Abstract
The transactivation response element (TAR) DNA-binding protein-43 (TDP-43) is a nuclear protein that normally regulates transcription and splicing. Abnormal accumulation of insoluble inclusions containing TDP-43 has been recently reported in the affected tissues of amyotrophic lateral sclerosis (ALS) patients. Here, we show that intracellular aggregation of TDP-43 can be triggered by transduction of fibrillar aggregates prepared from in vitro functional TDP-43. Sarkosyl is found to be incapable of solubilizing those intracellularly seeded aggregates of TDP-43, which is consistent with the observation that TDP-43 inclusions in ALS patients are sarkosyl-insoluble. In addition, intracellular seeding in our cell models reproduces ubiquitination of TDP-43 aggregates, which is another prominent feature of TDP-43 inclusions in ALS patients. Although it has been so far difficult to initiate disease-associated changes of TDP-43 using cultured cell models, we propose that a seeding reaction is a key to construct a model to monitor TDP-43 pathologies.
Keywords: Amyloid, Amyotrophic Lateral Sclerosis (Lou Gehrig's Disease), Neurodegeneration, Protein Folding, Protein-Protein Interactions, Protein Aggregation
Introduction
Misfolding of a protein molecule often leads to its insoluble aggregation, and the fibrillar aggregates rich in β-sheet structures are called amyloid, in particular (1). Although some amyloid-like aggregates exhibit physiological functions such as synthesis of melanin (2) and storage of peptide hormones (3), intra- and extracellular formation of amyloid-like protein aggregates is also a major pathological hallmark observed in many of the conformational diseases such as neurodegenerative disorders (4). Indeed, genetic as well as proteomic analyses on human diseases have revealed increasing numbers of proteins that form inclusions in the affected tissues (for example, Ref. 5); however, a pathomechanism of amyloid formation still remains to be clarified.
The TAR3 DNA-binding protein 43 (TDP-43) was recently identified as a constituent protein of inclusions in most sporadic cases of a neurodegenerative disorder, amyotrophic lateral sclerosis (ALS) (6, 7). Later, dominant mutations in TDP-43 have also been reported to cause a familial form of ALS (8–11). In the pathological inclusions, TDP-43 proteins form bundles of straight filaments (12, 13), which are resistant to be solubilized with a detergent called sarkosyl (6, 7). In cultured cells, however, simple overexpression of recombinant TDP-43 proteins did not produce sarkosyl-insoluble TDP-43 even with ALS-causing mutations, artificial mutations disturbing nuclear localization of TDP-43, or proteasomal inhibition (14). C-terminal fragments of TDP-43 have been reported to form sarkosyl-insoluble aggregates in the cell, which is, however, observed only in the presence of a GFP tag (15). Indeed, we have found that HA-tagged C-terminal fragments of TDP-43 form inclusions in mouse neuroblastoma cells, Neuro2a, but those are sarkosyl-soluble.4 Although the pathogenic roles of TDP-43 aggregation in diseases remain obscure, no cultured cell models are currently available in which sarkosyl-insoluble TDP-43 aggregates are reproduced.
In vitro TDP-43 is intrinsically aggregation-prone and is shown to form fibrillar aggregates (16), whereas detergent solubility of the fibrils has not been fully examined. It is notable that in many proteins that can form insoluble fibrils in a test tube, simple overexpression in cells did not lead to the formation of aggregates. Formation of amyloid-like fibrils has often been compared with a crystallization process, in which preformed fibrils or “seeds” trigger fibrillar aggregation (17). In other words, a preformed fibril functions as a structural template to facilitate the recruitment of soluble proteins into insoluble fibrils. Indeed, in vitro aggregation of purified proteins usually arises after initial lag time during which no detectable aggregation occurs, but the lag time is dramatically reduced when seeds are included in the solution. Although there are significant amounts of amyloid-forming proteins overexpressed, the cultured cells may stay in the lag phase within a timescale of ordinary cell experiments. The protein aggregation in the cell would, therefore, be efficiently reproduced by introducing preformed fibrils into the cell as seeds. Actually, α-synuclein, which is a fibril-forming protein causative for Parkinson disease, did not form any inclusions in cultured cells, but intracellular α-synuclein aggregates appeared when α-synuclein fibrils in vitro are transduced into the cell (18). It is hence possible that intracellular seeding reproduces and facilitates the formation of highly insoluble TDP-43 aggregates inside the cell.
Here, we have first prepared a soluble and functional TDP-43 protein from its bacterial inclusions and then revealed that amyloid-like fibrillation of a full-length TDP-43 protein occurs through sarkosyl-resistant interactions in its C-terminal regions. We have further found that transduction of in vitro sarkosyl-insoluble TDP-43 fibrils in cultured cells triggers the aggregation of intracellular TDP-43 proteins; that is, a seeding reaction of TDP-43 occurs inside the cell. More importantly, intracellular TDP-43 aggregates produced by this seeding reaction were sarkosyl-insoluble and ubiquitinated, which reproduces the pathological changes of TDP-43. We thus propose that a seeding reaction of TDP-43 fibrils plays a key role in constructing experimental models to describe the pathological changes of TDP-43 molecules in neurodegenerative disorders.
EXPERIMENTAL PROCEDURES
Preparation of Recombinant TDP-43 Proteins
For preparation of TDP-43 proteins with an N-terminal His tag, cDNA of TDP-43 was cloned in a multiple cloning site (XhoI and BamHI) of a plasmid vector, pET15b (Novagen). After transfection of the plasmids in Escherichia coli (Rosetta), expression of His-tagged TDP-43 proteins was induced by the addition of 1 mm isopropyl β-d-thiogalactoside. Following incubation at 37 °C for 6 h, all TDP-43 proteins in this study were obtained as inclusion bodies, which were redissolved in PBS containing 6 m guanidine hydrochloride (GdnHCl) and then purified with Ni2+ affinity chromatography using the Proteus IMAC Midi kit (Pro-Chem Inc.). The bound TDP-43 proteins were washed with a wash buffer (pH 8.0) containing 50 mm Tris, 500 mm NaCl, 25 mm imidazole, and 6 m GdnHCl and then eluted from a spin column with an elution buffer (50 mm Tris, 200 mm NaCl, 125 mm imidazole, 6 m GdnHCl, pH 8.0).
