Abstract
The Notch pathway is involved in cell-cell signaling during development and adulthood from invertebrates to higher eukaryotes. Activation of the Notch receptor by its ligands relies upon a multi-step processing. The extracellular part of the receptor is removed by a metalloprotease of the ADAM family and the remaining fragment is cleaved within its transmembrane domain by a presenilin-dependent γ-secretase activity. γ-Secretase processing of Notch has been shown to depend upon monoubiquitination as well as clathrin-mediated endocytosis (CME). We show here that AAK1, the adaptor-associated kinase 1, directly interacts with the membrane-tethered active form of Notch released by metalloprotease cleavage. Active AAK1 acts upstream of the γ-secretase cleavage by stabilizing both the membrane-tethered activated form of Notch and its monoubiquitinated counterpart. We propose that AAK1 acts as an adaptor for Notch interaction with components of the clathrin-mediated pathway such as Eps15b. Moreover, transfected AAK1 increases the localization of activated Notch to Rab5-positive endocytic vesicles, while AAK1 depletion or overexpression of Numb, an inhibitor of the pathway, interferes with this localization. These results suggest that after ligand-induced activation of Notch, the membrane-tethered form can be directed to different endocytic pathways leading to distinct fates.
Keywords: Endocytosis, Notch Pathway, Protein Turnover, Signal Transduction, Trafficking, Ubiquitination, AAK1, Numb
Introduction
The Notch gene encodes a receptor, matured by furin, and present at the cell surface as an heterodimer (1, 2). Notch activation is induced upon ligand binding and leads to its extracellular cleavage by TNF-α-converting enzyme (TACE)2 (3, 4), followed by a second cleavage within the transmembrane domain by a γ-secretase complex. The released intracellular domain of Notch translocates to the nucleus and induces transcription of target genes together with the CSL and Mastermind cofactors. Ligand-induced activation of Notch is strictly regulated at several levels, both at the extracellular and the intracellular levels (5–7).
An increasing number of data reveals that the activity of both Notch receptor and its ligands is regulated by internalization as well as ubiquitination events (8, 9). A large repertoire of Notch pathway regulators has been identified, mainly in invertebrates, and conflicting data have emerged regarding Notch activity and trafficking (5). On one hand, endocytosis appears important for Notch activation in the signal-receiving cell (10–13) while other studies point to an inhibitory effect (14–18). Our data suggest that, in mammalian cells, both monoubiquitination and endocytosis are required before γ-secretase cleavage of ligand-activated Notch (10).
As the Notch pathway is conserved throughout evolution, we scanned the literature for genetic modulators of the pathway in invertebrates to further understand the intracellular events that control Notch activation. Loss of worm sel-5 (SEL for suppressor/enhancer of Lin12) results in suppression of the constitutive activity of lin-12(d) (equivalent to the metalloprotease-cleaved Notch or ΔE construct) but not of lin-12(intra) (corresponding to the γ-secretase released nuclear form of Notch) (19). SEL-5 kinase is the homolog of Drosophila NAK, Numb-associated kinase (20) and human AAK1, adaptor-associated kinase 1 (21, 22). These serine/threonine kinases belong to the Ark1/Prk1 (actin-regulating kinase 1) family involved in cortical actin regulation in yeast and include BIKE (BMP-2-inducible kinase), MPSK1 (myristoylated and palmitoylated serine/threonine kinase 1) and GAK (23, 24). AAK1 and GAK, the cyclin G-associated kinase, phosphorylate the μ subunits of both AP-1 and AP-2 trafficking adaptors (21, 22, 25). AAK1, via phosphorylation of Thr-156 of the μ2 subunit, increases the affinity of AP2 for membranes and induces a conformational change enabling cargo recruitment by this adaptor (26, 27). Clathrin assembly stimulates AAK1 activity thus suggesting a role in clathrin-mediated cargo uptake (28, 29). Besides its function at the internalization step, AAK1 was recently shown to act at the level of early/sorting endosome and to colocalize with EEA1 (30). Based on these observations as well as on the genetic data obtained in invertebrates, we hypothesized that AAK1 could play a positive role in early Notch endocytosis.
Results presented here show that AAK1 directly interacts with Notch ΔE, a membrane-tethered and activated form of Notch that resembles the ligand-induced metalloprotease-processing product, but does not interact with the full-length receptor. Further investigation reveals that increasing AAK1 expression results in stabilization of Notch ΔE and its monoubiquitinated form. Moreover, shRNA-mediated AAK1 depletion decreases Notch transcriptional activity in coculture experiments. It has been shown that AAK1 can phosphorylate Numb, a negative regulator of the Notch pathway, and modify its subcellular localization (31). We demonstrate here that AAK1 targets Notch to Rab5-positive endosomes, whereas Numb interferes with this localization. Based on these results we propose that after activation by its ligand, AAK1 promotes Notch activation through stabilization of the monoubiquitinated Notch intermediate and sorting toward Rab5-positive endosomes. On the other hand, Numb interferes with the monoubiquitination step and the Rab5 localization, probably leading to Notch degradation through an alternate sorting pathway.
EXPERIMENTAL PROCEDURES
Materials
All Notch1 constructs, Notch FL, ΔE, ΔE+CT, and IC have been previously described (6, 10, 32–34). GFP-fused Rab5Q79L mutant was a kind gift from C. Schnatwinkel and M. Zerial (MPI, Dresden). His-tagged ubiquitin variants were provided by M. Treier and D. Bohmann (EMBL, Heidelberg). AAK1 was amplified by PCR (primers 5′-NAKSacII and 3′-NAKRI) from the human cDNA clone HH02560, KIAA1048 (Kazusa DNA Research Institute (35)) and cloned into a G-VSV/pcDNA3 vector. Site-directed mutagenesis of AAK1 was used to generate the KI-AAK1 (primers K74A) and the DPF (DPF forward) mutants. The AID-truncated AAK1 (ΔAID) was obtained by introducing a STOP codon in VSV-AAK1 (primer 3′-NAK631). All AAK1 constructs have an N-terminal VSV-tag and were checked by sequencing (Genome Express). shRNA against AAK1 and Numb were generated by cloning the corresponding sequences (shAAK1 forward/reverse and shNumb forward/reverse) into the pSUPER plasmid cloned into pcDNA3 for mammalian expression (pSUPER was a gift from R. Agami). Primer sequences are detailed in a supplemental Table S1.
Antibodies were purchased from Abcam (Numb 14140), Santa Cruz Biotechnology (clone K15, Eps15), Molecular Probes (Alexa-Fluor 488 conjugates), Sigma (actin), Jackson ImmunoResearch (Cy5- and Cy3-labeled secondary antibodies) or ZYMED (anti-transferrin receptor). The anti-Notch antibody has been described previously (2). A rabbit polyclonal antibody (AAK1 39174) was raised against amino acids 2–16 of hAAK1, coupled to KLH (keyhole limpet hemocyanin) via a C-terminal cysteine (Agro-Bio, France). Anti-VSV (P5D4) and anti-Myc (9E10) monoclonal antibodies were used for immunoprecipitations and isolated on protein G-Sepharose (Amersham Biosciences).
In Vitro Experiments: Protein Expression and Isolation
Notch IC, amino acids 1759–2306, was cloned into the pET28a+ vector and expressed in Rosetta(DE3) pLys bacterial strain (Novagen). Bacterial cells were lysed using 50 mm Na2HPO4, 300 mm NaCl, 1% Nonidet P-40, 10 mm imidazole, pH 8. The recombinant proteins were isolated using HIS-Select nickel affinity gel (Sigma). Recombinant GST and GST-AAK1 were isolated as described (21). For in vitro interaction experiments, baculovirus expressed GST-AAK1 was incubated with 6His-Notch IC immobilized on HIS-Select Nickel affinity gel (Sigma) for 1 h at room temperature in binding buffer (20 mm HEPES, 20 mm imidazole, 120 mm potassium acetate, 0.1% Triton X-100, pH 8.0). Beads were washed three times (50 mm Na2HPO4, 300 mm NaCl, 20 mm imidazole, pH 8.0) and bead-bound proteins solubilized with protein sample buffer before being resolved by SDS-PAGE and immunoblotted for GST-AAK1 or GST alone with AAK1-specific antisera (AAK1-SC (36)).
