Abstract
During gene transcription, the RNA polymerase (Pol) active center can catalyze RNA cleavage. This intrinsic cleavage activity is strong for Pol I and Pol III but very weak for Pol II. The reason for this difference is unclear because the active centers of the polymerases are virtually identical. Here we show that Pol II gains strong cleavage activity when the C-terminal zinc ribbon domain (C-ribbon) of subunit Rpb9 is replaced by its counterpart from the Pol III subunit C11. X-ray analysis shows that the C-ribbon has detached from its site on the Pol II surface and is mobile. Mutagenesis indicates that the C-ribbon transiently inserts into the Pol II pore to complement the active center. This mechanism is also used by transcription factor IIS, a factor that can bind Pol II and induce strong RNA cleavage. Together with published data, our results indicate that Pol I and Pol III contain catalytic C-ribbons that complement the active center, whereas Pol II contains a non-catalytic C-ribbon that is immobilized on the enzyme surface. Evolution of the Pol II system may have rendered mRNA transcript cleavage controllable by the dissociable factor transcription factor IIS to enable promoter-proximal gene regulation and elaborate 3′-processing and transcription termination.
Keywords: Gene Expression, RNA Polymerase, RNA Polymerase II, RNA Synthesis, Transcription
Introduction
The eukaryotic RNA polymerases Pol3 I, II, and III share a highly conserved active center (1–3) that catalyzes DNA-dependent RNA synthesis during gene transcription. The same active center also catalyzes cleavage of the nascent transcript during proofreading and recovery from transcription arrest (4, 5). Transcript cleavage is essential for cell viability (6). Despite the active center conservation, the strength of this intrinsic cleavage activity greatly differs between polymerases. Although the cleavage activity is very strong for Pol I (3) and Pol III (7, 8), it is very weak for Pol II. The molecular basis for this phenomenon remains unknown.
Intrinsic transcript cleavage requires the homologous subunits A12.2, Rpb9, and C11 in Pol I, II, and III, respectively (3, 9, 10). Archaea contain a related protein, TFS, which is required for RNA cleavage by the polymerase (11, 12). All these proteins consist of two zinc-binding β-ribbon domains. Rpb9 resides on the enzyme surface, where its N-terminal zinc ribbon (N-ribbon) forms part of the Rpb1/9 jaw, and its C-terminal zinc ribbon (C-ribbon) binds between the Rpb1 funnel domain and the Rpb2 domains lobe and external-1 (Fig. 1). The very weak cleavage activity of Pol II is greatly stimulated by TFIIS, which contains a Pol II-binding domain that is located at the Rpb1/9 jaw and a C-ribbon (5, 13). The TFIIS C-ribbon binds the pore beneath the Pol II active center and reaches the active site with a hairpin containing the invariant residues Arg-287, Asp-290, and Glu-291 that are required for function (5, 14–18). Although A12.2 and C11 contain these three hairpin residues, Rpb9 lacks the residue corresponding to Glu-291. It remains unclear how the ribbon domains are related evolutionarily and mechanistically and how this may result in different cleavage activities. Here we used a combination of mutagenesis, cleavage assays, and x-ray crystallography to unravel the molecular basis for differential intrinsic RNA cleavage activities of Pol II and Pol III and suggest how the C-ribbon domains are related evolutionarily and how different cleavage activities arose during evolution.
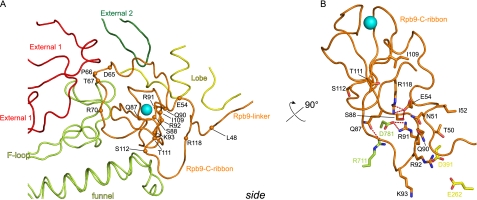
FIGURE 1.
Structure of the Pol II-TFIIS complex with nucleic acids (18) viewed from the side (A) and proposed evolutionary relationship between zinc ribbon domains in TFS, A12.2, C11, Rpb9, and TFIIS (B). Highlighted are Rpb9 (orange) and TFIIS (green).