For preparation of soluble and refolded TDP-43, TDP-43 proteins unfolded in the elution buffer (400 μm) were 40-fold diluted with a buffer (pH 8.0) containing 100 mm Na-Pi, 100 mm NaCl, 5 mm EDTA, 5 mm DTT, and 10% glycerol (buffer A), which, however, produced significant amounts of precipitates. These insoluble materials were removed by ultracentrifugation at 110,000 × g for 30 min, and we thus recovered the soluble TDP-43 proteins in a supernatant fraction. Protein concentration was spectroscopically determined from the absorbance at 280 nm by using the following extinction coefficients: 44,920 cm−1 m−1, TDP-43FL; 26,930 cm−1 m−1, TDP-43N; 17,990 cm−1 m−1, TDP-43C; 25,440 cm−1 m−1, TDP-43N-1; 19,480 cm−1 m−1, TDP-432-C.
Filter Binding Assay
TG- or AC-repeated DNA was modified with a biotin at the 5′-end (Operon Biotechnologies, Tokyo). To reduce retention of free single-stranded DNA, a nitrocellulose membrane (PROTRAN, 0.2 μm, Schleicher & Schuell) was presoaked for 10 min in 0.4 m KOH, rinsed in H2O, and then equilibrated in buffer A for an hour before use (19). The biotin-labeled TG- or AC-repeated DNA (biotin-(TG)12 or biotin-(AC)12, 0.1 μm) was incubated with soluble TDP-43 proteins (0.1–1.0 μm) in buffer A. After an hour at 37 °C, the mixture was filtered through a pretreated nitrocellulose membrane overlaid on a nylon membrane (Hybond-N+, 0.45 μm, Amersham Biosciences) in a 96-well slot-blot apparatus. After extensive washing of membranes with buffer A, the bound DNAs were cross-linked to the membranes by shortwave ultraviolet radiation (UV Stratalinker, Stratagene). Followed by blocking with 3% bovine serum albumin (BSA) in TBS with 0.1% Tween 20, the membranes were incubated with streptavidin-HRP (Promega), and the biotin-labeled DNAs on the membranes were developed with the SuperSignal West Dura chemiluminescent substrate (Pierce).
Biochemical and Morphological Analysis of in Vitro TDP-43 Aggregates
To examine aggregation of TDP-43 proteins, 1 μm soluble TDP-43 protein in buffer A was set in a 96-well plate and shaken at 37 °C at 1,200 rpm (BioShaker MBR-024, TAITEC). Turbidity was monitored at every 30 min in a plate reader (ARVO MX, PerkinElmer Life Sciences) by measuring absorbance around 405 nm. After agitation for 27 h, 25 μm thioflavin T was then added to the reaction mixture, and the fluorescence intensity was recorded by using a plate reader (ARVO MX) with a CW lamp filter (440 nm cutoff) and an emission filter (486 nm cutoff). Thioflavin T fluorescence before sample agitation was also examined. Final TDP-43 aggregates were obtained as a pellet fraction after ultracentrifugation of the reaction mixtures at 110,000 × g for 30 min.
For a seeding reaction, TDP-43 aggregates were resuspended in H2O, sheared with ultrasonication, and then added to a solution of 3 μm soluble TDP-43 proteins in buffer A containing 25 μm thioflavin T. The fluorescence was monitored by using SpectraMax M2 (Molecular Devices) at intervals of 2 min with 442 and 485 nm of excitation and emission wavelength, respectively. No plate shaking was performed during the reactions.
For morphological analysis, TDP-43 aggregates were first adsorbed on 400-mesh grids coated by a glow-charged supporting membrane. After washing with pure water, negative staining with 1% uranyl acetate was performed, and then images were obtained using an electron microscope (1200EX, JEOL).
Identification of Core Regions in TDP-43 Aggregates
22.5 μg of TDP-43 aggregates were first resuspended in 100 μl of a buffer (pH 8.0) containing 50 mm Tris, 100 mm NaCl, and 5 mm CaCl2 and mixed with 5 μg of Pronase (Calbiochem). After incubation at 37 °C for an hour, the mixture was ultracentrifuged at 110,000 × g for 30 min, and the supernatant fraction was discarded. The pellets were washed once with 150 μl of 50 mm Tris, 100 mm NaCl, pH 8.0 and redissolved in 20 μl of 50 mm Tris, 200 mm NaCl, 6 m GdnHCl, 5 mm EDTA, 5 mm DTT, pH 8.0. After desalting by using NuTip C-18 (Glygen Corp.), samples were mixed with a matrix, α-cyano-4-hydroxycinnamic acid, and MALDI-MS and MS/MS spectra were acquired using a 4800 Plus MALDI-TOF/TOF analyzer (Applied Biosystems). Identification of peptides based upon the observed m/z values was performed using MS-NonSpecific.
Plasmid Construction and Cell Culture
All plasmids for expression of TDP-43 proteins in cultured cells were constructed using pIRESneo3 (Clontech). Namely, cDNAs of full-length TDP-43 (TDP-43FL) and a C-terminal truncate of TDP-43 (TDP-432-C) that were fused with a C-terminal HA tag were cloned in a multiple cloning site using AgeI and BamHI sites.
Transduction of protein aggregates into cultured cells was performed by using the BioPORTER protein delivery reagent (Genlantis). On day 1, 1.2 × 105 cells of human embryonic kidney cells, HEK293T, in DMEM, 10% FBS were seeded on each well of a 12-well plate coated with polyethyleneimine and incubated at 37 °C. On the next day (day 2), 1.6 μg of plasmids was transfected per each well using Lipofectamine 2000 (Invitrogen). After 4 h of transfection, a culture medium was replaced with fresh DMEM, 10% FBS and incubated at 37 °C. On day 3, a culture medium was changed to Opti-MEM 1 h before aggregate transduction. A dried film of BioPORTER was first dissolved with 50 μl of PBS containing 10 μm protein aggregates and then mixed with 450 μl of Opti-MEM. By using this aggregate/BioPORTER-containing Opti-MEM, the transfected cells were further incubated at 37 °C for 4 h. The culture medium was then changed to DMEM, 0.5% FBS and again incubated at 37 °C overnight. On day 4, cells were harvested by PBS containing 1% sarkosyl with a protease inhibitor mixture, Complete, EDTA-free (Roche Diagnostics). Following lysis with ultrasonication, lysates were ultracentrifuged at 110,000 × g for 30 min to separate into supernatant (a soluble fraction) and pellets (an insoluble fraction). The pellets were redissolved in a 2% SDS-containing PBS with the same volume with that of the corresponding supernatant. The protein concentration in supernatant was determined by BCA assay using BSA as a standard.