Cell Culture, Extracts, Immunoprecipitations, and Immunoblots
HEK 293T and HeLa cells were cultured as described previously (10). 293T cells were transiently transfected, lysed 40 h later, and immunoprecipitations carried out (10). Samples were resuspended in Laemmli buffer for SDS-PAGE resolution on 7.5% polyacrylamide gels, and immunoblots performed (3). 5% of total protein lysates were loaded for direct immunoblotting.
Interaction with Endogenous AAK1
HeLa cells were stably transfected with empty pcDNA3-pSUPER (CTL cell line) or shAAK1 using jetPEI reagent as per instructions (PolyPlus-transfection). Cells were G418-selected and clones were checked for AAK1 knockdown by immunofluorescence with the 39174 antibody. CTL and shAAK1 HeLa cell lines were transiently transfected with Notch ΔE and protein extracts were immunoprecipitated with the 39174 antibody or the pre-immune serum from the same rabbit. SDS-PAGE analysis on 8% polyacrylamide gels, and immunoblots were performed with the anti-Myc antibody. The same blot was reprobed for AAK1 (AAK1-SC, (36)). 10% of total protein lysates were loaded for direct immunoblotting.
Cell Surface Labeling
HEK 293T transfected with plasmids encoding Notch ΔE, AAK1, or both plasmids were labeled with Sulfo-NHS-LC biotin (Pierce) as described (2). The extracts were either directly immunoprecipitated with the anti-VSV antibody or incubated with streptavidin-agarose beads. The VSV-precipitates were eluted with 2%SDS and heated at 70 °C. Diluted elutes (0,2% SDS) were then fixed on streptavidin-agarose beads for 1 h. The bound proteins were eluted in Laemmli buffer before SDS-PAGE resolution. At each step, 5% of total protein lysate or precipitate was kept for control immunoblotting.
Ubiquitination Assay
HEK 293T transfected with plasmids encoding Notch ΔE, AAK1, Numb, and/or His-tagged Ub variants were lysed in 8 m urea buffer as described (37). Ubiquitinated proteins were purified on nickel-agarose columns according to manufacturer's instructions (GE Healthcare) and analyzed by immunoblotting. 2.5% of total protein lysate was loaded for direct immunoblotting.
Half-life Monitoring with Cycloheximide
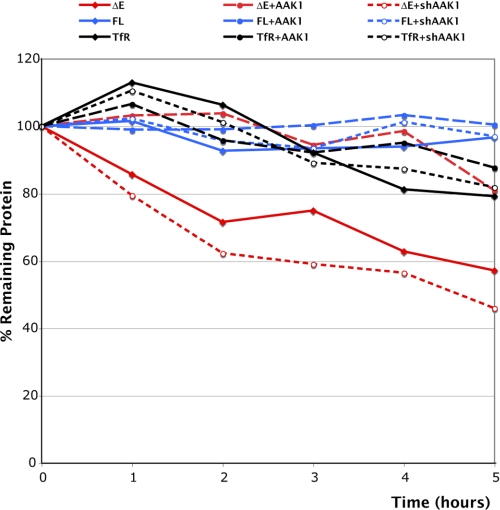
48 h after transfection, 293T cells were incubated in complete DMEM with 50 μg/ml cycloheximide (Sigma). Cells were harvested each hour, lysed, and protein extracts quantified by Bradford. A same amount of each extract was analyzed by SDS-PAGE. Quantity One software (Bio-Rad) was used to assess the amount of ΔE, FL, or transferrin receptor (TfR) remaining at each time point; Actin was used as a loading control.
Reporter Assays
CTL and shAAK1 HeLa cell lines were transiently transfected with CSL-luciferase reporter (38, 39), pRL-TK (Promega) and Notch ΔE. Luciferase-assay was performed 24 h post-transfection using the dual-luciferase kit (Promega) following the manufacturer's instructions using a Berthold luminometer.
Coculture Assay
40,000 U2OS cells stably transfected with Notch1-HA, UN1, were seeded in 24-well plates and transiently transfected with Fugene HD (Roche). 200 ng of CSL-reporter plasmid was completed with 300 ng of the gene/shRNA to be tested and pcDNA3. 24 h after transfection, cells were cocultured with 70,000 OP9 cells expressing or not the Delta-1 ligand of Notch (40). Cells were lysed 20 h later, and a luciferase assay performed. DAPT (50 μm) was added 20 h before cell lysis when indicated.
Immunofluorescence, Confocal Microscopy, and Quantitative Image Analysis
HeLa cells were grown on glass coverslips in 24-well plates, transfected with FUGENE 6 (Roche) and processed for immunofluorescence 48 h after transfection (10). Cells were pre-treated with 0.02% saponin. Images were acquired using a confocal laser imaging system (0.2 μm sections, LSM 510, Zeiss). Colocalization quantification was performed using the Icy software (from Quantitative Image Analysis Unit). Spot detection (41) based on Isotropic Undecimated Wavelet Transform (IUWT) was performed on the green channel (GFP-Rab5Q). The created mask area was used to compute the mean intensity in the red channel (ΔE). A spot was colocalized when the ratio of intensity (ΔE/Rab5Q) was higher than 50%.
RESULTS
AAK1 Interacts with the Activated Membrane-anchored Form of Notch, ΔE
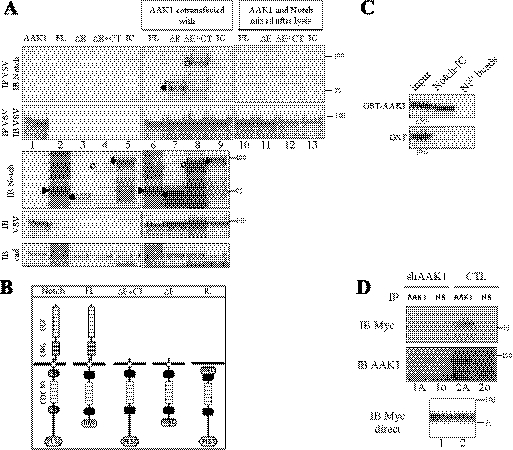
Considering published data on Notch endocytosis and AAK1 orthologs, we investigated a possible role for AAK1, the adaptor-associated kinase 1, in Notch activation in mammals. We cotransfected VSV-tagged AAK1 with different Notch constructs and tested their interactions in 293T cells. The Notch constructs used represent either inactive (FL) or active (ΔE/ΔE+CT and IC) forms of the receptor (illustrated Fig. 1B). ΔE and ΔE+CT constructs encode truncated forms of mNotch1 (Δ21–1703) that resemble the TACE cleavage product and can be constitutively processed by γ-secretase, thus releasing the active nuclear form (3, 33, 42). ΔE also lacks the C-terminal part as this deletion (at amino acid 2186) removes the PEST domain and thus stabilizes nuclear Notch (6). In transiently transfected 293T cells, both ΔE and ΔE+CT demonstrated interaction with VSV-tagged AAK1 (Fig. 1A, lanes 7 and 8, IP VSV IB Notch). No interaction could be observed with the non-activated full-length Notch (FL, lane 6). The intracellular form of Notch, IC (lane 9), did not interact either, but a mutant form that remains cytoplasmic due to the deletion of its NLS sequences did interact with AAK1 (data not shown). These interactions did not take place in the test tube as an AAK1-immunoprecipitation (lanes 10–13) done on cell extracts from Notch-transfected cells (shown lanes 2–5), mixed, after cell lysis, with extracts from AAK1-transfected cells showed no interaction. The lack of interaction observed with IC is probably due to a difference in the subcellular localization of the two molecules. To make sure that the AAK1/ΔE interaction was not due to the higher expression level of ΔE in comparison with the other Notch constructs (because of ΔE stabilization in the presence of AAK1, see for example Fig. 2A), the specificity of the coimmunoprecipitation was confirmed on “normalized” Notch expression levels (supplemental Fig. S1). Taken together, these data restrict the interaction domain of Notch1 with AAK1 to the ankyrin repeats spanning amino acids 1840–2142.
