Abstract
SET domain lysine methyltransferases (KMTs) are S-adenosylmethionine (AdoMet)-dependent enzymes that catalyze the site-specific methylation of lysyl residues in histone and non-histone proteins. Based on crystallographic and cofactor binding studies, carbon-oxygen (CH···O) hydrogen bonds have been proposed to coordinate the methyl groups of AdoMet and methyllysine within the SET domain active site. However, the presence of these hydrogen bonds has only been inferred due to the uncertainty of hydrogen atom positions in x-ray crystal structures. To experimentally resolve the positions of the methyl hydrogen atoms, we used NMR 1H chemical shift coupled with quantum mechanics calculations to examine the interactions of the AdoMet methyl group in the active site of the human KMT SET7/9. Our results indicated that at least two of the three hydrogens in the AdoMet methyl group engage in CH···O hydrogen bonding. These findings represent direct, quantitative evidence of CH···O hydrogen bond formation in the SET domain active site and suggest a role for these interactions in catalysis. Furthermore, thermodynamic analysis of AdoMet binding indicated that these interactions are important for cofactor binding across SET domain enzymes.
Keywords: Calorimetry, Chromatin Histone Modification, Histone Methylation, NMR, Protein Methylation, C-H..O, CH..O, Lysine Methyltransferase, S-Adenosylmethionine, Chemical Shift
Introduction
Post-translational modifications in proteins are now well recognized as important players in many biological processes. Among these modifications, site-specific lysine methylation by SET domain KMTs3 is known to be critical to a diverse set of processes within the nucleus, including gene expression, cell cycle progression, and DNA damage response (1, 2). In particular, the human KMT SET7/9 has been shown to methylate lysine residues on many histone and non-histone proteins and is now considered to be important in many cellular pathways (3). Furthermore, SET7/9 has emerged as an archetype for the specificity and catalytic mechanism of the SET domain family due to multiple high resolution crystal structures, NMR analyses, and computational studies on its structure and function (4–13). Despite these studies, many aspects regarding its methyl transfer reaction mechanism remain unclear, including the possibility that unconventional CH···O hydrogen bonds participate in catalysis (6).
CH···O hydrogen bonding has been recognized as an important interaction in proteins and other biological macromolecules dating back 40 years (14–17). For example, it has been estimated that 17% of the energy percentage at protein-protein surfaces is due to CH···O hydrogen bonding, and at some protein surfaces, that percentage is as high as 40–50% (18). These hydrogen bonds also have been implicated in enzyme catalysis (19–23), stabilizing nucleic acid structure (24–27), as well as interactions with methyl groups in small molecules (28–32). Despite their importance, experimental characterization of CH···O hydrogen bonds in proteins remains challenging. Current methods for identifying CH···O hydrogen bonds (33–35) are difficult to employ for many proteins, including SET7/9. However, NMR spectroscopy holds promise in identifying CH···O hydrogen bonds in proteins and other macromolecules via 1H chemical shift (23, 36, 37).
Recent structural and functional studies have suggested that CH···O hydrogen bonds play pivotal roles in substrate binding and catalysis in SET domain KMTs (6, 38, 39). These interactions were identified in x-ray crystal structures, in which C···O distances between the AdoMet methyl carbon, structurally conserved carbonyl oxygen atoms, and an invariant tyrosine were appropriate (<3.7 Å) for CH···O hydrogen bonding to occur (6) (supplemental Fig. S1). These hydrogen bonds can form because the electron withdrawing character of the sulfonium cation polarizes the methyl group of AdoMet. Isothermal calorimetry demonstrated that AdoMet displays higher binding affinity to a SET domain KMT than AdoMet analogues that are incapable of forming CH···O hydrogen bonds (6), indicating the importance of these interactions in substrate binding. In addition, it was postulated that the CH···O hydrogen bonds align the AdoMet methyl group in the requisite linear geometry with the substrate lysine ϵ-amine group for the SN2 methyl transfer reaction. (supplemental Fig. S2) (2, 5, 6, 40, 41). However, as hydrogen atoms are not observed in all but the highest resolution x-ray crystal structures, it is possible that the AdoMet methyl protons occupy positions that preclude CH···O hydrogen bond formation, despite close C···O contacts within SET domain crystal structures. Thus, the presence of CH···O hydrogen bonding can only be inferred from these structures. Here, we address the fundamental question, is there direct experimental evidence for CH···O hydrogen bonding between the AdoMet methyl group and the SET domain active site in solution?
EXPERIMENTAL PROCEDURES
Expression and Purification of MetK and SET7/9
A plasmid encoding the AdoMet synthetase (MetK) gene from Methanococcus jannaschii was generously provided by George D. Markham. The gene was subcloned into pHT4, a variant on the pET15b vector (Novagen) that contains an N-terminal His6 tag with a tobacco etch virus protease cleavage site to facilitate protein purification. MetK was expressed in Escherichia coli BL21 DE3 cells grown in LB media by induction with 0.1 mm isopropyl β-d-thiogalactopyranoside for 4 h at 37° C. The enzyme was purified on a Talon cobalt affinity column (Clontech) followed by Superdex 200 (GE Healthcare) gel filtration chromatography. Following gel filtration, MetK was concentrated to ∼20 mg/ml as determined by its absorbance at 280 nm, flash-frozen in liquid nitrogen, and stored at −80 °C.