Electrophoresis
Each of a soluble fraction containing 3 μg of total proteins and an insoluble fraction (10× volume of the corresponding soluble fraction) was mixed with an SDS-PAGE sample buffer containing β-mercaptoethanol and loaded on a 12.5% SDS-PAGE gel (PAGEL, ATTO) after boiling for 5 min. Following electrophoresis, proteins were electroblotted on a 0.2-μm PVDF membrane (Bio-Rad) and analyzed by Western blotting using rabbit polyclonal anti-HA (Y-11, Santa Cruz Biotechnology, 1:1,000), rabbit polyclonal anti-His probe (H-15, Santa Cruz Biotechnology, 1:1,000), rabbit polyclonal anti-ubiquitin (DAKO, 1:1,000), or mouse anti-TDP-43 phospho-Ser-409/410 antibody (Cosmo Bio, 1:1,000). Corresponding secondary antibodies that are conjugated with HRP were used as follows: anti-rabbit (1:5,000) and anti-mouse (1:1,000) antibodies. Blots were developed with the SuperSignal West Dura chemiluminescent substrate (Pierce), and images were obtained using LAS1000 (Fujifilm).
Immunocytochemistry
For immunostaining of HA-tagged TDP-43 proteins, cells were fixed with 4% paraformaldehyde containing 0.12 m sucrose in PBS for 10 min, permeabilized with 0.5% Triton X-100 in PBS for 5 min, and blocked with 0.1% BSA in PBS for 30 min. Cells were then incubated with anti-HA-fluorescein, high affinity (3F10) (1:100 dilution, Roche Diagnostics) in PBS containing 0.1% BSA for an hour, washed once with 0.1% Triton X-100 in PBS, and washed twice with 0.1% BSA in PBS. For detection of ubiquitin, rabbit polyclonal anti-ubiquitin mouse polyclonal ubiquitin (DAKO, 1:300) and anti-rabbit IgG conjugated with Alexa Fluor 555 (Invitrogen, 1:1,000) were used. Cells were incubated with Hoechst 33342 (Invitrogen, 1:3,000 dilution) in PBS for 15 min and then washed twice with PBS and mounted onto glass slides.
For visualization of TDP-432-C seeds added to cells, TDP-432-C aggregates were incubated in the presence of 1 mm tris(2-carboxyethyl)phosphine with 1 mm Alexa Fluor 555 maleimide (Invitrogen) for an hour at room temperature. Modified aggregates were washed three times with PBS by ultracentrifugation at 110,000 × g for 30 min and used for transduction experiments. When modified aggregates were resolubilized in PBS containing 2% SDS and loaded on a SDS-PAGE gel, a red fluorescent band with a molecular weight corresponding to that of TDP-432-C was found (data not shown), which confirms the successful modification.
Confocal images with a slice thickness of ∼1 μm were obtained by a laser scanning microscope of LSM5 Exciter system (Carl Zeiss, Germany) using a 40× objective lens. Obtained images were processed and analyzed by LSM image examiner (Carl Zeiss).
RESULTS AND DISCUSSION
Preparation of Functional TDP-43 from Bacterial Inclusion Bodies
Overexpression of recombinant human full-length TDP-43 (TDP-43FL) in E. coli has led to the formation of insoluble inclusion bodies, which can then be resolubilized in a buffer containing 6 m GdnHCl. Following 40-fold dilution and ultracentrifugation of resolubilized TDP-43FL in a GdnHCl-free buffer (see “Experimental Procedures”), we obtained at most a 2 μm solution of soluble TDP-43FL.
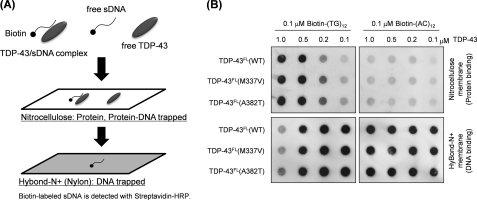
To test whether our soluble TDP-43FL retains its physiological functions, the DNA binding ability of TDP-43FL was examined by filter binding assays (20) with some modifications (Fig. 1A). It has been known that TDP-43 is a DNA/RNA-binding protein and preferentially recognizes TG/UG repeats of single-stranded DNA/RNA (21). Increasing amounts (0.1–1.0 μm) of soluble TDP-43FL were first mixed with either 0.1 μm biotin-labeled TG (biotin-(TG)12) or 0.1 μm biotin-labeled AC (biotin-(AC)12) repeats. Following incubation at 37 °C for an hour, the mixture was then filtered through a nitrocellulose membrane overlaid on a nylon membrane (Hybond-N+). In this assay method, free proteins and protein-DNA complexes are trapped on a nitrocellulose membrane, whereas free unbound DNAs flow through a nitrocellulose membrane but are trapped on a nylon membrane (Fig. 1A). As shown in Fig. 1B, increasing amounts of TDP-43FL shifted biotin-(TG)12 from a nylon to a nitrocellulose membrane, supporting the complex formation between TDP-43FL and biotin-(TG)12. In contrast, biotin-(AC)12 was detected only on a nylon membrane even in the highest TDP-43FL concentration (1 μm) among those tested, which is consistent with the fact that AC repeats are not the physiological target of TDP-43. It is also notable that no drastic changes of DNA binding were observed in TDP-43FL with ALS mutations (M337V and A382T). Based upon the intensities of blots, equal fractions of free and protein-bound TG repeats are expected between 0.1 and 0.5 μm TDP-43 (Fig. 1B). The dissociation constant of TDP-43FL with TG repeats can hence be roughly estimated as several hundred nm, which is comparable with that previously reported between truncated mouse TDP-43 and single-stranded (TG)6 repeats (∼100 nm) (20). Taking these results together, our soluble WT as well as ALS mutant TDP-43FL proteins are functional to the extent that TDP-43FL selectively recognizes and binds a physiological substrate, TG-repeated DNA.
FIGURE 1.
An assay for DNA binding of in vitro refolded TDP-43 proteins. A, a schematic representation of our filter binding assay using biotinylated single-stranded DNA (sDNA) as a substrate for TDP-43 proteins. A nitrocellulose membrane traps proteins including DNA-bound and unbound TDP-43, whereas unbound DNAs are recovered on a nylon membrane. B, 0.1 μm of either biotinylated (TG)12 (left) or biotinylated (AC)12 (right) was mixed with TDP-43FL (wild type as well as ALS mutant) at indicated concentrations. These mixtures were filtered through a nitrocellulose membrane (upper) overlaid on a nylon membrane (lower), and DNAs trapped on each of membranes were probed with Streptavidin-HRP.