FIGURE 1.
AAK1 directly interacts with the activated form of membrane-anchored Notch. A, AAK1 interacts with Notch in mammalian cells. 293T cells were transiently transfected with plasmids encoding different Notch constructs with or without AAK1 as indicated. Symbols on the left of the lanes indicate the expected Notch forms: Notch FL (furin-cleaved form, ▶), ΔE+CT (○), IC (♦; migrates slower than ΔE+CT because of the 6 N-terminal Myc tags)) or ΔE (●). In lanes 10–13, 500 μg of lysate from AAK1-transfected cells (extract loaded in lane 1) was mixed with 500 μg of lysate from cells transfected with different Notch constructs as indicated (corresponding to extracts loaded in lanes 2–5), and the mixed extracts were immunoprecipitated with anti-VSV. Cell lysates were analyzed by Western blotting for Notch (IB Notch) or AAK1 (IB VSV) following or not immunoprecipitation with anti-VSV (IP VSV). Cadherin was used as a loading control (IB cad); an asterisk indicates a slower migrating form of ΔE and ΔE+CT (lanes 7, 8). B, Notch constructs. Full-length Notch receptor domains are depicted: EGF-, LNG-, ankyrin/CDC10-repeats, NLS, and PEST domains. The ΔE constructs represent extracellular-truncated and constitutively active forms of the receptor, with (ΔE+CT) or without (ΔE) the PEST domain (deleted from amino acids 2186). IC is a construct mimicking the γ-secretase cleavage product of Notch; FL is the inactive full-length receptor, deleted of the C-terminal region including the PEST domain. FL and ΔE constructs are C-terminally truncated; N- or C-terminal 6-Myc tags are indicated. C, AAK1 directly interacts with the Notch intracellular domain. In vitro interaction assay: Purified GST-AAK1 or GST alone were incubated in the presence (Notch-IC) or absence (Ni2+ beads) of immobilized intracellular His-tagged Notch IC. AAK1 binding was detected using an AAK1-specific antiserum (upper panel); GST binding was tested in parallel (lower panel). D, Endogenous AAK1 interacts with Notch ΔE. HeLa cells stably transfected with a vector expressing an shRNA specific for AAK1 (shAAK1) or the empty pSUPER vector (CTL) were transiently transfected with Myc-tagged ΔE. Cell lysates (500 μg) were analyzed after immunoprecipitation with a pre-immune (IP NS, lanes 1o and 2o) or anti-AAK1 serum (IP AAK1, lanes 1A and 2A), followed by immunoblotting for Notch (IB Myc, upper panel) and then reblotted for AAK1 (IB AAK1). A direct Western blot shows the expression of ΔE in 10% of extract (IB Myc direct).
FIGURE 2.
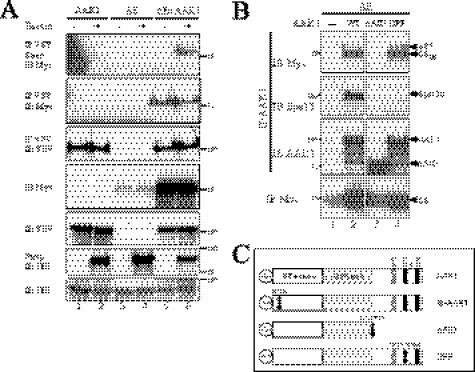
AAK1 acts as an endocytic adaptor for Notch ΔE. A, AAK1 interacts with Notch at the cell surface. 293T cells were transiently transfected with plasmids encoding Myc-tagged ΔE and VSV-tagged AAK1 as indicated. Live cells were labeled with biotin (+) at 4 °C before lysis. 5% of each cell lysate was analyzed by Western blotting for Notch (IB Myc) or AAK1 (IB VSV). AAK1-bound proteins were pulled-down (IP VSV), eluted in 2% SDS and fixed on streptavidin-agarose (Strep) to isolate biotin-labeled Notch (IP VSV/Strep/IB Myc). VSV- immunoprecipitates were tested for AAK1 (IP VSV, IB VSV) and Notch (IP VSV, IB Myc) pull-down. Transferrin receptor was used as a loading (IB TfR) and biotin-labeling control (Strep/IB TfR). B, mapping of the interaction domain of AAK1 with ΔE and Eps15b. 293T cells were transfected with plasmids encoding ΔE and different VSV-tagged AAK1 mutants figured in C: truncated (ΔAID) or DPF777AAA mutated (DPF). Lysates were immunoprecipitated with anti-VSV (IP AAK1) and resolved by SDS-PAGE. The same membrane was blotted for Notch (IB Myc), stripped and reblotted for Eps15 (IB Eps15, K15 antibody) and then for AAK1 (IB AAK1, antibody 39174). ΔE expression was controlled by direct immunoblotting with anti-Myc antibody (5% load, IB Myc); an arrow indicates an ubiquitinated form of ΔE, named ΔEU (lanes 2, 4). C, AAK1 constructs. Wild type (AAK1), AID-deleted (ΔAID), kinase-inactive (K74A mutate, KI-AAK1), or DPF777AAA-mutated (DPF) forms of AAK1 were all VSV-tagged at their N terminus.
The Notch/AAK1 interaction observed could be indirect and require the recruitment of an adaptor protein. To clarify this issue, we used recombinant proteins for both Notch (6His-tagged Notch IC) and AAK1 (GST-AAK1) and tested the interaction in vitro (Fig. 1C). Recombinant intracellular Notch could pull down GST-AAK1 but not GST alone, indicating that the interaction was direct. We then checked for an interaction between endogenous AAK1 and overexpressed and activated Notch in vivo. We generated HeLa cell lines stably expressing an AAK1-specific shRNA (shAAK1). As shown in Fig. 1D, AAK1 was efficiently knocked down by this shRNA. The knocked-down cell line (shAAK1) and a control cell line (CTL) were transiently transfected with ΔE (Myc-tagged, Fig. 1B). 500 μg of each cell lysate was immunoprecipitated with an anti-AAK1 antibody (IP AAK1, 1A and 2A) or the corresponding pre-immune serum (IP NS, 1o and 2o), and blotted for ΔE (IB Myc). ΔE expression was controlled (IB Myc direct). ΔE pulldown was reduced to background levels when endogenous AAK1 was knocked down (lane 1A versus 1o). The AAK1 band observed (lane 2A) migrates around 140 kDa and probably represents AAK1L, the long isoform present in HeLa cells (30). These results thus show that endogenous AAK1 interacts with the membrane-tethered, activated form of Notch.