EXPERIMENTAL PROCEDURES
Purification of RNA Polymerases
The Δrpb9 strain was created from strain BJ5464 Rpb3 His-Bio (19) by homologous recombination. The natNT2 cassette was amplified by polymerase chain reaction (PCR) from pFA6a-natNT2 (20) and was used to replace the RPB9 ORF. The gene replacement was verified by PCR. Pol IIΔ9 was purified essentially as described (21). Pyrococcus furiosus RNA polymerase was purified as described (22).
Mutagenesis
Fragments containing wild-type Rpb9 coding regions or C11 coding regions were amplified by PCR using NdeI and XhoI restriction sites at the 5′- and 3′-terminus, respectively. The same strategy was used for other fusion proteins. Each PCR product was digested with restriction endonucleases and inserted into the pET 28b(+) expression vector (Novagen) to generate a fusion protein with an N-terminal hexahistidine tag. All plasmids were verified by sequencing.
Preparation of Recombinant Proteins
BL21(DE3) cells containing the plasmid were grown to A600 = 0.6 at 37 °C. Expression was induced overnight at 18 °C by the addition of 40 μm isopropyl-1-thio-β-d-galactopyranoside. Cells were sonicated in PBS buffer containing 10 μm ZnCl2. All subsequent steps were performed at 4 °C. The supernatant of the resulting crude extract was collected by centrifugation and applied to a 1-ml HisTrap column (GE Healthcare), charged with 100 mm NiSO4, and equilibrated in purification buffer (50 mm Hepes, pH 7.5, 10 μm ZnCl2, 10% glycerol) containing 100 mm NaCl. The column was washed with 10 column volumes of purification buffer containing 2 m NaCl and 40 mm imidazole followed by another 10 column volumes of purification buffer containing 100 mm NaCl and 100 mm imidazole. The fusion protein was eluted from the column with purification buffer containing 100 mm NaCl and 300 mm imidazole. The eluate was concentrated and further purified on a gel filtration column (Superdex 75, GE Healthcare), equilibrated with transcription buffer (20 mm Hepes, pH 7.6, 60 mm (NH4)2SO4, 8 mm MgCl2, 10 μm ZnCl2, 10% glycerol, 10 mm DTT), and stored at −80 °C. For archaeal TFS and its TFS-DE/AA variant, heat treatment (90 °C, 20 min) was performed after cell lysis. After ultracentrifugation (1 h at 30,000 rpm), the proteins remained in the supernatant and were purified as above. The final preparation for each protein had a protein concentration between 0.2 and 0.5 mg/ml.
Transcript Cleavage Assay
5 pmol of Pol IIΔ9 were assembled on a hybrid of the DNA template strand annealed to the RNA (2-fold molar excess) in transcription buffer for 15 min at 20 °C followed by the biotinylated non-template DNA strand (4-fold molar excess) for 10 min at 25 °C and then by recombinant Rpb4/7 (5-fold molar excess) for 10 min at 25 °C. The bead-based cleavage assay was carried out as described (21, 23). Briefly, after elongation complex assembly, streptavidin-coated beads (Dynabeads MyOne Streptavidin T1, Invitrogen) were added (20 μl of beads per reaction, 30 min at 25 °C). Beads were resuspended in transcription buffer, and the purified recombinant proteins (5-fold molar excess) were added followed by incubation at 28 °C. For assays with P. furiosus RNA polymerase, purified TFS and its variants (5-fold molar excess) were added to the reaction mixture followed by incubation at 70 °C for 10 min. The reactions were stopped by adding an equal volume of 100 mm EDTA. Samples were loaded on a 20% polyacrylamide gel containing 7 m urea. The FAM 5′-labeled RNA products were visualized with a Typhoon 9400 scanner (GE Healthcare). All gel bands were quantified using ImageQuant7 (GE Healthcare).