Functional TDP-43 Forms Insoluble Fibrillar Aggregates after Vigorous Agitation
Given that TDP-43 constitutes pathological inclusions observed in ALS patients (6, 7), we have next examined in vitro aggregation propensities of soluble WT as well as ALS mutant (M337V and A382T) TDP-43FL proteins. Constant agitation (shaking at 1,200 rpm) of a solution containing TDP-43FL increased its turbidity without a discernable lag phase (Fig. 2A), showing aggregation of TDP-43FL. Although effects of ALS mutations on TDP-43 aggregation are still controversial (16), mutational differences could not be confirmed in the in vitro aggregation kinetics of TDP-43FL in our experimental conditions (Fig. 2A). After the turbidity change reached plateau, sample solutions were separated by ultracentrifugation into supernatant and pellets. As shown in Fig. 2B, WT as well as ALS mutant TDP-43FL proteins were detected only in the pellet fraction; furthermore, these TDP-43FL aggregates exhibited limited solubility in the presence of a detergent, sarkosyl. Soluble and functional TDP-43FL molecules are hence converted into sarkosyl-insoluble aggregates after constant agitation.
FIGURE 2.
Formation of sarkosyl-insoluble fibrillar aggregates of TDP-43FL proteins. A, aggregation kinetics of 1 μm refolded TDP-43FL proteins monitored by the increase of solution turbidity at 405 nm: open circles, WT; filled circles, M337V; open squares, A382T. Three independent experiments were performed to estimate errors. Error bars indicate S.E. B, formation of TDP-43FL aggregates with limited solubility in a buffer containing 1% sarkosyl. After the turbidity changes reached plateau (∼1,600 min in A), ultracentrifugation separated the soluble supernatant (lane 1, soluble fraction) from insoluble pellets, which were resuspended and sonicated in PBS containing 1% sarkosyl with the same volume as that of the corresponding supernatant. Ultracentrifugation again separated supernatant (lane 2, sarkosyl-soluble fraction) from the insoluble pellets, which were resolubilized in PBS containing 2% SDS with the same volume as that of the corresponding supernatant (lane 3, sarkosyl-insoluble fraction). Equal volumes of the samples were boiled in the presence of β-mercaptoethanol, loaded on a 12.5% SDS-PAGE gel, and stained with Coomassie Brilliant Blue. C, 1 μm TDP-43 before (open bars) and after (∼1,600 min in A, filled bars) aggregation was mixed with 25 μm thioflavin T. Fluorescence intensity was measured as described under “Experimental Procedures.” Three independent experiments were done to estimate errors. Error bars indicate S.E. D–F, electron micrograms of TDP-43FL aggregates: WT (D), M337V (E), and A382T (F). The bar in each panel represents 50 nm.
We have further found that not soluble but aggregated TDP-43FL increases the fluorescence intensity of thioflavin T (ThT) (Fig. 2C). Although ThT becomes fluorescent upon binding to amyloid-like fibrillar structures (22), the increase in its fluorescence intensity seems much smaller in TDP-43FL aggregates than those in the other protein aggregates such as SOD1 (23). This would confirm the previous observation that pathological TDP-43 inclusions are not well stained by amyloid binding dyes (24), although their morphology is expected to be fibrillar. Indeed, we have confirmed fibrillar morphologies of in vitro TDP-43FL aggregates by electron microscopic observation, where thin fibers appear to stack together and form big bundles (Fig. 2, D–F). There were no major differences in the aggregate morphologies between WT and ALS mutant (M337V and A382T) TDP-43FL. These data thus show that soluble and functional TDP-43FL forms insoluble amyloid-like fibrillar aggregates.
A C-terminal Half of TDP-43 Assumes a Core in the Fibrillar Aggregates
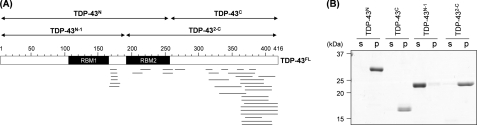
In general, structures of fibrillar aggregates are composed of a protease-resistant core(s) and the associated “fuzzy coat” that is susceptible to proteolysis (23, 25). To get structural insight into TDP-43 aggregates, we have attempted to identify a core region(s) of TDP-43FL aggregates. For that purpose, TDP-43FL aggregates were first treated with a nonspecific protease, Pronase, to remove the fuzzy coat regions. Then, resultant insoluble materials, which contain protease-resistant core(s) of TDP-43FL aggregates, were collected by ultracentrifugation, resolubilized with 6 m GdnHCl, and analyzed by MALDI-TOF mass spectrometry (supplemental Fig. S1). TDP-43 is a multidomain protein with two central RNA recognition motifs (RRMs) flanked by N-terminal and C-terminal domains; based upon the observed mass with MS/MS analysis, the identified peptides are found to be mapped on a half of a TDP-43FL primary sequence from the end of the first RRM to the C terminus (Fig. 3A, supplemental Table S1). Although this is consistent with previous studies suggesting a role of the C-terminal auxiliary domain in TDP-43 aggregation in vitro (16), these results suggest that a protease-resistant core in TDP-43FL aggregates extends to the region containing the second RRM domain.
FIGURE 3.
Identification of a core structure of TDP-43FL fibrillar aggregates. A, peptides corresponding to the mass peaks detected (see supplemental Fig. S1) are mapped on the primary sequence of TDP-43. Details are also summarized in supplemental Table S1. Numbers indicated above the primary sequence of TDP-43 (shown as open bars) represent the amino acid numbers. Four TDP-43 truncates examined here (TDP-43N, TDP-43N-1, TDP-43C, and TDP-432-C) are also indicated in the figure. B, an SDS-PAGE analysis of reduced solubility of TDP-43 truncates after agitation with 1,200 rpm at 37 °C for 21 h. Samples were ultracentrifuged at 110,000 × g for 30 min to separate soluble supernatant (s) and insoluble pellets (p). Insoluble pellets were redissolved in PBS containing 2% SDS before loading an SDS-PAGE gel.
To further examine a primary role of a C-terminal half of TDP-43 in the aggregate formation, we have examined in vitro aggregation propensities of the following four TDP-43 truncates (Fig. 3A): TDP-43N, Met1–Lys251; TDP-43C, Gly252–His416; TDP-43N-1, Met1–Lys192; TDP-432-C, Val193–His416. As shown in Fig. 3B, all truncates except TDP-43N-1 become insoluble pellets after shaking at 1,200 rpm for 21 h, whereas TDP-43N-1 remains completely soluble. It is notable that a region between the first and second RRMs is found to be a part of the protease-resistant core of in vitro TDP-43FL aggregates (Fig. 3A); nonetheless, no aggregation of TDP-43N-1 (Fig. 3B), which covers a region between the first and second RRMs, suggests a limited role of this region in the aggregate formation. Instead, efficient aggregation of TDP-43N (Fig. 3B) supports our notion that, in addition to a C-terminal auxiliary domain, the second RRM is also involved in the aggregate formation of TDP-43FL.
