass = 42 kDa). We previously observed a HER2-dependent increase in SK1 expression in the MCF-7 HER2 cells (HER218 cells, stably expressing HER2) (10). These cells remain sensitive to SKi, FTY720, or (S)-FTY720 vinylphosphonate. Indeed, all three compounds induced the MG132-sensitive proteasomal degradation of SK1 to an extent similar to that with MCF-7 Neo cells (Fig. 4).
FIGURE 4.
Proteasomal degradation of SK1. MCF-7 Neo or MCF-7 HER2 cells were treated with SKi or FTY720, or (S)-FTY720 vinylphosphonate ((S)-vinyl Pn) (all at 10 μm final concentration) for 24 h. MCF-7 HER2 cells were also pretreated with MG132 (10 μm) for 30 min prior to addition of SK1 inhibitors. Western blots were probed with anti-SK1, anti-ERK-2, and anti-actin antibodies. Results are representative of three independent experiments. Bar graph shows quantification of the effect of the SK1 inhibitors on the proteasomal degradation of SK1 in MCF-7 Neo or MCF-7 HER2 cells by calculating the SK1:actin ratio for each treatment (p < 0.05 for control versus SK1 inhibitor-treated cells, n = 3 experiments).
Effects of FTY720, (S)-FTY720 Vinylphosphonate, and SKi on Actin Rearrangement in MCF-7 Neo Cells
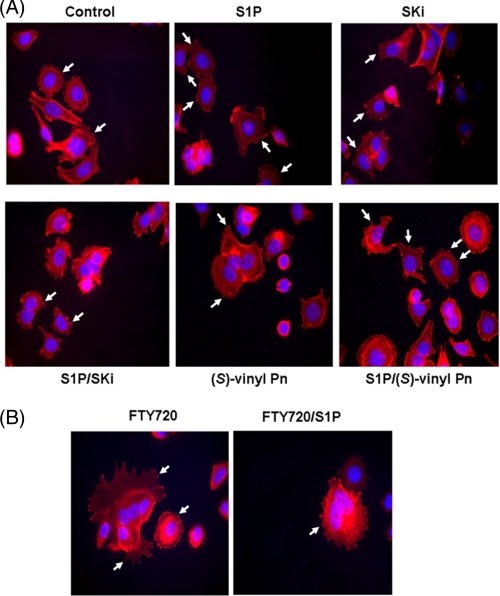
We have previously shown that siRNA knockdown of SK1 reduced the S1P-induced formation of a migratory phenotype in MCF-7 Neo cells, as assessed by inhibition of S1P-stimulated actin accumulation into membrane ruffles/lamellipodia and inhibited migration of cells in scratch assays; this is mediated by S1P3 (10). Actin is clustered into focal adhesions in MCF-7 Neo cells. Treatment of these cells with S1P stimulates the rearrangement of actin from focal adhesions into membrane ruffles/lamellipodia (10) (Fig. 5). Treatment of MCF-7 Neo cells with SKi, (S)-FTY720 vinylphosphonate (Fig. 5A), or FTY720 (Fig. 5B) for 15 min reduced the S1P-stimulated rearrangement of actin into lamellipodia/membrane ruffles and reestablished actin focal adhesions, thereby demonstrating activity that is consistent with these compounds functioning as SK1 inhibitors.
FIGURE 5.
Actin rearrangement in MCF-7 cells. MCF-7 Neo cells were treated with (A) SKi or (S)-FTY720 vinylphosphonate ((S)-vinyl Pn) or (B) FTY720 (both at 10 μm final concentration) for 15 min prior to stimulation with and without S1P (1 μm, 5 min). Actin was detected using phalloidin red staining. Arrows in the panel for S1P treatment identify actin localized to lamellipodia/membrane ruffles, whereas arrows in the other panels identify actin clustered into focal adhesions. Nuclei were stained with DAPI. Results are representative of three independent experiments.