Crystal Structure Determination
Pol IIΔ9 was incubated with 5-fold molar excess of recombinant Rpb4/7 for 20 min at 20 °C. The complex was purified on a Superose 6 gel filtration column (GE Healthcare), which was equilibrated with buffer (5 mm HEPES, pH 7.25, 40 mm (NH4)2SO4, 10 μm ZnCl2, 5 mm DTT) to generate the 11-subunit Pol IIΔ9. For buffer exchange, a single gel filtration run was performed with Rpb9-C11-1. Equal molar amounts of 11-subunit Pol IIΔ9 and Rpb9-C11-1 were mixed and incubated for 30 min at 20 °C and concentrated to a final concentration of 4.5 mg/ml. Crystals were grown at 20 °C using the hanging drop vapor diffusion method by mixing 2.5 μl of the protein mixture solution with 1 μl of reservoir solution (50 mm Hepes, pH 7.0, 200 mm NH4Ac, 300 mm NaAc, 5–6% w/v PEG 6000, 5 mm tris(2-carboxyethyl)phosphine). Crystals grew to a maximum size of ∼0.1 × 0.1 × 0.1 mm3. For cryo-protection, crystals were transferred stepwise over 5 h to the reservoir solution containing additionally 0–22% (v/v) glycerol. After incubation at 8 °C for 24 h in the presence of cryo-protectant, crystals were flash-cooled by plunging them into liquid nitrogen. Diffraction data were collected as consecutive series of 0.25° rotation images at cryo-temperature at the beamline X06SA at the Swiss Light Source. Diffraction data were processed with XDS and scaled with XSCALE (24). The structure was solved by molecular replacement with PHASER (25) using the structure of the 12-subunit Pol II (Protein Data Bank (PDB) code 3HOU (21)) with Rpb9 omitted as a search model. The structure was refined with PHENIX (26), using additional hydrogen-bond distance restraints for secondary structure elements (27), against the observed data that were sharpened with a B-factor of −80 Å2. Coordinates and structure factors were deposited in the Protein Data Bank under accession code 3QT1.
RESULTS
A Pol II Variant with Strong Intrinsic RNA Cleavage
Previous studies suggested that the weak intrinsic transcript cleavage activity of Pol II depends on Rpb9 and would thus be allosteric (9). We aimed at investigating the basis for intrinsic RNA cleavage by preparing Pol II lacking Rpb9 (Pol IIΔ9), complementing the Pol IIΔ9 enzyme with Rpb9 variants, and investigating the resulting Pol II variants for their cleavage activity. We prepared Pol IIΔ9 from a yeast strain lacking the rpb9 gene (20) (“Experimental Procedures”). As expected, Pol IIΔ9 was inactive in cleaving the RNA 3′-end in reconstituted elongation complexes with a 3′-RNA-DNA G-G mismatch (Fig. 2A and “Experimental Procedures”) (3, 21), whereas the addition of Rpb9 led to mild cleavage stimulation (Fig. 2B, lanes 2 and 3, and Fig. 3).
FIGURE 2.
Pol II cleavage activity strongly depends on the addition of protein variants shown in Fig. 3. A, nucleic acid scaffold used for the assays. Non-template and template DNA are shown in cyan and blue, respectively, and RNA is in red. A mismatched nucleotide at the RNA 3′-end is shown in brown. B, electrophoretic separation of RNA products in cleavage assay using different protein variants (compare “Experimental Procedures” and compare Fig. 3). RNA bands obtained after cleavage of two or four nucleotides are indicated by arrows (−2 and −4, respectively). React. time, reaction time.
FIGURE 3.
Protein variants used in functional and structural analysis. A, top, an alignment of amino acid sequences of the C-ribbons in Saccharomyces cerevisiae (Sc) Rpb9, A12.2, C11, and TFIIS and P. furiosus (Pfu) TFS is shown. Secondary structure elements in Rpb9 and TFIIS are in orange and green, respectively. Below the alignment, the C-terminal sequences of the fusion protein variants are shown. B, quantification of cleavage activities determined in Fig. 2. For each reaction, the amounts of uncleaved RNA and −2 and −4 cleavage products were quantified. The cleavage activity was calculated as the percentage of −2 and −4 cleavage products with respect to total RNA observed. Reaction times (React.time) of 10 and 60 min are indicated as red and dark green bars, respectively. Average values for two independent experiments are shown. Experiments were highly reproducible.
To explore whether the Pol III subunit C11 may replace Rpb9 function in Pol II, we prepared a Rpb9-C11 fusion protein that contains the Rpb9 N-ribbon fused to the C11 C-ribbon. We fused Rpb9 residues 1–74 to C11 residues 69–110 (protein variant Rpb9-C11-0, Fig. 3) because the Rpb9 N-ribbon and the linker between the two ribbon domains, including the conserved residues 65–70, interact with Pol II (28). Surprisingly, this fusion protein conferred very strong RNA cleavage activity to Pol II (Fig. 2B, lane 4, and Fig. 3).