Indeed, TDP-432-C covering both the second RRM and a C-terminal domain can reproduce the aggregation behavior of TDP-43FL. Namely, a similar aggregation kinetics was observed between TDP-43FL (Fig. 2A) and TDP-432-C (supplemental Fig. S2A), both of which became completely insoluble and exhibited limited solubility in a sarkosyl-containing buffer at the end of the reaction (supplemental Fig. S2B). As in TDP-43FL, any mutational effects were also not evident on the aggregation kinetics of TDP-432-C. Aggregates of TDP-432-C (WT and ALS mutants) increased the intensity of ThT fluorescence, although its intensity was much higher (∼10-fold) than that of TDP-43FL aggregates (supplemental Fig. S2C). Aggregates of TDP-43FL and TDP-432-C, however, resemble each other in their morphologies; thin fibers are stacked to form big bundles (supplemental Fig. S2, D–F). A C-terminal half of TDP-43 (i.e. TDP-432-C) thus serves as a model to recapitulate the aggregation propensities of the full-length protein.
A Seeding Reaction Facilitates Aggregation of TDP-43 in Vitro and in Vivo
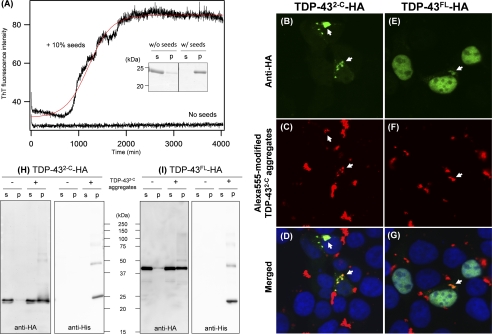
One of the notable features of amyloid-like fibrils is a seeding reaction, in which preformed fibrils act as seeds that accelerate the integration of soluble proteins into insoluble fibrils (17). In this study, we have first confirmed that a seeding reaction does facilitate the fibrillation of WT as well as ALS mutant TDP-43 proteins in vitro. Without any agitation, TDP-432-C exhibited no increase in the intensity of ThT fluorescence (Fig. 4A) and remained soluble (Fig. 4A, inset) for at least 3 days, showing no fibrillar aggregation. In contrast, even in the absence of agitation, the addition of 10% amounts of preformed TDP-432-C fibrils was sufficient to trigger the fibrillation of soluble TDP-432-C, as evidenced by increases in the ThT fluorescence intensity (Fig. 4A). After a seeding reaction, furthermore, all TDP-432-C molecules were detected in an insoluble pellet fraction (Fig. 4A, inset), suggesting the seeded fibrillation of TDP-432-C in vitro.
FIGURE 4.
A seeded fibrillation of TDP-43 proteins in vitro and in vivo. A, a seeded fibrillation of refolded TDP-432-C monitored by the increase of ThT fluorescence intensity. In the presence or absence of 0.3 μm (monomer base) sonicated TDP-432-C aggregates as seeds, 3 μm TDP-432-C was incubated at 37 °C without constant agitation (see “Experimental Procedures”). In this experimental condition, ThT fluorescence exhibited no changes until at least 4,000 min in the absence of added seeds but increased after ∼1,000 min of lag time in the presence of seeds. A red curve represents a sigmoidal fit to the data. As shown in the inset, an SDS-PAGE analysis has shown that all TDP-432-C molecules remain soluble without added seeds but become completely insoluble by seeding after incubation for 4,000 min. Soluble supernatant (s) and insoluble pellets (p) after ultracentrifugation are shown. B–G, intracellular seeded aggregation of TDP-432-C-HA (B–D) and TDP-43FL-HA (E–G) overexpressed in HEK293T cells by transduction of exogenous TDP-432-C aggregates modified with Alexa Fluor 555 (red in C and F). Cells were fixed, stained with an anti-HA-fluorescein antibody (green in B and E), and observed using a confocal microscope. Merged images are shown in D and G, and clear representative co-localization of intracellular TDP-43 proteins (green) with transduced TDP-432-C aggregates (red) is shown by white arrows. Nuclei were counterstained by Hoechst 33342 (blue in D and G). H and I, a seeding reaction by transduction of His-TDP-432-C aggregates reduces solubility of intracellular TDP-432-C-HA (H) and TDP-43FL-HA (I) in the presence of 1% sarkosyl. HEK293T cells expressing either TDP-432-C-HA (H) or TDP-43FL-HA (I) were transduced with exogenous His-TDP-432-C aggregates, lysed in the presence of 1% sarkosyl, and ultracentrifuged to prepare soluble supernatant (s) and insoluble pellets (p). The sample volume of insoluble fractions that are loaded on a gel is 10 times more than that of soluble fractions (see “Experimental Procedures”). Western blotting using an anti-HA antibody (left panels) has shown increased amounts of TDP-43FL/2-C proteins in the insoluble pellets after transduction of aggregates. Transduced TDP-432-C aggregates were also probed by using an anti-His antibody (right panels).
Although in vitro seeding reactions have been reported in many other amyloid-forming proteins (17), it should be noted that a seeding reaction of TDP-432-C can also proceed inside the cell. When transiently overexpressed in human cultured cells (HEK293T), TDP-432-C with a C-terminal HA tag (TDP-432-C-HA) remains diffused throughout the cell without formation of any inclusions (Fig. 5A). In contrast, TDP-432-C-HA formed cytoplasmic inclusions after intracellular transduction of TDP-432-C fibrils (Fig. 4B). Exogenous TDP-432-C fibrils were transduced into the cells after being modified with a thiol-specific fluorescent dye, Alexa Fluor 555; clear co-localization of intracellular TDP-432-C-HA with Alexa Fluor 555-modified TDP-432-C fibrils was observed with a confocal microscope (Fig. 4, B–D). We have also examined transduction of TDP-432-C fibrils in human neuroblastoma cells, SH-SY5Y, which was, however, found to be quite inefficient in our current experimental conditions. Nonetheless, these data obtained using HEK293T cells suggest that exogenous TDP-432-C fibrils are taken up by cells and then function as seeds to trigger aggregation of intracellular TDP-432-C-HA.
FIGURE 5.