SET7/9 (residues 110–366) was expressed and purified as described previously (39), with the following exceptions. The enzyme was purified by denaturation and refolding while immobilized on a nickel-Sepharose column (GE Healthcare). SET7/9 was unfolded by washing the column with 7–10 column volumes of 6 m guanidinium chloride, refolded with a gradient into the lysis buffer, and subsequently eluted using an imidazole gradient. The denaturation and refolding protocol were necessary to remove AdoMet that can co-purify with the recombinant enzyme expressed in bacteria (39). After gel filtration chromatography, SET7/9 was concentrated to at least 40 mg/ml as determined by its absorbance at 280 nm, flash-frozen, and stored at −80 °C.
Synthesis and Purification of AdoMet
[Methyl-13C]AdoMet was enzymatically synthesized from ATP and [methyl-13C]l-methionine (Sigma) using MetK as reported previously (43) with the following modifications. Reactions were performed in final volume of 5 ml for 5 h at 298 K. At the end of the reaction, AdoMet was purified using a Source 15S column (GE Healthcare), as described previously (44). The purity of the resulting AdoMet was verified by NMR and estimated to be ∼95% pure. Existing impurities showed neither chemical shift change nor intensity change upon addition of [methyl-13C]AdoMet to SET7/9.
Molecular Dynamics (MD) Simulation and Quantum Chemistry Calculations of AdoMet in Water
To accurately quantify the chemical shift of the AdoMet methyl group in solution, we first determined the number hydrogen bonds formed by the AdoMet methyl group in water, followed by quantum chemical calculations of the chemical shift of the AdoMet methyl group. The starting structure for the MD simulation was derived from a B3LYP/6–311+G(2d,p) optimized AdoMet structure. Partial charges were assigned to the starting AdoMet structure using B3LYP/6–311+G(2d,p) charges from electrostatic potentials calculation (45). All density functional theory computations were accomplished using Gaussian 03 (46). The MD simulation was run using the CHARMM36 force field (47) in a 35.2 × 25.5 × 22.2 Å TIP3P water box (48) for 10 ns, using a 2-fs time step. The simulation was conducted at 298 K with a Nose-Hoover Thermostat (49), and periodic boundary conditions were managed by the particle mesh Ewald method (50). Other AdoMet (bond, angle, and dihedral) parameters derived from the CHARMM General Force Field parameter set (51). Analysis of the MD trajectory in 20-fs steps revealed that the AdoMet methyl group forms, on average, 0.36 hydrogen bonds in solution per methyl group. Therefore, to correctly model the solution state of the AdoMet methyl group, one weak CH···O hydrogen bond was included in the chemical shift calculations.
The coordinates of the aqueous AdoMet solution structure were provided by George D. Markham (52). The methyl proton geometry was optimized using B3LYP/6–311+G(2d,p) (53, 54) with implicit water solvation by the Polarizable Continuum Model (55) with all other atoms frozen. The optimized geometry of the AdoMet methyl group formed one intramolecular CH···O hydrogen bond with the ribose 3′-hydroxyl group (supplemental Fig. S3). The geometry of this interaction (C-H···O angle = 131° and H···O interaction distance = 2.5 Å) was consistent with a weak hydrogen bond. Thus, this interaction satisfied our condition to include one weak CH···O hydrogen bond in the quantum chemistry calculations to correctly represent AdoMet in water, as determined by the MD simulation described above. Chemical shifts were calculated using the Gauge Independent Atomic Orbitals (56) method in implicit water for the reasons described above. The chemical shift of the AdoMet methyl group in this conformation yielded a chemical shift of 3.0 ppm, well within 0.1 ppm of the experiment. Breaking the CH···O hydrogen bond by rotating the AdoMet methyl group yielded a chemical shift of 2.9 ppm, confirming that the suboptimal geometry of the hydrogen bond resulted in a weak interaction (57). As a reference, tetramethylsilane was optimized in implicit water, and chemical shifts were calculated with the same methods as above.
Quantum Chemistry Calculations on the SET7/9·AdoMet Complex
The model SET7/9 active site included all atoms within 5 Å of the AdoMet methyl group from the SET7/9·AdoMet binary complex (4). The AdoMet molecule was truncated at the carbon positions adjacent to the sulfonium cation. Protons were added automatically using Chimera (58), and a single chlorine atom was placed in the position of the AdoMet carboxylic acid group to neutralize the system. All added hydrogen positions were optimized using B3LYP/3–21G* (59, 60), and methyl protons were subsequently optimized, and chemical shifts were calculated with B3LYP/6–311+G(2d,p) as described for free AdoMet. Using implicit solvent with a lower dielectric constant to reflect the hydrophobic core of proteins (ϵ = 4.9) for chemical shift calculations yielded no change in the 1H chemical shift of the AdoMet methyl group as compared with implicit water solvation. All calculated and measured chemical shifts are shown in supplemental Table S1. Methyl rotamers for chemical shift calculations were created by manually rotating the AdoMet methyl group. Structural figures were rendered using PyMOL software (Schrödinger, LLC).