To investigate this interesting gain-of-function mutation, we prepared and functionally analyzed a total of 27 fusion protein variants (variants Rpb9-C11-0 to -24, Rpb9-C11-26, -27, Fig. 3). These experiments revealed that the minimal C11 region required to transfer strong cleavage to Pol II comprised C11 residues 84–110 (Figs. 2B and 3, variants Rpb9-C11-1, -2, -3, -4, -7, -8). This region forms the core of the zinc-binding C-ribbon fold, suggesting that the C-ribbon domain must be structurally intact to induce strong cleavage. Indeed, the N- and C-ribbon are both required for strong cleavage (Figs. 2B and 3, variants Rpb9-C11-5, -6, -18, -19).
The Cleavage-active Pol II Variant Contains a Mobile C-ribbon
To investigate the structural basis for the observed strong RNA cleavage, we crystallized the Pol II variant containing the fusion protein Rpb9-C11-1. Despite extensive efforts, only poorly diffracting crystals could be obtained, but eventually we solved the structure at 4.3 Å resolution (“Experimental Procedures” and Table 1). The structure revealed that the conformation of Pol II around the active center was unchanged, with the bridge helix straight and the trigger loop open and mobile. The Rpb9-C11-1 N-ribbon and the Rpb9 linker strand β4 (residues 1–48) were located at the Rpb1 jaw as in wild-type Pol II. However, the C-ribbon was mobile and did not occupy the position of the Rpb9 C-ribbon on the surface (Fig. 4). These results indicated that strong cleavage was not due to enhanced allostery.
TABLE 1.
Crystallographic data and refinement statistics for the cleavage-inducing Pol II variant containing the fusion protein Rpb9-C11-1
| Data collection | |
| Space group | C2221 |
| Cell dimensions | 222.4, 393.4, 281.4 |
| a, b, c (Å) | |
| Resolution (Å) | 48.6-4.3 (4.4-4.3)a |
| Rsym (%) | 10.6 (103.3) |
| I/σ(I) | 8.6 (2.1) |
| Completeness (%) | 98.4 (99.0) |
| Redundancy | 3.8 (3.9) |
| Wavelength (Å) | 1.2664 |
| Refinement | |
| Resolution (Å) | 48.6-4.3 (4.4-4.3) |
| No. of reflections | 82,532 (6065) |
| Rwork/Rfree (%) | 23.5/28.1 |
| No. of atoms | 30,544 |
| r.m.s.b deviations | |
| Bond lengths (Å) | 0.011 |
| Bond angles (°) | 1.099 |
| Seven zinc peaks in anomalous difference Fourier (σ) | 9.4, 11.7, 9.9,c 9.2, 8.2, 13.0, 11.6 |
a Values in parentheses correspond to the highest resolution shell.
b r.m.s., root mean square.
c Peak for zinc ion in the N-ribbon of Rpb9-C11-1.
FIGURE 4.
Crystallographic analysis of the highly cleavage-active Pol II variant containing Rpb9-C11-1 (Fig. 3 and Table 1). Shown is the difference electron density map contoured at 2.5σ (green mesh) for the N-ribbon of Rpb9-C11-1 (orange ribbon model). A peak in the anomalous difference electron density map (magenta mesh) coincides with the position of the N-ribbon zinc ion Zn3 (cyan sphere).