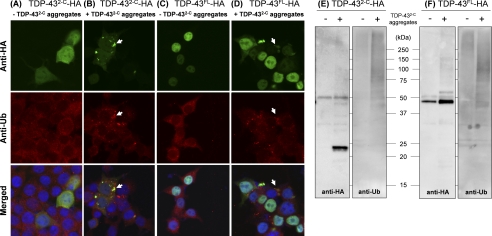
Intracellularly seeded TDP-43 aggregates with limited sarkosyl solubility are polyubiquitinated. A–D, immunocytochemistry of HEK293T cells was performed by double-staining with rat anti-HA-fluorescein antibodies (green, upper panels) and rabbit anti-Ub antibodies followed with Alexa Fluor 555-anti-rabbit antibodies (red, middle panels). Nuclei were counterstained with Hoechst 33342 (blue, lower panels). HEK293T cells transiently expressing TDP-432-C-HA (A and B) and TDP-43FL-HA (C and D) were examined with (B and D) or without (A and C) transduction of exogenous His-TDP-432-C aggregates. Representative clear co-localization of seeded TDP-43 aggregates (green) with ubiquitin (red) is shown by white arrows (see merged images in lower panels). E and F, sarkosyl-insoluble TDP-43 proteins (TDP-432-C-HA (E) and TDP-43FL-HA (F)) seeded by exogenous His-TDP-432-C aggregates were immunoprecipitated with anti-HA antibodies. A Western blotting analysis of immunoprecipitants by using anti-HA antibodies (E and F, left panels) confirms successful precipitation of sarkosyl-insoluble TDP-43 in the samples transduced with TDP-432-C aggregates. When these Western blots were probed with anti-Ub antibodies (E and F, right panels), smears in the high molecular weight region were observed in the seeded samples, suggesting polyubiquitination of seeded TDP-43FL/2-C aggregates.
TDP-43 Aggregates Formed by Intracellular Seeding Are Sarkosyl-insoluble
It has been reported that pathological TDP-43 aggregates are resistant to be solubilized in a 1% sarkosyl-containing buffer (6, 7). Indeed, we have found that both TDP-43FL and TDP-432-C fibrils prepared in vitro show limited solubility in the presence of 1% sarkosyl (Fig. 2B and supplemental Fig. S2B). After transduction of exogenous TDP-432-C fibrils, therefore, we performed the cell lysis in the presence of 1% sarkosyl and found significant amounts of intracellular TDP-432-C-HA in the insoluble pellets (Fig. 4H, left panel). Without the fibril transduction, however, intracellular TDP-432-C-HA remained almost completely soluble (Fig. 4H, left panel). Because exogenously added TDP-432-C seeds do not possess an HA tag, the blots probed with an anti-HA antibody in the pellet fraction (Fig. 4H, left panel) represent intracellularly expressed TDP-432-C-HA but not the exogenously added TDP-432-C fibrils. On the other hand, the transduced fibrils were prepared from TDP-432-C with an N-terminal His tag (His-TDP-432-C) and hence detected with an anti-His antibody (Fig. 4H, right panel); thereby, His-TDP-432-C seeds were found to be recovered in the pellets and distinguished from intracellular TDP-432-C-HA with its slightly slower electrophoretic mobility. Furthermore, a seeded aggregation is confirmed to occur inside the cell but not during the cell lysis, given that TDP-432-C-HA remains soluble after the addition of His-TDP-432-C fibrils in the lysates (supplemental Fig. S3A, left). Taking these results together, therefore, the intracellular aggregation of TDP-432-C-HA is triggered by exogenously transduced TDP-432-C fibrils.
We have also confirmed that the observed intracellular seeding is not a nonspecific process; intracellular aggregation of TDP-432-C-HA was not triggered by transducing the fibrils of an unrelated protein, SOD1 (23) (supplemental Fig. S3A, right). Nonetheless, we have found that TDP-432-C fibrils possess a “cross-seeding” activity to facilitate the intracellular aggregation of full-length TDP-43 (Fig. 4, E–G and I). In HEK293T cells, overexpressed TDP-43FL with an HA tag (TDP-43FL-HA) localized at the nucleus without any inclusions (Fig. 5C); upon transduction of TDP-432-C fibrils, however, some fractions of intracellular TDP-43FL-HA were co-localized with exogenously added TDP-432-C fibrils within cytoplasmic inclusions (Fig. 4, E–G). Concomitantly, furthermore, significant amounts of TDP-43FL-HA formed the sarkosyl-insoluble pellets (Fig. 4I). These changes were again not observed when TDP-432-C aggregates were added during the cell lysis (supplemental Fig. S3B, left) and did not occur with the transduction of SOD1 aggregates (supplemental Fig. S3B, right). Based upon these results, such a cross-seeding interaction between TDP-43FL and TDP-432-C may be relevant in describing the pathological observation that TDP-43 inclusions are found in the cytoplasm and composed of full-length TDP-43 and its C-terminal fragments.
As also performed in this study, a biochemical diagnosis for the TDP-43 aggregation is to test its decreased solubility in detergent-containing buffers. So far, several models using cultured cells have reproduced insoluble TDP-43 proteins; however, most of those have examined relatively mild solubilizing conditions such as a RIPA buffer (26–28) and a buffer containing Triton X-100 (29). Instead, pathological TDP-43 aggregates remain insoluble in more stringent conditions containing 1% sarkosyl (6, 7). To the extent that our seeded aggregates in the cell are insoluble after being treated with sarkosyl, a seeding reaction would well reproduce the formation of pathological TDP-43 aggregates.
In Vivo Seeding Recapitulates Molecular Pathological Changes of TDP-43
Another pathological feature observed in TDP-43 aggregates is ubiquitination (6, 7). By introducing in vitro TDP-432-C fibrils into cells expressing TDP-432-C-HA, we have again confirmed the seeded aggregation of intracellular TDP-432-C-HA and also found that the seeded aggregates were immunostained by anti-ubiquitin (Ub) antibodies (Fig. 5B). No inclusions, which are immunoreactive to anti-HA and anti-Ub antibodies, were observed in the absence of exogenously added fibrils (Fig. 5A). To test whether TDP-432-C-HA itself is ubiquitinated, furthermore, the seeded aggregates of TDP-432-C-HA were recovered as sarkosyl-insoluble pellets and redissolved in PBS containing 1% SDS. After ∼10-fold dilution with PBS to make final SDS concentration below 0.1%, resolubilized TDP-432-C-HA was then immunoprecipitated by incubation with the anti-HA antibodies immobilized on agarose. Successful immunoprecipitation of TDP-432-C-HA was confirmed by Western blotting using an anti-HA antibody, which detected a distinct band (∼25 kDa) with smears in the high molecular weight region (Fig. 5E, left). These smears were also probed with an anti-Ub antibody (Fig. 5E, right), consistent with polyubiquitination of TDP-432-C-HA.