NMR Spectroscopy
All NMR experiments were performed on a 600-MHz Avance Bruker NMR spectrometer equipped with a triple resonance cryoprobe at 298 K. Spectra were referenced using the water signal. Data were processed and analyzed using NMRPipe/NMRDraw and Sparky, respectively (61, 62). The assignment of the AdoMet methyl group was confirmed by recording control 13C two-dimensional heteronuclear single quantum coherence (HSQC) spectra of three separate samples: SET7/9 with no added AdoMet, SET7/9 plus stoichiometric quantities of unlabeled AdoMet, and [methyl-13C]AdoMet with no added SET7/9. The resonance assigned as the bound AdoMet methyl group was not present in any of these three control spectra, whereas it was apparent in the [methyl-13C]AdoMet·SET7/9 two-dimensional HSQC spectrum. Upon addition of slight stoichiometric excess of SET7/9 to 13C-labeled AdoMet, the peak corresponding to the free AdoMet [13C]methyl group completely shifted to the enzyme-bound peak in the two-dimensional HSQC spectrum, indicating that the AdoMet was fully saturated by the enzyme. Spectral overlays of SET7/9 in cofactor bound and free states were reported previously (8, 9).
Isothermal Titration Calorimetry (ITC)
ITC experiments were performed using a Microcal VP-ITC (GE Healthcare). All experiments were performed at 293 K in 20 mm sodium phosphate, pH = 7.0, and 100 mm sodium chloride. Varying concentrations of ligand (0.07–7.7 mm) and protein (0.006–0.193 mm) were used due to the large range of binding constants measured. Data analysis was accomplished using Microcal Origin (GE Healthcare). All binding curves had N-values between 0.8–1.0. Errors reported in Table 1 are from curve-fitting errors. Previous crystallographic studies have shown that the binding modes of AdoMet, sinefungin, and S-adenosylhomocysteine (AdoHcy) to SET7/9 are highly homologous (supplemental Fig. S4).
TABLE 1.
Dissociation constants and binding enthalpies of AdoMet, sinefungin, and AdoHcy to SET7/9
| Ligand | KD | ΔH |
|---|---|---|
| nm | kcal/mol | |
| AdoMet | 53 ± 3 | −26.7 ± 0.1 |
| Sinefungin | 330 ± 24 | −23.0 ± 0.2 |
| AdoHcy | 60,000 ± 5000 | −24.4 ± 1.2 |
RESULTS
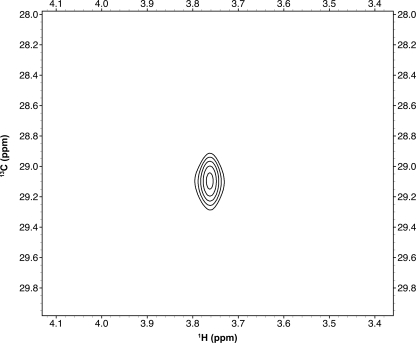
To probe CH···O hydrogen bonding between SET7/9 and the AdoMet methyl group, we examined the NMR chemical shift produced by the AdoMet methyl group while bound to SET7/9. To measure the bound 1H chemical shift for the AdoMet methyl group and distinguish it from resonances arising from the enzyme, we synthesized [13C]methyl-labeled AdoMet using AdoMet synthase and [methyl-13C]methioine (43, 63, 64) (for experimental details, please see “Experimental Procedures”). We recorded two-dimensional HSQC spectra of [methyl-13C]AdoMet in the presence of stoichiometric quantities of the unlabeled catalytic domain of SET7/9 (Fig. 1). The 1H chemical shift of the AdoMet methyl group was recorded as 3.8 ppm. This chemical shift was unusually far downfield for a methyl group and also represented a large downfield change relative to the reported chemical shift for the AdoMet methyl group free in solution (3.0 ppm) (65). Unlike 13C, downfield 1H chemical shift changes often qualitatively indicate hydrogen bond formation (23, 66–69); thus, we sought to verify our experimental chemical shift change using quantum chemistry calculations. Although this combination of techniques has been used to identify CH···O hydrogen bonds in small organic molecules (37, 70, 71) and in computational biology (68), it has, to our knowledge, not yet been applied experimentally in biological macromolecules. Using this combination of techniques, we reasoned that it should be possible to solve for the hydrogen positions and, thus hydrogen bonding patterns, of the AdoMet methyl group within the SET7/9 active site.
FIGURE 1.
Two-dimensional HSQC of the SET7/9·[methyl-13C]AdoMet complex.