AAK1 Interaction with ΔE Occurs at the Cell Surface
As AAK1 is an endocytic protein involved in CME, we used a biotinylation assay to test whether AAK1 could interact with ΔE at the cell surface. Extracts from biotin-labeled 293T cells transiently transfected with AAK1, ΔE or both constructs were first immunoprecipitated with an anti-VSV antibody to pull down AAK1-associated proteins. The eluted precipitate was then fixed on streptavidin to isolate the biotinylated (i.e. surface-expressed) proteins. AAK1 was equally immunoprecipitated from cells incubated (+) or not (−) with biotin (Fig. 2A, lanes 1, 2, 5, and 6, IP VSV/IB VSV). ΔE could be isolated from the VSV-precipitated eluates by streptavidin fixation (lane 6, IP VSV Strep, IB Myc), indicating that ΔE interaction with AAK1 occurs (at least) at the cell surface. The endogenous transferrin receptor was used as a control for biotinylation and streptavidin purification (IB TfR and Strep IB TfR).
The AP2-interacting Domain of AAK1 Is Required for Interaction with ΔE and Eps15b
The kinase activity and the C-terminal domains of both SEL-5 and NAK appear critical for Notch activation in worms (19) and flies (20). Several interaction motifs for endocytic factors can be found in the C terminus of AAK1 and its orthologs (Ref. 21 and Fig. 2C): the DLL clathrin-binding motif (43), two DPF α-adaptin-interaction motifs (44) and the NPF, EH- (45) or AP2-binding motif (46). AAK1 could thus act as an adaptor protein for the recruitment of different endocytic partners to the Notch complex. To identify the domain(s) of AAK1 that may be involved in the interaction with Notch, we generated different mutants (Fig. 2C): a mutant lacking the entire AP2-interacting domain (ΔAID, stop codon at amino acid 631) and a point mutant in one of the DPF motifs (DPF, DPF777AAA). In transiently transfected 293T cells, WT AAK1 as well as the DPF mutant pulled-down both the membrane-anchored form, ΔETM, and the monoubiquitinated form (10), ΔEU, of ΔE (Fig. 2B, IP AAK1 IB Myc, respectively, lanes 2 and 4). The ΔAID mutant did not interact with ΔE (lane 3) implying that, in cells, the C-terminal part of AAK1 is important for interaction with Notch.
Eps15 is a UIM-containing protein known to interact with monoubiquitinated receptors and might thus be involved in the Notch endocytic complex (47). We had previously shown that a dominant-negative form of Eps15 inhibits Notch endocytosis (10). Eps15 is also known to bind NPF-containing proteins through EH-domains (45). One NPF motif being present in the AID of AAK1, we searched for Eps15 in AAK1 immunoprecipitates (IP AAK1 IB Eps15). We could identify only the small isoform of Eps15, recently described as Eps15b (48). This interaction was abolished when AAK1 was deleted of its AID or mutated in the DPF motif (IB Eps15, respectively, lanes 3 and 4) but not when the NPF motif was mutated (data not shown). These results indicate that a DPF motif is required for interaction between Eps15b and AAK1.
Active AAK1 Preferentially Interacts with Monoubiquitinated ΔE
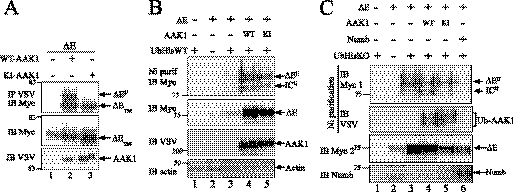
AAK1 seemed to pull down a slow-migrating product (asterisked lanes 7 and 8 in Fig. 1A, IP VSV IB Notch; ΔEU in Fig. 2B) co-migrating with a previously described monoubiquitinated form of ΔE, which is barely visible under normal conditions (10). Fig. 3A indicates that AAK1 seems to preferentially interact with this form of Notch (compare IB Myc with IP VSV/IB Myc) or to favor its existence. To determine whether the kinase activity of AAK1 is required for this interaction, we used a kinase-inactive mutant of AAK1 (KI, obtained by mutating Lys-74 of the catalytic site to Ala, see Fig. 2C). Contrary to WT-AAK1, immunoprecipitation with KI-AAK1 only pulled down ΔETM (Fig. 3A, compare lanes 2 and 3), despite a similar if not higher level of expression (IB VSV). As the amount of ΔE is identical in the presence of WT AAK1 or its KI mutant (IB Myc), it can be concluded that AAK1 preferentially interacts with the ΔEU form, and that this interaction requires its catalytic activity (see “Discussion”). As AAK1 is involved in receptor endocytosis and directly interacts with Notch, we wondered whether Notch could be one of its phosphorylation substrates. In vitro kinase assays using immunoprecipitated or recombinant AAK1 showed that this was not the case (data not shown).
FIGURE 3.
Differential effect of AAK1 and Numb on Notch ΔE. A, active AAK1 preferentially interacts with monoubiquitinated ΔE (ΔEU). 293T cells were transiently transfected with plasmids encoding ΔE and wild-type (WT-AAK1) or kinase-inactive (KI-AAK1) AAK1 as indicated. Cell lysates were analyzed by immunoblotting for Notch before (IB Myc) or after AAK1 immunoprecipitation (IP VSV/IB Myc). The different forms of ΔE (ΔEU and ΔETM) are indicated. Expression of AAK1 (IB VSV) was verified in the same extracts on 5% of the total lysate. B, cotransfection of AAK1 increases the level of Notch ΔE and its monoubiquitinated form. 293T cells were transiently transfected with ΔE along with AAK1 wild-type (WT, lane 4), kinase-inactive (KI, lane 5) and His-tagged ubiquitin (UbHisWT) as indicated. Cell lysates were analyzed by Western blotting for Notch after purification of ubiquitinated species on Nickel beads (Ni purif, IB Myc). Monoubiquitinated forms of Notch either membrane-anchored (ΔEU) or γ-secretase cleaved (ICU) can be visualized (lanes 4–5, upper panel). 5% of the total lysate was directly blotted for detection of ΔE (IB Myc), AAK1 (IB VSV), and actin (IB actin) as a loading control. C, cotransfection of Numb decreases the level of Notch ΔE and its monoubiquitinated form. 293T cells were transfected with plasmids encoding ΔE, Numb, AAK1 (WT or KI) or His-tagged lysine-less Ub (UbHisKO) as indicated. Cell lysates were analyzed by SDS-PAGE and blotted directly for Notch (IB Myc 2), or after purification on nickel beads to visualize monoubiquitinated ΔE (Ni purification IB Myc 1) or AAK1 (Ni purification IB VSV). Ubiquitinated forms of Notch (ΔEU, ICU) and AAK1 (Ub-AAK1) are indicated. The expression of Numb was also verified (IB Numb).
To fully exclude the possibility that the kinase activity of AAK1 is required for the ΔE monoubiquitination, we cotransfected His-tagged ubiquitin along with ΔE and WT- or KI-AAK1, and purified the ubiquitin-conjugated proteins on Nickel beads. This purification in urea enables dissociation of any bound interactors, allowing to only visualize the ubiquitination of the protein of interest. ΔEU can be detected in the presence of either WT- or KI-AAK1 (Fig. 3B, lanes 4 and 5), indicating that the kinase activity of AAK1 is not directly involved in ΔE monoubiquitination. As ΔE is constitutively cleaved by γ-secretase, we could also observe monoubiquitinated IC (ICU) with the anti-Myc antibody. The fact that this species has not been detected before is probably the consequence of the overexpression procedure used here. This band was no longer visible when a ΔE construct mutated at the γ-secretase cleavage site (LLFF mutant) was used in a similar assay (data not shown).