DISCUSSION
We demonstrate here for the first time that (S)-FTY720 vinylphosphonate binds to a putative allosteric site(s) in SK1 contingent on formation of a SK1-sphingosine complex. This conclusion is based on the finding that (S)-FTY720 vinylphosphonate inhibited SK1 activity in an uncompetitive manner with a Kiu = 14.5 ± 4.4 μm. We also demonstrate that SK1 is an oligomer, consistent with a model that is conducive to allosterism. Moreover, SK1 appears to contain an allosteric site(s) that exerts an autoinhibitory effect on the catalytic sites. This conclusion is based on the finding that FLAG-tagged D178N SK1 mutant inhibited the catalytic activity of the WT myc-tagged SK1 in an oliogmeric arrangement. There are three alternative explanations for these findings, all of which can be excluded. First, we tested whether minimal dimerization is required to form two competent catalytic sites in a dimer by domain swapping. If this were the case, then mutagenesis of Asp178 might disable one of the catalytic sites formed between WT SK1 and D178N mutant SK1. The ATP binding site in monomer 1 might form a competent active site with the sphingosine binding site in monomer 2. Equally, the ATP binding site in monomer 2 might form a competent active site with the sphingosine binding site in monomer 1. Therefore, the formation of two competent active sites in a dimeric arrangement predicts that co-expression of the two catalytically deficient mutants to equal levels should reconstitute 50% of the wild-type activity. However, we found that co-expression of each mutant did not result in a catalytically competent oligomer (activity of the oligomer (∼10% of WT SK1) was equivalent with that of the D178N mutant SK1). Therefore, oligomerization is not required for formation of two competent catalytic sites by domain swapping. Second, it is possible that formation of oligomers is required for co-operation between catalytic sites. Thus, formation of WT SK1-D178N SK1 oligomers might disrupt this cooperation and reduce SK1 activity. However, SK1 exhibits simple Michaelis-Menten kinetics, and thus, the catalytic sites function independently of each other in the oligomeric arrangement. Third, oligomerization might enable functionalization of the catalytic sites, and mutagenesis of Asp178 might impair oligomerization. However, the D178N mutant SK1 is still able to form oligomers with WT SK1. Because the concentration of competent WT catalytic sites is maintained constant in the experiment, we can, therefore, attribute inhibition of WT SK1 catalytic activity by the D178N mutant to putative allosteric site(s) that exhibit autoinhibitory activity.
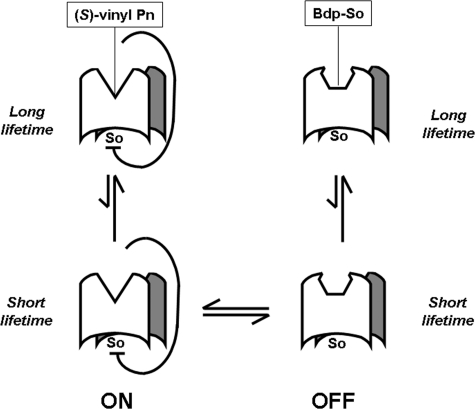
The allosteric site(s) might exist in an “on” and “off” state in equilibrium (Fig. 6). Therefore, binding of (S)-FTY720 vinylphosphonate to SK1 conforms to a model in which (S)-FTY720 vinylphosphonate might stabilize the on state (which autoinhibits SK1 activity) of the allosteric site (which is predicted to have a longer lifetime than the unbound allosteric site), thereby increasing the lifetime of the autoinhibitory effect of the site on SK1 activity. It follows that the activators of SK1, e.g. Bdp-So and (S)-FTY720 regioisomer, might stabilize the off state, thereby relieving inhibition of SK1 activity by the allosteric site and resulting in activation of the enzyme (Fig. 6). We also demonstrated a Hill coefficient of 1.9–2.2 for the inhibition of SK1 catalytic activity by (S)-FTY720 vinylphosphonate. Thus, stabilization of the on allosteric site conformation by (S)-FTY720 vinylphosphonate will induce equilibrium transition such that the concentration of the on site conformation will increase by mass action. A Hill coefficient of ∼2 suggests that a dimer is likely to contain at least two allosteric sites that function in a cooperative manner. For Bdp-So, we obtained a Hill coefficient of 4.5, suggesting that binding of Bdp-So to the allosteric sites is also cooperative.