Previous studies of CH···O hydrogen bonding in small organic molecules showed that the chemical shift calculations of hydrogen were usually accurate to 0.1 ppm of the experiment (37, 70, 71). Error in biological molecules could arise from many sources, including but not limited to implicit solvation modeling and large or truncated molecules used in calculations. Therefore, to validate the accuracy of our calculations for the AdoMet methyl group, we first calculated the chemical shift of the AdoMet methyl group using the solution state NMR structure of free AdoMet in water. Methyl proton geometry was optimized, allowing for the appropriate number of hydrogen bonds formed in solution by the methyl group, followed by chemical shift calculation (see “Experimental Procedures” for computational details and supplemental Table S1 for a list of all chemical shifts). By averaging all three values together to reproduce the single experimental methyl resonance, the difference in chemical shift between the experimental (3.0 ppm) (65) and calculated values was <0.1 ppm. This level of accuracy prompted us to attempt to locate the methyl protons of AdoMet within the active site of SET7/9.
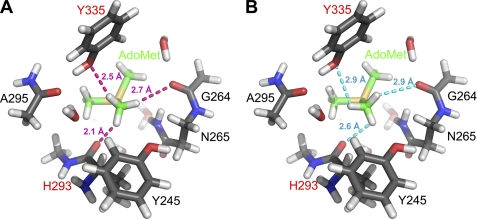
To compare with our experimental data, we then modeled the active site of SET7/9 using its crystal structure bound to AdoMet at 1.7 Å resolution (4). Hydrogens were added to this model, and the geometry of the hydrogen positions was optimized (see “Experimental Procedures” for details). Chemical shifts were then calculated for the methyl group protons and averaged to a single value, as performed for free AdoMet. The geometry-optimized structure of the AdoMet methyl group shows CH···O hydrogen bond formation from the AdoMet methyl group to the hydroxyl group of Tyr-335 and the main chain carbonyl oxygen of His-293, with H···O distances of 2.5 and 2.1 Å, respectively (Fig. 2A). Moreover, the C-H···O angles of 146° and 140°, respectively, are acceptable hydrogen bonding angles (16, 17, 72). As predicted, the calculations showed that protons participating in CH···O hydrogen bonds experienced significant downfield changes in chemical shift. The proton engaged in close hydrogen bonding with His-293 had a calculated chemical shift of 5.0 ppm, whereas the proton forming hydrogen bonds with Tyr-335 had a calculated chemical shift of 3.3 ppm. The chemical shift of the third proton was calculated to be 2.6 ppm. Averaging all three values together yielded a calculated chemical shift of 3.7 ppm, which is within 0.1 ppm of the experimental value of 3.8 ppm.
FIGURE 2.
Optimized active site with bound AdoMet (A) and manually rotated geometry (B) to eliminate CH···O hydrogen bonds. Truncated AdoMet and the protein are depicted with green and gray carbon atoms, respectively. Residues labeled in red designate CH···O acceptors. H···O distances from methyl protons to nearest oxygen atom for optimized and broken geometry are shown in magenta and cyan, respectively. The crystal structure of the SET7/9·AdoMet complex (Protein Data Bank code 1N6A) was used as the model for the calculations.
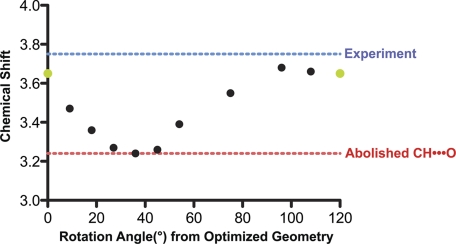
To further confirm the presence of CH···O hydrogen bonding, the methyl group was rotated manually through a range of 120° (example shown in Fig. 2B), and chemical shifts were recalculated at intermediate geometries to explore whether alternative hydrogen positions could agree with the experimental data. The rotation of the AdoMet methyl group resulted in a smooth functional change in the calculated chemical shift, passing through a minimum at 36° (Fig. 3). The minimum of this plot corresponded to the structure for which the model and the experimentally measured chemical shift differed by 0.6 ppm. This difference represents the following: 1) the largest deviation between the experiment and model, 2) the most upfield of the calculated chemical shifts, and 3) the model in which the methyl rotamer forms minimal CH···O hydrogen bonds in the active site (Fig. 2B). These calculations indicated that AdoMet methyl rotamers precluding CH···O hydrogen bond formation were inconsistent with the experimental chemical shift data. All of the calculated chemical shifts that agreed closely with experimental chemical shifts represented structures that optimized CH···O hydrogen bonding for at least one AdoMet methyl hydrogen atom. Collectively, the experimental data and calculations yielded direct evidence via NMR chemical shift for CH···O hydrogen bond formation between the AdoMet methyl group and oxygen atoms within the active site of SET7/9.
FIGURE 3.
Chemical shift of the AdoMet methyl group as a function of rotation angle. 0° and 120°, rendered in green, are equivalent geometry optimized positions (displayed in Fig. 2A), whereas all other points were derived from manual rotation of the AdoMet methyl group. The point at 36° represents the chemical shift calculated from the AdoMet methyl conformation shown in Fig. 2B. The blue and red dashed lines indicate the experimental chemical shift and the structure calculated to contain the least CH···O hydrogen bonding, respectively.