Numb has been shown to inhibit Notch signaling (49) and to increase polyubiquitination of the Notch intracellular domain, leading to its degradation (50, 51). To get a better insight into the respective effects of AAK1 and Numb on ubiquitin-mediated regulation of ΔE, we cotransfected ΔE with a lysine-less mutant of ubiquitin (UbHisKO) in the presence of cotransfected AAK1 or Numb (Fig. 3C). UbKO enables the conjugation of a single ubiquitin moiety via its C-terminal glycine residue but prevents further branched polyubiquitination (52). The use of this mutated ubiquitin molecule allows the detection of monoubiquitinated ΔE but prevents Numb-mediated proteasomal degradation. This may confuse the interpretation of the experiment as the detection of monoubiquitinated Notch might either be due to activation-induced bona fide monoubiquitination, or to limitation of the degradation-linked polyubiquitin chains caused by Numb. However the results in panel C indicate that under these conditions the presence of Numb does not generate monoubiquitinated Notch. It can thus be concluded that overexpressing UbKO increased the level of ΔE (compare lanes 2 and 3, IB Myc2). The presence of AAK1 did not modify the ubiquitination pattern of ΔE (IB Myc1, lanes 3 and 4), but the KI mutant of AAK1 seemed to reduce the overall level of expression of ΔE (IB Myc2, compare lane 5 with 3 and 4). AAK1 itself seemed to be ubiquitinated as revealed by the VSV blot on Nickel-purified extracts (lanes 4 and 5). On the contrary, Numb coexpression prevented the generation of ΔEU (lane 6, IB Myc1) although ΔE was present in the extract at a level similar to that observed in the KI-AAK1 extract (IB Myc2, compare lanes 6 and 5).
AAK1 and Numb Drive Differential Trafficking of ΔE
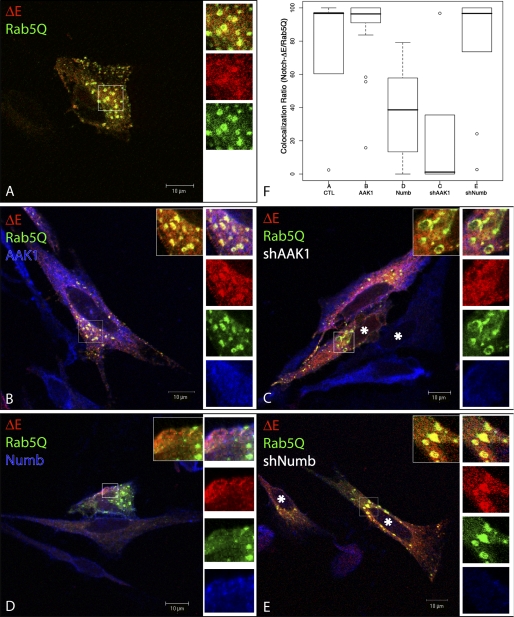
To address the question of ΔE trafficking in the presence of AAK1 or Numb, we transfected HeLa cells with the constitutively active mutant of the small GTPase Rab5, Rab5Q79L (Fig. 4, Rab5Q79L (Rab5Q) is fused to GFP, green). This Rab5 mutant induces the formation of enlarged early endosomes exhibiting distinguishable endosomal microdomains (53). Cotransfected Notch ΔE (red) could be partially detected in the enlarged Rab5Q-positive structures, mostly at the outer side of the vesicles (panel A, overlap in yellow; see enlarged inset). In the presence of transfected AAK1 (panel B, blue) or shNumb (panel E, only the marked cells (*) express the shRNA), enhanced colocalization between ΔE and Rab5Q could be observed (insets show the yellow staining indicating colocalization); the quantification data (panel F) showed more than 90% colocalization. The colocalization between Rab5Q and ΔE tends to disappear in the presence of shAAK1 (less than 10%) and the endosomes become more donut-shaped (panel C, only the marked cells (*) express shAAK1). When cotransfected with Numb (panel D, blue), ΔE could be observed in Rab5Q-endocytic structures only to a minor extent (median at 39%). These results suggest that AAK1 and Numb target ΔE to different sorting routes: AAK1 favors the Rab5 endocytic route while Numb diverts it to a different pathway, probably associated with degradation.
FIGURE 4.
AAK1 and Numb differentially regulate ΔE trafficking. HeLa cells were transiently transfected with ΔE and GFP-fused Rab5Q79L, a constitutively active mutant of Rab5, (Rab5Q, green, A) along with AAK1 (B), shAAK1 (C), Numb (D), or shNumb (E). Cells were fixed, permeabilized, and incubated with anti-Myc to visualize ΔE (red), anti-AAK1 (B and C, blue) or anti-Numb (D and E, blue) antibodies. A blow-up of the boxed region is shown on the right of each panel, top to bottom: merge of the 3 colors, red, green, blue, along with a merged version of Rab5Q/ΔE only (inset). An asterisk (*) indicates the cells that have received shAAK1 (panel C), or shNumb (panel E), as these cells are negative for the corresponding staining (blue). Acquisition was done by confocal microscopy (0.2-μm sections, Zeiss LSM510). F, quantitative analysis of colocalization. The colocalization quantification results using Icy software are summarized by the boxplot graph. Spot detection based on Isotropic Undecimated Wavelet Transform (IUWT) was performed on the green channel (GFP-Rab5Q vesicles). The mask area thus created was used to compute the mean intensity in the red channel (ΔE) in the selected cell expressing or not AAK1 or Numb. A spot was colocalized when the ratio of intensity (ΔE/Rab5Q) was higher than 50%; the percentage of colocalization per cell was expressed as the ratio of colocalized vesicles over the total number of vesicles detected. The mean intensity was determined for each immunofluorescence experiment described in panels A–E; at least 200 vesicles were analyzed. Boxes indicate 25, 50 (median), and 75% quartiles; whiskers extend to 1.5× the interquartile range; dots indicate outliners beyond this range.
AAK1 Stabilizes ΔE but Not Notch FL
To determine whether AAK1 stabilizes Notch as suggested by our previous data, we monitored its half-life in a cycloheximide experiment. We transiently expressed ΔE (red lines) or FL (blue lines) in 293T cells (Fig. 5A), alone or together with AAK1 or shAAK1 and quantified the amount of Notch at different time points after cycloheximide treatment. Actin was used as an internal loading control for each time point and transferrin receptor (TfR, black lines) was monitored in parallel. 30–40% of ΔE was degraded within two hours while cotransfection of AAK1 resulted in a half-life longer than 5 h (● broken lines). Conversely, cotransfection of shAAK1 decreased the stability of ΔE during the time course of cycloheximide treatment (○ dotted lines). On the contrary, both Notch FL and TfR were not affected by the presence or absence of AAK1. These data indicate that AAK1 is involved in the stabilization of Notch ΔE but it does not affect the stability of Notch FL or the transferrin receptor.
FIGURE 5.
Influence of AAK1 on Notch stability. 293T cells were transiently transfected with plasmids encoding ΔE (red) or Notch FL (blue) alone (♦ solid lines), along with AAK1 (● broken lines), or shAAK1 (○ dotted lines). Cells were lysed at the indicated time (hours) after cycloheximide treatment. The same amount of extract (40 μg for all transfections except for ΔE+AAK1 loaded with only 10 μg and ΔE+ shAAK1 loaded with 70 μg of extract) was loaded for each time point and analyzed by SDS-PAGE. The endogenous transferrin receptor (TfR, black lines) was monitored in parallel; AAK1 expression was verified and actin was used as an internal loading control for quantification. The intensity of the bands was quantified using the Quantity One software (Bio-Rad).