FIGURE 6.
Schematic showing the on or off allosterism model for interaction of SK1 with sphingosine (So), (S)-FTY720 vinylphosphonate ((S)-vinyl Pn), or Bdp-So.
The mutation of Gly113 to alanine in SK1 results in constitutive activation of the lipid kinase (22), suggesting that Gly113 might be part of the allosteric site and that the mutation disables its function. Indeed, one can predict that mutation might trap the allosteric site in the off state, thereby converting SK1 into a form that is resistant to inhibition by (S)-FTY720 vinylphosphonate and activation by Bdp-So. However, the G113A mutant was still sensitive to inhibition by (S)-FTY720 vinylphosphonate and activation by Bdp-So. Therefore, Gly113 is unlikely to be involved in allosterism.
We also report that the inhibition of SK1 by SKi conforms to a mixed inhibition model with a Kic = 17 ± 3.5 μm and a Kiu = 48.3 ± 11.5 μm. Therefore, SKi appears to bind to both the catalytic site and the putative allosteric site. The latter is consistent with the observation made by others that inhibition of SK1 activity by SKi is not competitive with the second substrate, ATP (18). These authors demonstrated that SKi is relatively specific for SK, with no activity against ERK-2, PI3K, or PKCα (18, 23) and that SKi inhibits proliferation and induces apoptosis of various tumor cell lines (18). Indeed, our findings confirm that SKi induces the onset of apoptosis of MCF-7 Neo cells (12). A role for SK1 in regulating the survival of MCF-7 breast cancer cells was also evident by studies of Taha et al. (24), who demonstrated that siRNA knockdown of SK1 in MCF-7 breast cancer cells induced apoptosis. This involves caspase activation and is associated with increased ceramide formation and Bax oligomerization.
Treatment of MCF-7 Neo and MCF-7 HER218 cells with SKi, FTY720, or (S)-FTY720 vinylphosphonate induced the proteasomal degradation of SK1. The ability of SK1 inhibitors to induce proteasomal degradation of SK1 and to exhibit high efficacy killing of cancer cells is likely dependent on the specific adopted inhibitor-SK1 conformation. This conformation might result in different extents of unfolding of SK1 required for polyubiquitination and proteasomal degradation. This proteasomal degradation is also stimulated in a ceramide-dependent manner due to acute inhibition of SK1 catalytic activity by the inhibitor as we demonstrated previously (12).
SKi is an effective anticancer agent in vivo and inhibits tumor growth (syngenic tumor model with mammary adenocarcinoma cells) and inflammation (ulcerative colitis model) (23, 25). In addition, FTY720 inhibits the growth of various tumor mouse models (bladder, kidney, prostate, breast, liver, etc.) (26–29) and also reduces tumor vascularization and angiogenesis (30). In light of our current findings, it remains to be determined whether the in vivo efficacy in cancer models of SKi and FTY720 is related to the ability of these compounds to inhibit SK1 catalytic activity and/or to induce proteasomal degradation of SK1.
In conclusion, the removal of SK1 from cancer cells by SK1 inhibitors suggests that the efficacy and duration of action of these compounds in inducing apoptosis might be significantly superior compared with compounds that simply reversibly inhibit enzyme catalytic activity and are therefore short acting. Moreover, the presence of a putative allosteric site(s) in SK1 offers opportunities for the implementation of new drug discovery programs. Taken together, these findings provide impetus for developing SK1 inhibitors, e.g. by using (S)-FTY720 vinylphosphonate as the prototype, as putative anticancer agents, not least because their mechanism of action should enable fewer administrations to the patient and development of resistance might be impaired, thereby opening new avenues for combination therapies.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant HL083187 (to R. B.). This work was also supported by two Strathclyde University scholarships (to K. G. L. and F. T.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2, Experimental Procedures, and additional references.