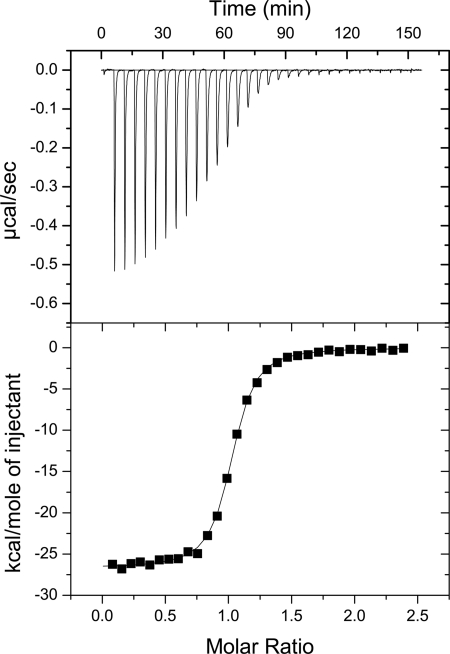
These findings prompted us to examine the thermodynamic parameters for cofactor binding by SET7/9 using ITC (see “Experimental Procedures” for experimental details). Binding affinities and enthalpies for SET7/9 were measured with AdoMet (Fig. 4), its methyl transfer product AdoHcy, and sinefungin, an AdoMet analogue in which the methyl sulfonium cation is substituted by an amine-methylene group that can participate in conventional NH···O hydrogen bonding. Crystal structures of SET7/9 and other SET domain enzymes illustrated that these cofactors share a structurally homologous binding mode (supplemental Fig. S4) (4, 5, 7). The ITC data revealed that SET7/9 displayed nanomolar affinities for AdoMet and sinefungin, whereas its affinity for AdoHcy was ∼1000-fold weaker (Table 1). These data were analogous to those obtained for the binding of these ligands to the SET domain protein Rubisco large subunit methyltransferase (LSMT), demonstrating consistency in rank order of binding affinities across different SET domain enzymes (6). In addition, SET7/9 and LSMT displayed comparable differences in binding enthalpy between AdoMet and sinefungin (∼3 kcal/mol). The high affinity that these enzymes displayed for AdoMet is presumably due to the ability of its methyl group to engage in CH···O hydrogen bonding, emphasizing the importance of these interactions in cofactor binding by SET domain KMTs.
FIGURE 4.
ITC analysis of AdoMet binding to SET7/9. The top panel represents the titration of AdoMet into SET7/9, whereas the bottom panel represents the binding isotherm with fitted curve.
DISCUSSION
The identification of CH···O hydrogen bonding between the AdoMet methyl group and oxygen atoms within the SET domain active site has implications for these interactions in lysine methyl transfer reactions. As proposed previously, CH···O hydrogen bonds appear to play roles in binding AdoMet, positioning its methyl group in an appropriate geometry for transfer, and stabilizing the SN2 transition state (6). These data suggest that the CH···O hydrogen bonds confer a specific orientation for the methyl group to align it during catalysis and could potentially limit its motion within the active site. Future studies will further define the specific roles that CH···O hydrogen bonds may play in promoting the methyl transfer reaction catalyzed by SET domain enzymes.
The thermodynamic analyses of SET7/9 bound to AdoMet and its analogues are consistent with previous findings on LSMT (6). The consistency of rank order in binding affinity indicates that CH···O hydrogen bonds are important in cofactor binding by multiple KMTs due to the structural conservation of the SET domain active site. Moreover, these CH···O interactions may in part explain the importance of the evolutionarily invariant Tyr-335 to enzyme function (Fig. 2A), given the propensity of its hydroxyl group to form CH···O hydrogen bonds with the AdoMet methyl group. It is also interesting to note that the change in binding affinity between AdoMet and its analogues is substantially more dramatic in SET7/9 than in LSMT. This effect could be a function of the plasticity of the SET7/9 active site (9), as compared with the preformed active site of LSMT (42). Future studies may address how conformational flexibility within the SET domain family influences CH···O hydrogen bonding to AdoMet.
In terms of methodology, our results have demonstrated that chemical shift can be used as a structural parameter for determining hydrogen positions and hydrogen bonding patterns within an enzyme active site. To our knowledge, these chemical shift data and calculations provide the first direct, quantitative evidence of CH···O hydrogen bonding in an enzyme active site in solution. One advantage of chemical shift as a probe to examine CH···O hydrogen bonding in biomolecular structure is the relative ease of data acquisition. In the future, this methodology could be applied broadly to characterize CH···O hydrogen bonding in proteins, nucleic acids, and other biological molecules, expanding our understanding of the functional importance of these interactions in macromolecular structure, ligand binding, and enzyme catalysis.