AAK1 Is Involved in Ligand-induced Activation of Notch
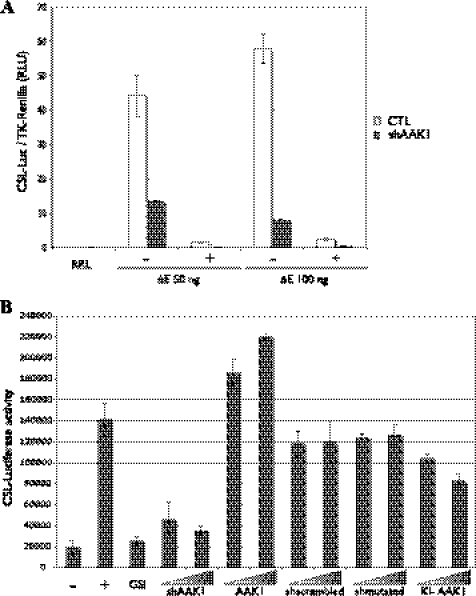
If AAK1 controls the stability of the ligand-activated form of Notch, the transcriptional activity of Notch should also be affected by its presence/absence. To address this issue we performed a reporter gene assay using a CSL-firefly luciferase reporter (38, 39). HeLa cells stably expressing shAAK1 and its control counterpart (see Fig. 1D) were transiently transfected with the CSL-reporter gene along with different concentrations of ΔE+CT. A Renilla luciferase construct was cotransfected to normalize the transfection levels. In control cells, ΔE+CT increased the activity of the reporter gene by 40–60-fold, in a γ-secretase-dependent manner (Fig. 6A, with (+) or without (−) DAPT, a γ-secretase inhibitor). In the presence of shAAK1, this activation was strongly reduced (from 3–7-fold).
FIGURE 6.
AAK1 is a positive regulator of Notch activity. A, shAAK1 inhibits γ-secretase-dependent transcriptional activity of Notch. HeLa cells stably transfected with an shRNA specific for AAK1 (shAAK1) or the empty pSUPER vector (CTL) were transiently transfected with a CSL-firefly luciferase reporter (CSL-Luc), pRL-TK Renilla luciferase (dual luciferase noted RRL) and different amounts of ΔE+CT (50 or 100 ng) as indicated. A γ-secretase inhibitor (DAPT) was added (+) 20 h before cell lysis. Results from four experiments were averaged; error bars represent standard deviation. B, AAK1 is a positive regulator of Notch transcriptional activity. A U2OS cell-line stably expressing full-length Notch1 (UN1) was transiently transfected with the CSL-firefly luciferase reporter, increasing amounts of shAAK1, a control scrambled AAK1 shRNA (shscrambled), a mutated shAAK1 (shmutated), wild-type (AAK1), or kinase-inactive (KI-AAK1) AAK1 plasmids as indicated. UN1 cells were cocultured for 20 h with empty OP9 cells (−) or Dll1-expressing OP9 cells (+ and all other points). DAPT (GSI) was added along with the OP9-Dll1 cells when indicated. Results from three experiments were averaged; error bars represent standard deviation.
To verify that this reduced activation was not due to a clonal effect, we tested Notch activity in a coculture assay using a stable U2OS Notch1-expressing cell line (UN1). The UN1 cells were transiently transfected with the CSL-reporter and either shAAK1, control shRNAs (sh-scrambled, see supplemental Fig. S2 or a 3-nucleotide-mutated shAAK1, sh-mutated), WT AAK1 or KI-AAK1, and cocultured with OP9 cells expressing or not the Notch ligand Delta1 (40). The effect of the different shRNA constructs on AAK1 expression was verified in transfected UN1 cells (supplemental Fig. S2). We observed a 7-fold increase (Fig. 6B) in reporter activity when the UN1 cells were cocultured with Delta1-expressing cells (+) compared with control OP9 cells (−). Here again the presence of DAPT (GSI) reduced Notch-dependent activity. Increasing amounts of shAAK1 led to a transcriptional reduction similar to that observed in the presence of GSI, whereas AAK1 overexpression increased reporter activation. Both control shRNAs (sh-scrambled or sh-mutated) led to a non-significant decrease of Notch activity regardless of the amount used in the assay. Inactive AAK1, KI-AAK1, reduced Notch activity in a dose-dependent manner. We can therefore conclude that AAK1 is a positive regulator of Notch activity.
DISCUSSION
Increasing evidence indicates that Notch signaling is controlled by ubiquitination and endocytic events (8, 11, 54). Dynamin is clearly involved in Notch receptor signaling (12) but the exact molecular mechanisms underlying Notch trafficking are still under investigation. Different proteins belonging to the CME pathway have been involved in Notch trafficking in mammals: α-adaptin, Numb, Eps15, and clathrin (10, 55). Recently, signaling by Notch was confirmed to be clathrin-dependent but not AP2-dependent in Drosophila ovary (56).
Our previous study had shown that, in mammalian cells, a monoubiquitination step on a juxtamembrane lysine was required for further cleavage of Notch by γ-secretase and activation of the pathway (10). We now demonstrate that the human ortholog of Drosophila NAK and worm SEL-5 (19, 20), AAK1, acts as a positive regulator of Notch signaling in mammalian cells, similar to its invertebrate counterparts. AAK1 is involved in CME through phosphorylation of the μ2 subunit of the AP2 adaptor (21, 22), but silencing of this kinase interferes with some AP2-independent events: accumulation of transferrin receptor in perinuclear endosomes, reduced viral entry of vesicular stomatitis virus and inhibition of LDL uptake (30, 57). We demonstrate here that AAK1 directly interacts with ligand-activated Notch (represented here by the ΔE molecule) but not with the inactive full-length receptor. AAK1 interacts with a domain spanning the ankyrin repeats of Notch. AAK1 action on the Notch pathway is unconventional in the sense that its kinase activity is required, but not through direct phosphorylation of the receptor itself.
AAK1 overexpression results in stabilization of activated Notch but not of the inactive receptor. This stabilization results in an increased level of the monoubiquitinated form of ΔE (ΔEU), which precedes the critical γ-secretase cleavage (10). HeLa cells predominantly express the AAK1L splice variant of AAK1 (30), which exhibits an additional clathrin-binding domain but is functionally equivalent to AAK1. We observed an interaction between endogenous AAK1L and transfected Notch ΔE. The AAK1 kinase activity does not seem to be required for ΔE monoubiquitination, but a transfected kinase dead mutant interacts less efficiently with the monoubiquitinated form of ΔE and leads to reduced Notch activity. A conformational switch between the active and the inactive forms of the kinase may explain these results, as the inactive kinase may not enable interaction with other endocytic adaptors (28) and consequent stabilization of monoubiquitinated Notch. Clathrin binding is known to stimulate AAK1 kinase activity and promote cargo recruitment into coated pits (28, 29). AAK1 also exhibits multiple binding sites for accessory endocytic components such as AP2 and EH-containing proteins (21, 22) and thus acts as a scaffolding hub for the recruitment of several endocytic partners. Notch could thus be targeted to clathrin-coated pits by AAK1. AAK1 fulfills all the requirements of a bona fide endocytic adaptor (58) of Notch: it can bind directly the Notch cargo at the cell surface and interacts with clathrin and other endocytic partners.
We have previously shown that dominant-negative Eps15 inhibits the cleavage of Notch ΔE. It now appears that Eps15b, an Eps15 isoform that lacks EH-domains (48), interacts with AAK1 in fibroblasts. Eps15b is the only Eps15 isoform found in AAK1 pull-down, as antibodies against Eps-15 EH-domains did not reveal any additional partner (6G4 antibody, (59)). Whereas one of the DPF-motifs of AAK1 seems to be directly or indirectly involved in AAK1-Eps15b interaction, mutation of the EH domain-binding NPF motif did not interfere with this interaction (45). Both Eps15 and Eps15b contain an ubiquitin-interacting motif that directly recognizes monoubiquitinated cargos (47, 48, 60). Eps15 colocalizes with AP2 at the plasma membrane but Eps15b is localized in EEA1-positive early endosomes (48). Monoubiquitinated Notch could thus interact with Eps15b and be further sorted to late endosomal structures. Interestingly, AAK1 is itself ubiquitinated (Fig. 3C) and colocalizes with EEA1 (30), thus suggesting that an AAK1/Eps15b/Notch ΔE complex may form in such structures. Further experiments will determine if Eps15b functions as a sorting signal for monoubiquitinated Notch cargo toward late endosomal structures.