Inspection of the Pol II structure suggested that detachment of the C-ribbon from the lobe requires weakening or loss of several contacts (Fig. 5). First, a contact of Rpb9 residue Arg-92 with the lobe residues Glu-262 and Asp-391 is lost in the Rpb9-C11 variant because the arginine is replaced by a serine. Second, Rpb9 residue Lys-93 is within contact distance with Rpb2 residue Asp-391 in the lobe, but this lysine is not present in C11, leading to loss of a potential salt bridge. Third, the C-ribbon residue Arg-91 forms a salt bridge with Rpb1 residue Asp-781 (Fig. 5). This arginine is invariant in all C-ribbons; thus the salt bridge could in principle be maintained. However, the preceding Rpb9 residues Ser-88 and Gln-90 buttress C-ribbon residues that interact with the Pol II surface. In particular, residue Gln-90 buttresses Rpb9 residues 50–52, which bind the Pol II lobe, and the preceding residue Gln-87 interacts with the Pol II funnel domain (Fig. 5). Because the counterparts of Ser-88 and Gln-90 are hydrophobic in C11 (Leu-85 and Ile-87) and also in the C-ribbons of TFIIS, A12.2, and TFS (Fig. 3), C-ribbon binding to the polymerase surface is apparently weakened. Consistent with this prediction, replacing the two hydrophobic residues in the cleavage-inducing variant Rpb9-C11-1 with valines retained strong cleavage (Figs. 2B and 3, variant Rpb9-C11-9). These results help rationalize why the C-ribbon is detached from the polymerase surface and mobile in the cleavage-active Pol II variant.
FIGURE 5.
Interface between the Rpb9 C-ribbon and linker (orange) and Pol II domains in different colors as indicated (PDB 1WCM). A, side view as in Figs. 1 and 4. Orange spheres indicate the location of Rpb9 amino acid residues referred to under “Results.” B, view rotated by 90 degrees with respect to that in A as indicated. Important interface residues in Rpb9 and Rpb1 are depicted. Dashed lines indicate salt bridges or hydrogen bonds.
Evidence That the C-ribbon Is Catalytic and Enters the Pol II Pore
The above results suggested that cleavage stimulation by the Rpb9-C11 fusion protein is not due to enhanced allostery but that a switch in cleavage mechanism occurred and the C-ribbon transiently inserted into the pore to directly stimulate cleavage by complementation of the active center with catalytic residues in the hairpin. This model predicted that the hairpin residues are required for cleavage stimulation, just like the corresponding catalytic residues in TFIIS. Indeed, mutation of C11 residues Asp-91 and Glu-92 in the β6-β7 hairpin of the fusion protein or just residue Asp-91 to alanine abolished cleavage (Figs. 2B and 3, variants Rpb9-C11-16/17). The model also predicted that the residue Lys-108 in the C-ribbon forms a salt bridge with Rpb1 pore residue Asp-1359, as observed in the Pol II-TFIIS complex structure (5). Indeed, mutation of Lys-108 leads to a strong reduction in cleavage stimulation (Figs. 2B and 3, variant Rpb9-C11-20,-21,-24). In addition, the conserved residue Glu-109 in the C-ribbon forms a salt bridge with the Rpb1 residue Lys-619 that is also located in the pore and is invariant in Pol III enzymes. Consistent with this proposal, deletion of the C-terminal C11 residue Glu-109 leads to a strong reduction in cleavage stimulation (Figs. 2B and 3, variant Rpb9-C11-22, -23). Variants that do not contain this residue also lost activity (Figs. 2B and 3, variant Rpb9-C11-13,-14).
We next asked whether and how the C-ribbon could reach the pore and active center. Modeling showed that the Rpb9 linker residues 48–53 are not long enough to link the Rpb9-C11 N-ribbon located on the jaw with a C-ribbon located in the pore. However, residues 54–85 could additionally be used to link the domains if their limited interactions with the Rpb9 C-ribbon would be broken in the Rpb9-C11-1 variant. This is apparently achieved in the variant because Rpb9 C-ribbon residues Ile-109 and Thr-111, which interact with linker residues, are replaced with arginine and lysine, respectively, in the fusion protein, which apparently breaks the hydrophobic contacts between the linker and C-ribbon. Consistent with this proposal, mutations in the cleavage-inducing variant Rpb9-C11-1 that were predicted to prevent detachment of the Rpb9 linker from the C-ribbon could not stimulate strong cleavage (Figs. 2B and 3, Rpb9-C11-3, -7, -8, -10, -11, -12, -20, -21, -24).
We also tested whether shortening of the linker between the two ribbons would abolish cleavage because the C-ribbon could not reach the active center. Indeed, variants with shorter linkers did not induce strong RNA cleavage (Figs. 2B and 3, variants Rpb9-C11-26, -27). In addition, Rpb9 contains a salt bridge between the linker residue Glu-54 and Arg-118 in the C-ribbon (Fig. 5), but this is lost in cleavage-inducing variants that lack the C-terminal arginine. In variants that could form this interaction, strong cleavage activity was lost (Figs. 2B and 3, variant Rpb9-C11-3, -7, -10, -11, -12, -15).