Supplementary Material
Acknowledgments
We thank George D. Markham for the gift of MetK DNA, coordinates for the free AdoMet structure, and advice on AdoMet synthesis and purification. We also acknowledge Samuel Krimm for useful discussions on CH···O hydrogen bonds and Charles L. Brooks III for the use of a computing cluster. The authors also acknowledge the University of Michigan LS&A computing cluster, as well as Alex Kurochkin for NMR expertise. Finally, the authors acknowledge Jeffrey Brender and Noemi Mirkin for advice and useful discussion of density functional theory methods.
This work was supported by National Institutes of Health Grant R01-GM073839 (to R. C. T.) and a National Science Foundation CAREER award (MCB 0644278) (to H. M. A.). The UCSF Chimera package is developed and distributed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by National Institutes of Health Grant P41 RR001081).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1, Figs. S1–S4, and additional references.
- KMT
- lysine methyltransferase
- AdoMet
- S-adenosylmethionine
- CH···O
- carbon-oxygen hydrogen bond
- B3LYP
- Becke, three-parameter, Lee-Yang-Parr
- MD
- molecular dynamics
- HSQC
- heteronuclear single quantum coherence
- ITC
- isothermal titration calorimetry
- AdoHcy
- S-adenosylhomocysteine
- LSMT
- Rubisco large subunit methyltransferase.
REFERENCES
- 1. Albert M., Helin K. (2010) Semin. Cell Dev. Biol. 21, 209–220 [DOI] [PubMed] [Google Scholar]
- 2. Dillon S. C., Zhang X., Trievel R. C., Cheng X. D. (2005) Genome Biol. 6, 227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pradhan S., Chin H. G., Esteve P. O., Jacobsen S. E. (2009) Epigenetics 4, 282–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kwon T., Chang J. H., Kwak E., Lee C. W., Joachimiak A., Kim Y. C., Lee J., Cho Y. (2003) EMBO J. 22, 292–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Xiao B., Jing C., Wilson J. R., Walker P. A., Vasisht N., Kelly G., Howell S., Taylor I. A., Blackburn G. M., Gamblin S. J. (2003) Nature 421, 652–656 [DOI] [PubMed] [Google Scholar]
- 6. Couture J. F., Hauk G., Thompson M. J., Blackburn G. M., Trievel R. C. (2006) J. Biol. Chem. 281, 19280–19287 [DOI] [PubMed] [Google Scholar]
- 7. Subramanian K., Jia D., Kapoor-Vazirani P., Powell D. R., Collins R. E., Sharma D., Peng J., Cheng X., Vertino P. M. (2008) Mol. Cell 30, 336–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jacobs S. A., Harp J. M., Devarakonda S., Kim Y., Rastinejad F., Khorasanizadeh S. (2002) Nat. Struct. Biol. 9, 833–838 [DOI] [PubMed] [Google Scholar]
- 9. Xiao B., Jing C., Kelly G., Walker P. A., Muskett F. W., Frenkiel T. A., Martin S. R., Sarma K., Reinberg D., Gamblin S. J., Wilson J. R. (2005) Gene Dev. 19, 1444–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hu P., Zhang Y. K. (2006) J. Am. Chem. Soc. 128, 1272–1278 [DOI] [PubMed] [Google Scholar]
- 11. Guo H. B., Guo H. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 8797–8802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhang X., Bruice T. C. (2007) Biochemistry 46, 14838–14844 [DOI] [PubMed] [Google Scholar]
- 13. Hu P., Wang S., Zhang Y. (2008) J. Am. Chem. Soc. 130, 3806–3813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ramachandran G. N., Sasisekharan V. (1965) Biochim. Biophys. Acta 109, 314–316 [DOI] [PubMed] [Google Scholar]
- 15. Krimm S. (1967) Science 158, 530–531 [DOI] [PubMed] [Google Scholar]
- 16. Desiraju G. R. (1991) Acc. Chem. Res. 24, 290–296 [Google Scholar]
- 17. Derewenda Z., Lee L., Kobos P., Derewenda U. (1995) FASEB J. 9, A1246–1246 [Google Scholar]
- 18. Jiang L., Lai L. H. (2002) J. Biol. Chem. 277, 37732–37740 [DOI] [PubMed] [Google Scholar]
- 19. Derewenda Z. S., Derewenda U., Kobos P. M. (1994) J. Mol. Biol. 241, 83–93 [DOI] [PubMed] [Google Scholar]
- 20. Bond C. S., Zhang Y., Berriman M., Cunningham M. L., Fairlamb A. H., Hunter W. N. (1999) Structure 7, 81–89 [DOI] [PubMed] [Google Scholar]