Numb is another endocytic protein that acts as a Notch inhibitor via binding to AP2 in coated pits (14, 55, 61). The C terminus of Numb was shown to be essential for Notch repression in Drosophila cells (49), in agreement with the presence of conserved endocytic NPF- and DPF-motifs (55). We present data showing that Numb and AAK1 differentially regulate ΔE. Contrary to AAK1, Numb inhibits ΔE monoubiquitination probably favoring polyubiquitination. Of note, in cells cotransfected with AAK1 and Numb, ΔEU was successfully pulled-down with AAK1 but not with Numb (data not shown). These results correlate with published data showing that Numb promotes Notch degradation through AIP4-mediated polyubiquitination (50, 51, 62). AAK1 and Numb act antagonistically, probably by inducing different sorting events through recruitment of alternate accessory factors. AAK1 could have a double role in these events: phosphorylation and redistribution of Numb to recycling endosomes on one hand (31); recruitment of Eps15b and stabilization of ΔE/ΔEU on the other hand. Numb phosphorylation releases it from clathrin-coated pits (63, 64). Therefore, Numb phosphorylation may act as an important regulatory process in the Notch pathway. Use of Numb phospho-mutants will allow a better understanding of these events and their contribution to Notch signaling.
To understand the relationship between endocytosis and monoubiquitination of activated Notch, we used a lysine-mutated form of ΔE that was resistant to monoubiquitination (10). We observed an accumulation of this mutant in EEA1-positive endosomes (data not shown) suggesting that internalization can take place before monoubiquitination. In germline cells of flies, Rab5 overexpression results in the accumulation of Notch in enlarged Rab5-positive structures (65). We show here that AAK1 colocalizes with ΔE in Rab5Q79L endosomes whereas overexpression of Numb interferes with this routing. This observation was confirmed by knocking-down Numb or AAK1: Numb knockdown enhanced ΔE/Rab5Q79L colocalization (recovery of 90% colocalization) whereas AAK1 depletion disorganized the Rab5 structures and reduced the presence of ΔE in these endosomes (<10% ΔE/Rab5Q-positive vesicles).
We can therefore propose a model involving differential sorting of activated Notch, depending on the presence of specific endocytic factors (13, 54). Ligand-induced Notch cleavage by a metalloprotease (giving rise to a molecule similar to ΔETM) is followed by AAK1 binding. AAK1 then phosphorylates Numb, leading to its dissociation from the plasma membrane and from Notch. Actived Notch can then be endocytosed through clathrin-coated pits via active AAK1. Notch monoubiquitination then takes place in EEA1-positive early endosomes and the Notch/AAK1 complex probably includes additional factors such as Eps15b. Of note, Rab5Q79L-generated EEA1 positive enlarged endosomes were also shown to contain Eps15b (48) and may thus account for the presence of activated Notch, probably monoubiquitinated, there. Further processing of Notch by the γ-secretase can then occur and lead to Notch transcriptional activation. In the absence of AAK1, Numb leads to polyubiquitination and degradation of Notch, therefore precluding further Notch activation. An alternate model would be that AAK1 acts only after Notch has been endocytosed. Competition between AAK1 and Numb for Notch could then occur later in the EE and sort the receptor to late endosomal structures such as lysosomes or MVBs (via Numb) or another compartment where γ-secretase is present and active (via AAK1). In this model the observed AAK1 interaction restricted to activated Notch can only be explained by a different localization of the inactive Notch (at the plasma-membrane) versus the activated membrane-tethered form (endosomal).
Our results show that fine-tuning of the level of two endocytic adaptors can account for the differential sorting of the Notch receptor: AAK1 leading to activation and Numb involving degradation. Further investigation will determine the other components that regulate this differential sorting of activated Notch.
Supplementary Material
Acknowledgments
We thank F. Logeat for helpful discussions. We are grateful to M. Treier, C. Schnatwinkel, and M. Zerial for constructs. We thank PFID members for help with confocal imaging and A. Vautrin for establishing the shAAK1 and control cell lines.
This work was supported in part by the Rubicon EC FP6 Grant and a DIM Stem-Pole grant (to S. O.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1 and S2.
- TACE
- TNF-α-converting enzyme
- CME
- clathrin-mediated endocytosis
- CSL
- CBF1/Su(H)/Lag2
- DAPT
- N-[N-(3,5-difluorophenacetyl-l-alanyl]-S-phenylglycine tert-butyl ester
- EEA1
- early endosome antigen 1
- TfR
- transferrin receptor.
REFERENCES
- 1. Blaumueller C. M., Qi H., Zagouras P., Artavanis-Tsakonas S. (1997) Cell 90, 281–291 [DOI] [PubMed] [Google Scholar]
- 2. Logeat F., Bessia C., Brou C., LeBail O., Jarriault S., Seidah N. G., Israël A. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 8108–8112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brou C., Logeat F., Gupta N., Bessia C., LeBail O., Doedens J. R., Cumano A., Roux P., Black R. A., Israël A. (2000) Mol. Cell. 5, 207–216 [DOI] [PubMed] [Google Scholar]
- 4. Mumm J. S., Schroeter E. H., Saxena M. T., Griesemer A., Tian X., Pan D. J., Ray W. J., Kopan R. (2000) Mol. Cell. 5, 197–206 [DOI] [PubMed] [Google Scholar]
- 5. Tien A. C., Rajan A., Bellen H. J. (2009) J. Cell Biol. 184, 621–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gupta-Rossi N., Le Bail O., Gonen H., Brou C., Logeat F., Six E., Ciechanover A., Israël A. (2001) J. Biol. Chem. 276, 34371–34378 [DOI] [PubMed] [Google Scholar]
- 7. Kopan R., Ilagan M. X. (2009) Cell 137, 216–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bray S. J. (2006) Nat. Rev. Mol. Cell Biol. 7, 678–689 [DOI] [PubMed] [Google Scholar]
- 9. Le Borgne R. (2006) Curr. Opin. Cell Biol. 18, 213–222 [DOI] [PubMed] [Google Scholar]
- 10. Gupta-Rossi N., Six E., LeBail O., Logeat F., Chastagner P., Olry A., Israël A., Brou C. (2004) J. Cell Biol. 166, 73–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nichols J. T., Miyamoto A., Weinmaster G. (2007) Traffic 8, 959–969 [DOI] [PubMed] [Google Scholar]
- 12. Seugnet L., Simpson P., Haenlin M. (1997) Dev. Biol. 192, 585–598 [DOI] [PubMed] [Google Scholar]
- 13. Vaccari T., Lu H., Kanwar R., Fortini M. E., Bilder D. (2008) J. Cell Biol. 180, 755–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Berdnik D., Török T., González-Gaitán M., Knoblich J. A. (2002) Dev. Cell 3, 221–231 [DOI] [PubMed] [Google Scholar]
- 15. Childress J. L., Acar M., Tao C., Halder G. (2006) Curr. Biol. 16, 2228–2233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gallagher C. M., Knoblich J. A. (2006) Dev. Cell 11, 641–653 [DOI] [PubMed] [Google Scholar]