The C11 C-ribbon Functions in the Pol II Pore
All the above results support the model that in the Rpb9-C11-1 variant, the N-ribbon remains on the jaw, whereas the C-ribbon transiently occupies the pore to induce strong RNA cleavage. This requires that the C11 C-ribbon can function in the Pol II pore. To test this, we prepared TFIIS variants in which the TFIIS C-ribbon is replaced by the C11 C-ribbon. Indeed, such fusion proteins were as active as wild-type TFIIS (Figs. 2B and 3, variant TFIIS-C11-2/3). Further, the model predicted that replacing the C11 C-ribbon in the Rpb9-C11-1 variant by the TFIIS C-ribbon should also induce strong RNA cleavage. This was indeed observed, although cleavage was weaker when we replaced the TFIIS linker with the Rpb9 linker (Figs. 2B and 3, variants Rpb9-TFIIS-1–4). Weaker cleavage induction by the variants Rpb9-TFIIS-3/4 when compared with the variants Rpb9-TFIIS-1/2 can, however, be explained by a loss of TFIIS residues Asp-267 and Arg-268 that form salt bridges with Pol II at the entrance to the pore (5). These results show that the C11 C-ribbon can bind the Pol II pore and induce strong RNA cleavage and that a cleavage-inducing C-ribbon can reach the pore if tethered to the Rpb9 N-ribbon located on the jaw.
Catalytic C-ribbons Are Conserved between Archaea and Eukaryotes
The above analysis suggested a simple evolutionary relationship between A12.2, Rpb9, C11, and TFIIS (Fig. 1B). First, A12.2 and C11 correspond to the archaeal TFS. In A12.2, C11, and TFS, the N-ribbon corresponds to that of Rpb9, whereas the C-ribbon corresponds to that in TFIIS. To test this prediction, we performed cleavage assays with the archaeal RNA polymerase from P. furiosus (Pfu). The polymerase alone could not induce cleavage, but the addition of recombinant Pfu TFS enabled strong cleavage (Fig. 6), consistent with previous reports (11, 12). Mutagenesis revealed that cleavage required the TFS hairpin residues Asp-90 and Glu-91 as predicted (Figs. 3 and 6). The addition of a fusion protein in which the TFS N-ribbon was fused to the C11 C-ribbon (Figs. 3 and 6) also enabled cleavage, strongly arguing that the pore-binding cleavage-inducing function of the C-ribbon was conserved between archaea and eukaryotes during evolution and supporting our model for the domain relationships.
FIGURE 6.
The C11 C-ribbon functions in the archaeal system. A, electrophoretic analysis of RNA products in a cleavage assay with different protein variants (Fig. 3). RNA bands obtained after cleavage of two or four nucleotides are indicated by arrows (−2 and −4, respectively). Lane 1 shows the reactant RNA. B, quantification of cleavage activities as in Fig. 3B.
DISCUSSION
Our results unravel the molecular basis for the difference in RNA cleavage activities of Pol II and Pol III. We show that replacement of the Rpb9 C-ribbon by the C11 C-ribbon confers strong intrinsic cleavage to Pol II. This unexpected gain of function stems from a switch in the cleavage mechanism, as suggested by x-ray crystallography and mutagenesis. Although the Rpb9 C-ribbon acts allosterically from the polymerase surface, the C11 C-ribbon acts directly by binding the pore and complementing the active center with its catalytic hairpin. Thus two modes exist for polymerase-intrinsic RNA cleavage, an allosteric, weak mode used by Rpb9, and a direct, strong mode used by C11 and TFIIS.
Our results also suggest a model for how polymerase cleavage activities evolved (Fig. 1B). Pol I and Pol III have strong intrinsic cleavage activities because they contain homologues of archaeal TFS (A12.2 and C11, respectively) that contain C-ribbons with catalytic hairpins that can enter the pore to directly stimulate cleavage. In the Pol II system, the two domains are, however, part of two different polypeptides. Although the N-ribbon is part of Rpb9, the C-ribbon is part of TFIIS. During evolution, the C-ribbon likely duplicated and was altered in Rpb9 to attach the domain to the surface and to allow only for weak, allosteric cleavage induction.