- 21. Bach R. D., Thorpe C., Dmitrenko O. (2002) J. Phys. Chem. B 106, 4325–4335 [Google Scholar]
- 22. Meyer P., Schneider B., Sarfati S., Deville-Bonne D., Guerreiro C., Boretto J., Janin J., Véron M., Canard B. (2000) EMBO J. 19, 3520–3529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ash E. L., Sudmeier J. L., Day R. M., Vincent M., Torchilin E. V., Haddad K. C., Bradshaw E. M., Sanford D. G., Bachovchin W. W. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 10371–10376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Quinn J. R., Zimmerman S. C., Del Bene J. E., Shavitt I. (2007) J. Am. Chem. Soc. 129, 934–941 [DOI] [PubMed] [Google Scholar]
- 25. Benevides J. M., Thomas G. J., Jr. (1988) Biochemistry 27, 3868–3873 [DOI] [PubMed] [Google Scholar]
- 26. Ghosh A., Bansal M. (1999) J. Mol. Biol. 294, 1149–1158 [DOI] [PubMed] [Google Scholar]
- 27. Liu H., Matsugami A., Katahira M., Uesugi S. (2002) J. Mol. Biol. 322, 955–970 [DOI] [PubMed] [Google Scholar]
- 28. Mizuno K., Ochi T., Shindo Y. (1998) J. Chem. Phys. 109, 9502–9507 [Google Scholar]
- 29. Ratajczyk T., Czerski I., Kamienska-Trela K., Szymanski S., Wojcik J. (2005) Angew Chem. Int. Edit. 44, 1230–1232 [DOI] [PubMed] [Google Scholar]
- 30. Qingzhong L., Wang N., Zhiwu Y. (2007) J. Mol. Struct. 847, 68–74 [Google Scholar]
- 31. Scheiner S. (2005) J. Phys. Chem. B 109, 16132–16141 [DOI] [PubMed] [Google Scholar]
- 32. Desiraju G. R. (1996) Acc. Chem. Res. 29, 441–449 [DOI] [PubMed] [Google Scholar]
- 33. Cordier F., Barfield M., Grzesiek S. (2003) J. Am. Chem. Soc. 125, 15750–15751 [DOI] [PubMed] [Google Scholar]
- 34. Sheppard D., Li D. W., Godoy-Ruiz R., Brüschweiler R., Tugarinov V. (2010) J. Am. Chem. Soc. 132, 7709–7719 [DOI] [PubMed] [Google Scholar]
- 35. Sukumar N., Mathews F. S., Langan P., Davidson V. L. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 6817–6822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Scheiner S. (2009) J. Phys. Chem. B 113, 10421–10427 [DOI] [PubMed] [Google Scholar]
- 37. Yates J. R., Pham T. N., Pickard C. J., Mauri F., Amado A. M., Gil A. M., Brown S. P. (2005) J. Am. Chem. Soc. 127, 10216–10220 [DOI] [PubMed] [Google Scholar]
- 38. Couture J. F., Dirk L. M., Brunzelle J. S., Houtz R. L., Trievel R. C. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 20659–20664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Del Rizzo P. A., Couture J. F., Dirk L. M., Strunk B. S., Roiko M. S., Brunzelle J. S., Houtz R. L., Trievel R. C. (2010) J. Biol. Chem. 285, 31849–31858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Trievel R. C., Beach B. M., Dirk L. M., Houtz R. L., Hurley J. H. (2002) Cell 111, 91–103 [DOI] [PubMed] [Google Scholar]
- 41. Zhang X., Yang Z., Khan S. I., Horton J. R., Tamaru H., Selker E. U., Cheng X. D. (2003) Mol. Cell 12, 177–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Trievel R. C., Flynn E. M., Houtz R. L., Hurley J. H. (2003) Nat. Struct. Biol. 10, 545–552 [DOI] [PubMed] [Google Scholar]
- 43. Iwig D. F., Booker S. J. (2004) Biochemistry 43, 13496–13509 [DOI] [PubMed] [Google Scholar]
- 44. Schalk-Hihi C., Markham G. D. (1999) Biochemistry 38, 2542–2550 [DOI] [PubMed] [Google Scholar]
- 45. Chirlian L. E., Francl M. M. (1987) J. Comput. Chem. 8, 894–905 [Google Scholar]
- 46. Frisch M. J., Schlegel H. B., Scuseria G. E., Robb M. A., Cheeseman J. R., Montgomery J. A., Jr., Vreven T., Kudin K. N., Burant J. C., Millam J. M., Iyengar S. S., Tomasi J., Barone V., Mennucci B., Cossi M., Scalmani G., Rega N., Petersson G. A., Nakatsuji H., Hada M., Ehara M., Toyota K., Fukuda R., Hasegawa J., Ishida M., Nakajima T., Honda Y., Kitao O., Nakai H., Klene M., Li X., Knox J. E., Hratchian H. P., Cross J. B., Bakken V., Adamo C., Jaramillo J., Gomperts R., Stratmann R. E., Yazyev O., Austin A. J., Cammi R., Pomelli C., Ochterski J. W., Ayala P. Y., Morokuma K., Voth G. A., Salvador P., Dannenberg J. J., Zakrzewski V. G., Dapprich S., Daniels A. D., Strain M. C., Farkas O., Malick D. K., Rabuck A. D., Raghavachari K., Foresman J. B., Ortiz J. V., Cui Q., Baboul A. G., Clifford S., Cioslowski J., Stefanov B. B., Liu G., Liashenko A., Piskorz P., Komaromi I., Martin R. L., Fox D. J., Keith T., Al-Laham M. A., Peng C. Y., Nanayakkara A., Challacombe M., Gill P. M., Johnson B., Chen W., Wong M. W., Gonzalez C., Pople J. A. (2004) Gaussian 03, Revision C. 02 Gaussian, Inc., Wallingford, CT [Google Scholar]