- S1P
- sphingosine 1-phosphate
- Bdp-So
- Bodipy sphingosine
- HER2
- human epidermal growth factor receptor-2
- Kic
- competitive inhibition constant
- Kiu
- uncompetitive inhibition constant
- SK1
- sphingosine kinase 1
- SKi
- 2-(p-hydroxyanilino)-4-(p-chlorophenyl)thiazole.
REFERENCES
- 1. Pyne N. J., Pyne S. (2010) Nat. Rev. Cancer 10, 489–503 [DOI] [PubMed] [Google Scholar]
- 2. Brinkmann V., Davis M. D., Heise C. E., Albert R., Cottens S., Hof R., Bruns C., Prieschl E., Baumruker T., Hiestand P., Foster C. A., Zollinger M., Lynch K. R. (2002) J. Biol. Chem. 277, 21453–21457 [DOI] [PubMed] [Google Scholar]
- 3. Brinkmann V., Billich A., Baumruker T., Heining P., Schmouder R., Francis G., Aradhye S., Burtin P. (2010) Nat. Rev. Drug Discov. 9, 883–897 [DOI] [PubMed] [Google Scholar]
- 4. Sanchez T., Estrada-Hernandez T., Paik J. H., Wu M. T., Venkataraman K., Brinkmann V., Claffey K., Hla T. (2003) J. Biol. Chem. 278, 47281–47290 [DOI] [PubMed] [Google Scholar]
- 5. Gräler M. H., Goetzl E. J. (2004) FASEB J. 18, 551–553 [DOI] [PubMed] [Google Scholar]
- 6. Mandala S., Hajdu R., Bergstrom J., Quackenbush E., Xie J., Milligan J., Thornton R., Shei G. J., Card D., Koehane C., Rosenbach M., Hale J., Lynch C. L., Rupprecht K., Parsons W., Rosen H. (2002) Science 296, 346–349 [DOI] [PubMed] [Google Scholar]
- 7. Visentin B., Vekich J. A., Sibbald B. J., Cavalli A. L., Moreno K. M., Matteo R. G., Garland W. A., Lu Y., Yu S., Hall H. S., Kundra V., Mills G. B., Sabbadini R. A. (2006) Cancer Cell 9, 225–238 [DOI] [PubMed] [Google Scholar]
- 8. Hait N. C., Allegood J., Maceyka M., Strub G. M., Harikumar K. B., Singh S. K., Luo C., Marmorstein R., Kordula T., Milstien S., Spiegel S. (2009) Science 325, 1254–1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Alvarez S. E., Harikumar K. B., Hait N. C., Allegood J., Strub G. M., Kim E. Y., Maceyka M., Jiang H., Luo C., Kordulla T., Milstein S., Spiegel S. (2010) Nature 465, 1084–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Long J. S., Edwards J., Watson C., Tovey S., Mair K., Schiff R., Natarajan V., Pyne N. J., Pyne S. (2010) Mol. Cell. Biol. 30, 3827–3841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Watson C., Long J. S., Orange C., Tannahill C. L., Mallon E., McGlynn L. M., Pyne S., Pyne N. J., Edwards J. (2010) Am. J. Pathol. 177, 2205–2215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Loveridge C., Tonelli F., Leclecq T., Lim K. G., Long. S., Berdyshev E., Tate R. J., Natarajan V., Pitson, Pyne N. J., Pyne S. (2010) J. Biol. Chem. 285, 38841–38852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kawamori T., Osta W., Johnson K. R., Pettus B. J., Bielawski J., Tanaka T., Wargovich M. J., Reddy B. S., Hannun Y. A., Obeid L. M., Zhou D. (2006) FASEB J. 20, 386–388 [DOI] [PubMed] [Google Scholar]