Our results are consistent with published data. First, mutation of the C11 hairpin residues is lethal (10). Second, the C11 C-ribbon is not observed on the surface in a recent electron microscopic structure of Pol III, consistent with transient binding to the pore (29). Third, C11 and A12.2 are required for transcription termination by Pol I and Pol III (10, 30), and the termination mechanism likely resembles that of archaeal polymerase (31) but is different in Pol II. Fourth, the A12.2 and C11 C-ribbon domains may be able to swing between surface and pore locations because some density for the A12.2 C-ribbon was observed near the lobe in a Pol I EM reconstruction (3) and because the strong Pol III cleavage can be even further enhanced by a mutation of the largest subunit that is predicted to disrupt a salt bridge between the Pol III counterpart of Rpb1 residue Asp-781 in the funnel domain F-loop and the C11 residue Arg-88 (corresponding to Rpb9 Arg-91) (8). It remains to be confirmed that A12.2 uses the same mechanism as C11. Unfortunately, replacing the Rpb9 C-ribbon with the A12.2 C-ribbon did not confer strong cleavage to Pol II (not shown), likely because Pol I has diverged much more from Pol II than Pol III.
These results unveil the exceptional nature of Pol II, in contrast to Pol I, Pol III, and the archaeal polymerase, with respect to RNA cleavage. In the Pol II system, implementation of allosteric and direct cleavage stimulatory modes on two different proteins may have enabled new mechanisms of transcription regulation such as regulation by release of promoter-proximally stalled Pol II (18, 32–34). It may also have prevented premature Pol II termination at sites that would terminate Pol III and may have enabled elaborate 3′-end processing of Pol II transcripts. The weak intrinsic cleavage activity of Pol II may, however, suffice for proofreading after ubiquitous misincorporation events.
Acknowledgments
We thank members of the Cramer laboratory for help. Part of this work was performed at the Swiss Light Source (SLS) at the Paul Scherrer Institut, Villigen, Switzerland.
This article was selected as a Paper of the Week.
The atomic coordinates and structure factors (code 3QT1) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- Pol
- RNA polymerase
- TFS
- transcription factor S
- TFIIS
- transcription factor IIS.
REFERENCES
- 1. Cramer P., Bushnell D. A., Kornberg R. D. (2001) Science 292, 1863–1876 [DOI] [PubMed] [Google Scholar]
- 2. Jasiak A. J., Armache K. J., Martens B., Jansen R. P., Cramer P. (2006) Mol. Cell 23, 71–81 [DOI] [PubMed] [Google Scholar]
- 3. Kuhn C. D., Geiger S. R., Baumli S., Gartmann M., Gerber J., Jennebach S., Mielke T., Tschochner H., Beckmann R., Cramer P. (2007) Cell 131, 1260–1272 [DOI] [PubMed] [Google Scholar]
- 4. Sosunov V., Sosunova E., Mustaev A., Bass I., Nikiforov V., Goldfarb A. (2003) EMBO J. 22, 2234–2244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kettenberger H., Armache K. J., Cramer P. (2003) Cell 114, 347–357 [DOI] [PubMed] [Google Scholar]
- 6. Sigurdsson S., Dirac-Svejstrup A. B., Svejstrup J. Q. (2010) Mol. Cell 38, 202–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Alic N., Ayoub N., Landrieux E., Favry E., Baudouin-Cornu P., Riva M., Carles C. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 10400–10405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Thuillier V., Brun I., Sentenac A., Werner M. (1996) EMBO J. 15, 618–629 [PMC free article] [PubMed] [Google Scholar]