- 17. Shaye D. D., Greenwald I. (2002) Nature 420, 686–690 [DOI] [PubMed] [Google Scholar]
- 18. Sorensen E. B., Conner S. D. (2010) Traffic 11, 1234–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fares H., Greenwald I. (1999) Genetics 153, 1641–1654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chien C. T., Wang S., Rothenberg M., Jan L. Y., Jan Y. N. (1998) Mol. Cell. Biol. 18, 598–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Conner S. D., Schmid S. L. (2002) J. Cell Biol. 156, 921–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ricotta D., Conner S. D., Schmid S. L., von Figura K., Honing S. (2002) J. Cell Biol. 156, 791–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cope M. J., Yang S., Shang C., Drubin D. G. (1999) J. Cell Biol. 144, 1203–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Manning G., Whyte D. B., Martinez R., Hunter T., Sudarsanam S. (2002) Science 298, 1912–1934 [DOI] [PubMed] [Google Scholar]
- 25. Umeda A., Meyerholz A., Ungewickell E. (2000) Eur. J. Cell Biol. 79, 336–342 [DOI] [PubMed] [Google Scholar]
- 26. Collins B. M., McCoy A. J., Kent H. M., Evans P. R., Owen D. J. (2002) Cell 109, 523–535 [DOI] [PubMed] [Google Scholar]
- 27. Höning S., Ricotta D., Krauss M., Späte K., Spolaore B., Motley A., Robinson M., Robinson C., Haucke V., Owen D. J. (2005) Mol. Cell 18, 519–531 [DOI] [PubMed] [Google Scholar]
- 28. Conner S. D., Schröter T., Schmid S. L. (2003) Traffic 4, 885–890 [DOI] [PubMed] [Google Scholar]
- 29. Jackson A. P., Flett A., Smythe C., Hufton L., Wettey F. R., Smythe E. (2003) J. Cell Biol. 163, 231–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Henderson D. M., Conner S. D. (2007) Mol. Biol. Cell 18, 2698–2706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sorensen E. B., Conner S. D. (2008) Traffic 9, 1791–1800 [DOI] [PubMed] [Google Scholar]
- 32. Jarriault S., Brou C., Logeat F., Schroeter E. H., Kopan R., Israel A. (1995) Nature 377, 355–358 [DOI] [PubMed] [Google Scholar]
- 33. Kopan R., Schroeter E. H., Weintraub H., Nye J. S. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 1683–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schroeter E. H., Kisslinger J. A., Kopan R. (1998) Nature 393, 382–386 [DOI] [PubMed] [Google Scholar]
- 35. Kikuno R., Nagase T., Ishikawa K., Hirosawa M., Miyajima N., Tanaka A., Kotani H., Nomura N., Ohara O. (1999) DNA. Res. 6, 197–205 [DOI] [PubMed] [Google Scholar]
- 36. Conner S. D., Schmid S. L. (2003) J. Cell Biol. 162, 773–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Heuss S. F., Ndiaye-Lobry D., Six E. M., Israël A., Logeat F. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 11212–11217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Honjo T. (1996) Genes Cells 1, 1–9 [DOI] [PubMed] [Google Scholar]
- 39. Minoguchi S., Taniguchi Y., Kato H., Okazaki T., Strobl L. J., Zimber-Strobl U., Bornkamm G. W., Honjo T. (1997) Mol. Cell. Biol. 17, 2679–2687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Six E. M., Ndiaye D., Sauer G., Laâbi Y., Athman R., Cumano A., Brou C., Israël A., Logeat F. (2004) J. Biol. Chem. 279, 55818–55826 [DOI] [PubMed] [Google Scholar]
- 41. Olivo-Marin J. C. (2002) Pattern Recognit. 35, 1989–1996 [Google Scholar]
- 42. De Strooper B., Annaert W., Cupers P., Saftig P., Craessaerts K., Mumm J. S., Schroeter E. H., Schrijvers V., Wolfe M. S., Ray W. J., Goate A., Kopan R. (1999) Nature 398, 518–522 [DOI] [PubMed] [Google Scholar]
- 43. Morgan J. R., Prasad K., Hao W., Augustine G. J., Lafer E. M. (2000) J. Neurosci. 20, 8667–8676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Owen D. J., Luzio J. P. (2000) Curr. Opin. Cell. Biol. 12, 467–474 [DOI] [PubMed] [Google Scholar]
- 45. Salcini A. E., Confalonieri S., Doria M., Santolini E., Tassi E., Minenkova O., Cesareni G., Pelicci P. G., Di Fiore P. P. (1997) Genes Dev. 11, 2239–2249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Jha A., Agostinelli N. R., Mishra S. K., Keyel P. A., Hawryluk M. J., Traub L. M. (2004) J. Biol. Chem. 279, 2281–2290 [DOI] [PubMed] [Google Scholar]
- 47. Polo S., Sigismund S., Faretta M., Guidi M., Capua M. R., Bossi G., Chen H., De Camilli P., Di Fiore P. P. (2002) Nature 416, 451–455 [DOI] [PubMed] [Google Scholar]
- 48. Roxrud I., Raiborg C., Pedersen N. M., Stang E., Stenmark H. (2008) J. Cell Biol. 180, 1205–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Frise E., Knoblich J. A., Younger-Shepherd S., Jan L. Y., Jan Y. N. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 11925–11932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. McGill M. A., McGlade C. J. (2003) J. Biol. Chem. 278, 23196–23203 [DOI] [PubMed] [Google Scholar]
- 51. McGill M. A., Dho S. E., Weinmaster G., McGlade C. J. (2009) J. Biol. Chem. 284, 26427–26438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wu P. Y., Hanlon M., Eddins M., Tsui C., Rogers R. S., Jensen J. P., Matunis M. J., Weissman A. M., Wolberger C. P., Pickart C. M. (2003) EMBO. J. 22, 5241–5250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Stenmark H., Parton R. G., Steele-Mortimer O., Lütcke A., Gruenberg J., Zerial M. (1994) EMBO. J. 13, 1287–1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yamamoto S., Charng W. L., Bellen H. J. (2010) Curr. Top Dev. Biol. 92, 165–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Santolini E., Puri C., Salcini A. E., Gagliani M. C., Pelicci P. G., Tacchetti C., Di Fiore P. P. (2000) J. Cell Biol. 151, 1345–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Windler S. L., Bilder D. (2010) Curr. Biol. 20, 538–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Pelkmans L., Fava E., Grabner H., Hannus M., Habermann B., Krausz E., Zerial M. (2005) Nature 436, 78–86 [DOI] [PubMed] [Google Scholar]
- 58. Wendland B. (2002) Nat. Rev. Mol. Cell Biol. 3, 971–977 [DOI] [PubMed] [Google Scholar]
- 59. Benmerah A., Gagnon J., Bègue B., Mégarbané B., Dautry-Varsat A., Cerf-Bensussan N. (1995) J. Cell Biol. 131, 1831–1838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Shih S. C., Katzmann D. J., Schnell J. D., Sutanto M., Emr S. D., Hicke L. (2002) Nat. Cell Biol. 4, 389–393 [DOI] [PubMed] [Google Scholar]
- 61. Guo M., Jan L. Y., Jan Y. N. (1996) Neuron 17, 27–41 [DOI] [PubMed] [Google Scholar]
- 62. Chastagner P., Israël A., Brou C. (2008) PLoS. ONE 3, e2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Nishimura T., Kaibuchi K. (2007) Dev. Cell 13, 15–28 [DOI] [PubMed] [Google Scholar]
- 64. Tokumitsu H., Hatano N., Yokokura S., Sueyoshi Y., Nozaki N., Kobayashi R. (2006) FEBS. Lett. 580, 5797–5801 [DOI] [PubMed] [Google Scholar]
- 65. Wilkin M., Tongngok P., Gensch N., Clemence S., Motoki M., Yamada K., Hori K., Taniguchi-Kanai M., Franklin E., Matsuno K., Baron M. (2008) Dev. Cell 15, 762–772 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.