- 14. Tonelli F., Lim K. G., Loveridge C., Long J., Pitson S. M., Tigyi G., Bittman R., Pyne S., Pyne N. J. (2010) Cell. Signal. 22, 1536–1542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Alderton F., Rakhit S., Kong K. C., Palmer T., Sambi S., Pyne S., Pyne N. J. (2001) J. Biol. Chem. 276, 28578–28585 [DOI] [PubMed] [Google Scholar]
- 16. Delon C., Manifava M., Wood E., Thompson D., Krugmann S., Pyne S.., Ktistakis N. T. (2004) J. Biol. Chem. 279, 44763–44774 [DOI] [PubMed] [Google Scholar]
- 17. Cortés A., Cascante M., Cárdenas M. L., Cornish-Bowden A. (2001) Biochem. J. 357, 263–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. French K. J., Schrecengost R. S., Lee B. D., Zhuang Y., Smith S. N., Eberly J. L., Yun J. K., Smith C. D. (2003) Cancer Res. 63, 5962–5969 [PubMed] [Google Scholar]
- 19. Kihara A., Anada Y., Igarashi Y. (2006) J. Biol. Chem. 281, 4532–4539 [DOI] [PubMed] [Google Scholar]
- 20. Yokota S., Taniguchi Y., Kihara A., Mitsutake S., Igarashi Y. (2004) FEBS Lett. 578, 106–110 [DOI] [PubMed] [Google Scholar]
- 21. Pitson S. M., Moretti P. A., Zebol J. R., Xia P., Gamble J. R., Vadas M. A., D'Andrea R. J., Wattenberg B. W. (2000) J. Biol. Chem. 275, 33945–33950 [DOI] [PubMed] [Google Scholar]
- 22. Pitson S. M., Moretti P. A., Zebol J. R., Vadas M. A., D'Andrea R. J., Wattenberg B. W. (2001) FEBS Lett. 509, 169–173 [DOI] [PubMed] [Google Scholar]
- 23. French K. J., Upson J. J., Keller S. N., Zhuang Y., Yun J. K., Smith C. D. (2006) J. Pharm. Exp. Ther. 318, 596–603 [DOI] [PubMed] [Google Scholar]
- 24. Taha T. A., Kitatani K., El-Alwani M., Bielawski J., Hannun Y. A., Obeid L. M. (2006) FASEB J. 20, 482–484 [DOI] [PubMed] [Google Scholar]
- 25. Maines L. W., Fitzpatrick L. R., French K. J., Zhuang Y., Xia Z., Keller S. N., Upson J. J., Smith C. D. (2008) Dig. Dis. Sci. 53, 997–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ubai T., Azuma H., Kotake Y., Inamoto T., Takahara K., Ito Y., Kiyama S., Sakamoto T., Horie S., Muto S., Takahara S., Otsuki Y., Katsuoka Y. (2007) Anticancer Res. 27, 75–88 [PubMed] [Google Scholar]
- 27. Azuma H., Takahara S., Horie S., Muto S., Otsuki Y., Katsuoka Y. (2003) J. Urol. 169, 2372–2377 [DOI] [PubMed] [Google Scholar]
- 28. Chua C. W., Lee D. T., Ling M. T., Zhou C., Man K., Ho J., Chan F. L., Wang X., Wong Y. C. (2005) Int. J. Cancer 117, 1039–1048 [DOI] [PubMed] [Google Scholar]
- 29. Azuma H., Takahara S., Ichimaru N., Wang J. D., Itoh Y., Otsuki Y., Morimoto J., Fukui R., Hoshiga M., Ishihara T., Nonomura N., Suzuki S., Okuyama A., Katsuoka Y. (2002) Cancer Res. 62, 1410–1419 [PubMed] [Google Scholar]
- 30. Ho J. W., Man K., Sun C. K., Lee T. K., Poon R. T., Fan S. T. (2005) Mol. Cancer Ther. 4, 1430–1438 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.