- 9. Walmacq C., Kireeva M. L., Irvin J., Nedialkov Y., Lubkowska L., Malagon F., Strathern J. N., Kashlev M. (2009) J. Biol. Chem. 284, 19601–19612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chédin S., Riva M., Schultz P., Sentenac A., Carles C. (1998) Genes Dev. 12, 3857–3871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hausner W., Lange U., Musfeldt M. (2000) J. Biol. Chem. 275, 12393–12399 [DOI] [PubMed] [Google Scholar]
- 12. Lange U., Hausner W. (2004) Mol. Microbiol. 52, 1133–1143 [DOI] [PubMed] [Google Scholar]
- 13. Qian X., Jeon C., Yoon H., Agarwal K., Weiss M. A. (1993) Nature 365, 277–279 [DOI] [PubMed] [Google Scholar]
- 14. Jeon C., Yoon H., Agarwal K. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 9106–9110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Awrey D. E., Shimasaki N., Koth C., Weilbaecher R., Olmsted V., Kazanis S., Shan X., Arellano J., Arrowsmith C. H., Kane C. M., Edwards A. M. (1998) J. Biol. Chem. 273, 22595–22605 [DOI] [PubMed] [Google Scholar]
- 16. Kettenberger H., Armache K. J., Cramer P. (2004) Mol. Cell 16, 955–965 [DOI] [PubMed] [Google Scholar]
- 17. Wang D., Bushnell D. A., Huang X., Westover K. D., Levitt M., Kornberg R. D. (2009) Science 324, 1203–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cheung A. C., Cramer P. (2011) Nature 471, 249–253 [DOI] [PubMed] [Google Scholar]
- 19. Kireeva M. L., Komissarova N., Waugh D. S., Kashlev M. (2000) J. Biol. Chem. 275, 6530–6536 [DOI] [PubMed] [Google Scholar]
- 20. Janke C., Magiera M. M., Rathfelder N., Taxis C., Reber S., Maekawa H., Moreno-Borchart A., Doenges G., Schwob E., Schiebel E., Knop M. (2004) Yeast 21, 947–962 [DOI] [PubMed] [Google Scholar]
- 21. Sydow J. F., Brueckner F., Cheung A. C., Damsma G. E., Dengl S., Lehmann E., Vassylyev D., Cramer P. (2009) Mol. Cell 34, 710–721 [DOI] [PubMed] [Google Scholar]
- 22. Kusser A. G., Bertero M. G., Naji S., Becker T., Thomm M., Beckmann R., Cramer P. (2008) J. Mol. Biol. 376, 303–307 [DOI] [PubMed] [Google Scholar]
- 23. Dengl S., Cramer P. (2009) J. Biol. Chem. 284, 21270–21279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kabsch W. (1988) J. Appl. Crystallogr. 21, 67–71 [Google Scholar]
- 25. McCoy A. J., Grosse-Kunstleve R. W., Storoni L. C., Read R. J. (2005) Acta Crystallogr. D Biol. Crystallogr. 61, 458–464 [DOI] [PubMed] [Google Scholar]
- 26. Afonine P. V., Grosse-Kunstleve R. W., Adams P. D. (2005) Acta Crystallogr. D Biol. Crystallogr. 61, 850–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kostrewa D., Zeller M. E., Armache K. J., Seizl M., Leike K., Thomm M., Cramer P. (2009) Nature 462, 323–330 [DOI] [PubMed] [Google Scholar]
- 28. Hemming S. A., Edwards A. M. (2000) J. Biol. Chem. 275, 2288–2294 [DOI] [PubMed] [Google Scholar]
- 29. Fernández-Tornero C., Böttcher B., Rashid U. J., Steuerwald U., Flörchinger B., Devos D. P., Lindner D., Müller C. W. (2010) EMBO J. 29, 3762–3772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Prescott E. M., Osheim Y. N., Jones H. S., Alen C. M., Roan J. G., Reeder R. H., Beyer A. L., Proudfoot N. J. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 6068–6073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Spitalny P., Thomm M. (2008) Mol. Microbiol. 67, 958–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Adelman K., Marr M. T., Werner J., Saunders A., Ni Z., Andrulis E. D., Lis J. T. (2005) Mol. Cell 17, 103–112 [DOI] [PubMed] [Google Scholar]
- 33. Palangat M., Renner D. B., Price D. H., Landick R. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 15036–15041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nechaev S., Fargo D. C., dos Santos G., Liu L., Gao Y., Adelman K. (2010) Science 327, 335–338 [DOI] [PMC free article] [PubMed] [Google Scholar]