TDP-43FL-HA forms cytoplasmic inclusions after transduction of TDP-432-C fibrils, which are also immunoreactive to anti-Ub antibody (Fig. 5D). When compared with TDP-432-C-HA (Fig. 5B), both HA immunofluorescence and Ub immunofluorescence are notably weaker in seeded aggregates of TDP-43FL-HA (Fig. 5D, white arrow). Furthermore, Ub-positive but HA-negative dot-like structures are evident. These structures were actually also observed in transduced cells expressing TDP-432-C-HA (Fig. 5B); however, it remains unknown what these structures mean. Given that no inclusions immunoreactive to Ub and/or HA were seen in non-transduced cells (Fig. 5C), formation of Ub and/or HA-positive inclusions is considered to be caused by transduction of exogenous TDP-432-C fibrils. Besides, Western blotting analysis on HA-immunoprecipitated aggregates confirms that seeded aggregates of TDP-43FL-HA are polyubiquitinated (Ub-positive smears in a high molecular weight region in Fig. 5F).
In addition to high insolubility and ubiquitination, truncation and phosphorylation have been observed in pathological TDP-43 (12, 30). Indeed, sarkosyl-insoluble fractions of the affected tissues of patients accumulate C-terminal phosphorylated fragments of TDP-43 (7). Any truncations were, however, not identified in intracellularly seeded aggregates of TDP-43FL in our current experimental conditions (Fig. 4I, left), implying that truncation is not a post-aggregation process. In addition, transduction of in vitro His-TDP-43FL fibrils was also found to trigger the formation of sarkosyl-insoluble TDP-43FL-HA aggregates in the cell (supplemental Fig. S4). Truncation may thus be optional for intracellular aggregation of TDP-43, although we do not exclude a possibility that truncation somehow facilitates the intracellular aggregation of TDP-43 and that the aggregates of truncated TDP-43 act as seeds to recruit full-length proteins into insoluble aggregates.
Regarding phosphorylation, Ser409 and Ser410 are known to be abnormally phosphorylated in the pathological TDP-43 aggregates (12), and its phospho-specific antibody (anti-TDP-43 (phospho-Ser-409/410)) has been commercially available. We found that this phospho-specific antibody detected His-TDP-432-C seeds added to the cell culture (supplemental Fig. S5, B and C) but not the intracellularly seeded aggregates of TDP-43FL/2-C-HA (supplemental Fig. S5, A and C). In Western blotting analysis, furthermore, the anti-TDP-43 (phospho-Ser-409/410) antibody detected our in vitro His-tagged TDP-432-C (supplemental Fig. S5, D and E), which was purified from bacterial inclusions and thus not supposed to be phosphorylated. It is thus complicated in our current experiments to examine whether the seeded aggregates of TDP-43 in the cell are phosphorylated. In addition, the anti-TDP-43 (phospho-Ser-409/410) antibody detected TDP-432-C more efficiently than TDP-43FL (supplemental Fig. S5, D and E), implying that this antibody specifically recognizes the C-terminal truncates of TDP-43 even in the absence of phosphorylation. Although more detailed investigations are now underway in our groups, previous studies have suggested that phosphorylation at Ser409/410 is not required for inclusion formation or toxicity of TDP-43 (26, 31). We have also observed few changes in the intracellular aggregation propensities of TDP-43, even when all Ser residues in the C-terminal domain were mutated to either Ala or Glu.4 Phosphorylation would hence be a distinct process from the formation of sarkosyl-insoluble aggregates.
In summary, we have shown in vitro formation of sarkosyl-insoluble TDP-43 fibrils, in which a C-terminal half of the protein constitutes a protease-resistant core structure. TDP-43 fibrils were furthermore found to exhibit a seeding activity in vitro and in vivo, which would reproduce the pathological formation of sarkosyl-insoluble ubiquitinated TDP-43 inclusions in the cell. It remains controversial whether the aggregation of TDP-43 is a cause or a result of the disease (32–34); however, as recently proposed in the other neurodegenerative diseases (35, 36), a seeding activity of TDP-43 proteins may contribute to the propagation of pathological changes with the progression of diseases.
Supplementary Material
Acknowledgments
We thank the Support Unit for Bio-material Analysis, RIKEN Brain Science Institute Research Resources Center, especially Masaya Usui and Kaori Otsuki for mass analysis and Tsuyako Hayashida for electron micrographic observations.
This work was supported by Grants-in-Aid 22240037 (to N. N.), 22110004 for Scientific Research on Innovative Areas (to N. N.), 22770162 (to Y. F.), and 21390274 (to K. Y.) from the Ministry of Education, Culture, Sports, Science and Technology of Japan, a grant from Core Research for Evolutional Science and Technology of Japan Science and Technology Agency (to K. Y.) a Japanese Health and Labor Sciences Research Grant (to K. Y.), a Strategic Research Program for Brain Science (to N. N.), and a grant from the Naito Foundation (to Y. F.).
This article was selected as a Paper of the Week.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5 and Table S1.
Y. Furukawa and N. Nukina, unpublished data.
- TAR
- transactivation response element
- TDP-43
- the TAR DNA-binding protein-43
- ALS
- amyotrophic lateral sclerosis
- RRM
- RNA recognition motif
- GdnHCl
- guanidine hydrochloride
- Ub
- ubiquitin
- ThT
- thioflavin T.