- 47. Brooks B. R., Brooks C. L., 3rd, Mackerell A. D., Jr., Nilsson L., Petrella R. J., Roux B., Won Y., Archontis G., Bartels C., Boresch S., Caflisch A., Caves L., Cui Q., Dinner A. R., Feig M., Fischer S., Gao J., Hodoscek M., Im W., Kuczera K., Lazaridis T., Ma J., Ovchinnikov V., Paci E., Pastor R. W., Post C. B., Pu J. Z., Schaefer M., Tidor B., Venable R. M., Woodcock H. L., Wu X., Yang W., York D. M., Karplus M. (2009) J. Comput. Chem. 30, 1545–1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Jorgensen W. L., Chandrasekhar J., Madura J. D., Impey R. W., Klein M. L. (1983) J. Chem. Phys. 79, 926–935 [Google Scholar]
- 49. Nosé S., Klein M. L. (1986) Phys. Rev. B Condens. Matter 33, 339–342 [DOI] [PubMed] [Google Scholar]
- 50. Essmann U., Perera L., Berkowitz M. L., Darden T., Lee H., Pedersen L. G. (1995) J. Chem. Phys. 103, 8577–8593 [Google Scholar]
- 51. Vanommeslaeghe K., Hatcher E., Acharya C., Kundu S., Zhong S., Shim J., Darian E., Guvench O., Lopes P., Vorobyov I., Mackerell A. D., Jr. (2010) J. Comput. Chem. 31, 671–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Markham G. D., Norrby P. O., Bock C. W. (2002) Biochemistry 41, 7636–7646 [DOI] [PubMed] [Google Scholar]
- 53. Becke A. D. (1993) J. Chem. Phys. 98, 5648–5652 [Google Scholar]
- 54. Ditchfield R., Hehre W. J., Pople J. A. (1971) J. Chem. Phys. 54, 724–728 [Google Scholar]
- 55. Miertus S., Scrocco E., Tomasi J. (1981) Chem. Phys. 55, 117–129 [Google Scholar]
- 56. Wolinski K., Hinton J. F., Pulay P. (1990) J. Am. Chem. Soc. 112, 8251–8260 [Google Scholar]
- 57. Scheiner S. (2010) Int. J. Quantum Chem. 110, 2775–2783 [Google Scholar]
- 58. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 59. Binkley J. S., Pople J. A., Hehre W. J. (1980) J. Am. Chem. Soc. 102, 939–947 [Google Scholar]
- 60. Clark T., Chandrasekhar J., Spitznagel G. W., Schleyer P. V. (1983) J. Comput. Chem. 4, 294–301 [Google Scholar]
- 61. Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 62. Goddard T. G., Kneller D. G. (2006) SPARKY 3. University of California, San Francisco [Google Scholar]
- 63. Markham G. D., Hafner E. W., Tabor C. W., Tabor H. (1980) J. Biol. Chem. 255, 9082–9092 [PubMed] [Google Scholar]
- 64. Park J., Tai J. Z., Roessner C. A., Scott A. I. (1995) Bioorg. Med. Chem. Lett. 5, 2203–2206 [Google Scholar]
- 65. Seeger K., Lein S., Reuter G., Berger S. (2005) Biochemistry 44, 6208–6213 [DOI] [PubMed] [Google Scholar]
- 66. Peralta J. E., de Azua M. C. R., Contreras R. H. (1999) J. Mol. Struc-Theochem. 491, 23–31 [Google Scholar]
- 67. Harris R. K. (1983) Nuclear Magnetic Resonance Spectroscopy, Pitman Books Ltd., London [Google Scholar]
- 68. Scheiner S., Gu Y., Kar T. (2000) J. Mol. Struct. 500, 441–452 [Google Scholar]
- 69. Afonin A. V., Ushakov I. A., Kuznetsova S. Y., Petrova O. V., Schmidt E. Y., Mikhaleva A. I. (2002) Magn. Reson. Chem. 40, 114–122 [Google Scholar]
- 70. Sigalov M., Vashchenko A., Khodorkovsky V. (2005) J. Org. Chem. 70, 92–100 [DOI] [PubMed] [Google Scholar]
- 71. Afonin A. V., Ushakov I. A., Vashchenko A. V., Simonenko D. E., Ivanov A. V., Vasil'tsov A. M., Mikhaleva A. I., Trofimov B. A. (2009) Magn. Reson. Chem. 47, 105–112 [DOI] [PubMed] [Google Scholar]
- 72. Sarkhel S., Desiraju G. R. (2004) Proteins 54, 247–259 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.