REFERENCES
- 1. Chiti F., Dobson C. M. (2009) Nat. Chem. Biol. 5, 15–22 [DOI] [PubMed] [Google Scholar]
- 2. Fowler D. M., Koulov A. V., Alory-Jost C., Marks M. S., Balch W. E., Kelly J. W. (2006) PLoS Biol. 4, e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Maji S. K., Perrin M. H., Sawaya M. R., Jessberger S., Vadodaria K., Rissman R. A., Singru P. S., Nilsson K. P., Simon R., Schubert D., Eisenberg D., Rivier J., Sawchenko P., Vale W., Riek R. (2009) Science 325, 328–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chiti F., Dobson C. M. (2006) Annu. Rev. Biochem. 75, 333–366 [DOI] [PubMed] [Google Scholar]
- 5. Aguzzi A., O'Connor T. (2010) Nat. Rev. Drug Discov. 9, 237–248 [DOI] [PubMed] [Google Scholar]
- 6. Arai T., Hasegawa M., Akiyama H., Ikeda K., Nonaka T., Mori H., Mann D., Tsuchiya K., Yoshida M., Hashizume Y., Oda T. (2006) Biochem. Biophys. Res. Commun. 351, 602–611 [DOI] [PubMed] [Google Scholar]
- 7. Neumann M., Sampathu D. M., Kwong L. K., Truax A. C., Micsenyi M. C., Chou T. T., Bruce J., Schuck T., Grossman M., Clark C. M., McCluskey L. F., Miller B. L., Masliah E., Mackenzie I. R., Feldman H., Feiden W., Kretzschmar H. A., Trojanowski J. Q., Lee V. M. (2006) Science 314, 130–133 [DOI] [PubMed] [Google Scholar]
- 8. Gitcho M. A., Baloh R. H., Chakraverty S., Mayo K., Norton J. B., Levitch D., Hatanpaa K. J., White C. L., 3rd, Bigio E. H., Caselli R., Baker M., Al-Lozi M. T., Morris J. C., Pestronk A., Rademakers R., Goate A. M., Cairns N. J. (2008) Ann. Neurol. 63, 535–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kabashi E., Valdmanis P. N., Dion P., Spiegelman D., McConkey B. J., Vande Velde C., Bouchard J. P., Lacomblez L., Pochigaeva K., Salachas F., Pradat P. F., Camu W., Meininger V., Dupre N., Rouleau G. A. (2008) Nat. Genet. 40, 572–574 [DOI] [PubMed] [Google Scholar]
- 10. Sreedharan J., Blair I. P., Tripathi V. B., Hu X., Vance C., Rogelj B., Ackerley S., Durnall J. C., Williams K. L., Buratti E., Baralle F., de Belleroche J., Mitchell J. D., Leigh P. N., Al-Chalabi A., Miller C. C., Nicholson G., Shaw C. E. (2008) Science 319, 1668–1672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Van Deerlin V. M., Leverenz J. B., Bekris L. M., Bird T. D., Yuan W., Elman L. B., Clay D., Wood E. M., Chen-Plotkin A. S., Martinez-Lage M., Steinbart E., McCluskey L., Grossman M., Neumann M., Wu I. L., Yang W. S., Kalb R., Galasko D. R., Montine T. J., Trojanowski J. Q., Lee V. M., Schellenberg G. D., Yu C. E. (2008) Lancet Neurol 7, 409–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hasegawa M., Arai T., Nonaka T., Kametani F., Yoshida M., Hashizume Y., Beach T. G., Buratti E., Baralle F., Morita M., Nakano I., Oda T., Tsuchiya K., Akiyama H. (2008) Ann. Neurol 64, 60–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lin W. L., Dickson D. W. (2008) Acta Neuropathol. 116, 205–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nonaka T., Arai T., Buratti E., Baralle F. E., Akiyama H., Hasegawa M. (2009) FEBS Lett. 583, 394–400 [DOI] [PubMed] [Google Scholar]
- 15. Nonaka T., Kametani F., Arai T., Akiyama H., Hasegawa M. (2009) Hum. Mol. Genet. 18, 3353–3364 [DOI] [PubMed] [Google Scholar]
- 16. Johnson B. S., Snead D., Lee J. J., McCaffery J. M., Shorter J., Gitler A. D. (2009) J. Biol. Chem. 284, 20329–20339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Harper J. D., Lansbury P. T., Jr. (1997) Annu. Rev. Biochem. 66, 385–407 [DOI] [PubMed] [Google Scholar]
- 18. Luk K. C., Song C., O'Brien P., Stieber A., Branch J. R., Brunden K. R., Trojanowski J. Q., Lee V. M. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 20051–20056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wong I., Lohman T. M. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 5428–5432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kuo P. H., Doudeva L. G., Wang Y. T., Shen C. K., Yuan H. S. (2009) Nucleic Acids Res. 37, 1799–1808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Buratti E., Baralle F. E. (2001) J. Biol. Chem. 276, 36337–36343 [DOI] [PubMed] [Google Scholar]
- 22. LeVine H., 3rd (1999) Methods Enzymol. 309, 274–284 [DOI] [PubMed] [Google Scholar]
- 23. Furukawa Y., Kaneko K., Yamanaka K., Nukina N. (2010) J. Biol. Chem. 285, 22221–22231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kerman A., Liu H. N., Croul S., Bilbao J., Rogaeva E., Zinman L., Robertson J., Chakrabartty A. (2010) Acta Neuropathol. 119, 335–344 [DOI] [PubMed] [Google Scholar]
- 25. Wischik C. M., Novak M., Thøgersen H. C., Edwards P. C., Runswick M. J., Jakes R., Walker J. E., Milstein C., Roth M., Klug A. (1988) Proc. Natl. Acad. Sci. U.S.A. 85, 4506–4510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dormann D., Capell A., Carlson A. M., Shankaran S. S., Rodde R., Neumann M., Kremmer E., Matsuwaki T., Yamanouchi K., Nishihara M., Haass C. (2009) J. Neurochem. 110, 1082–1094 [DOI] [PubMed] [Google Scholar]
- 27. Igaz L. M., Kwong L. K., Chen-Plotkin A., Winton M. J., Unger T. L., Xu Y., Neumann M., Trojanowski J. Q., Lee V. M. (2009) J. Biol. Chem. 284, 8516–8524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Winton M. J., Igaz L. M., Wong M. M., Kwong L. K., Trojanowski J. Q., Lee V. M. (2008) J. Biol. Chem. 283, 13302–13309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhang Y. J., Xu Y. F., Dickey C. A., Buratti E., Baralle F., Bailey R., Pickering-Brown S., Dickson D., Petrucelli L. (2007) J. Neurosci. 27, 10530–10534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Igaz L. M., Kwong L. K., Xu Y., Truax A. C., Uryu K., Neumann M., Clark C. M., Elman L. B., Miller B. L., Grossman M., McCluskey L. F., Trojanowski J. Q., Lee V. M. (2008) Am. J. Pathol 173, 182–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhang Y. J., Xu Y. F., Cook C., Gendron T. F., Roettges P., Link C. D., Lin W. L., Tong J., Castanedes-Casey M., Ash P., Gass J., Rangachari V., Buratti E., Baralle F., Golde T. E., Dickson D. W., Petrucelli L. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 7607–7612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Igaz L. M., Kwong L. K., Lee E. B., Chen-Plotkin A., Swanson E., Unger T., Malunda J., Xu Y., Winton M. J., Trojanowski J. Q., Lee V. M. (2011) J. Clin. Invest. 121, 726–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wegorzewska I., Bell S., Cairns N. J., Miller T. M., Baloh R. H. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 18809–18814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wils H., Kleinberger G., Janssens J., Pereson S., Joris G., Cuijt I., Smits V., Ceuterick-de Groote C., Van Broeckhoven C., Kumar-Singh S. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 3858–3863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Aguzzi A., Rajendran L. (2009) Neuron 64, 783–790 [DOI] [PubMed] [Google Scholar]
- 36. Brundin P., Melki R., Kopito R. (2010) Nat. Rev. Mol. Cell Biol. 11, 301–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.