Abstract
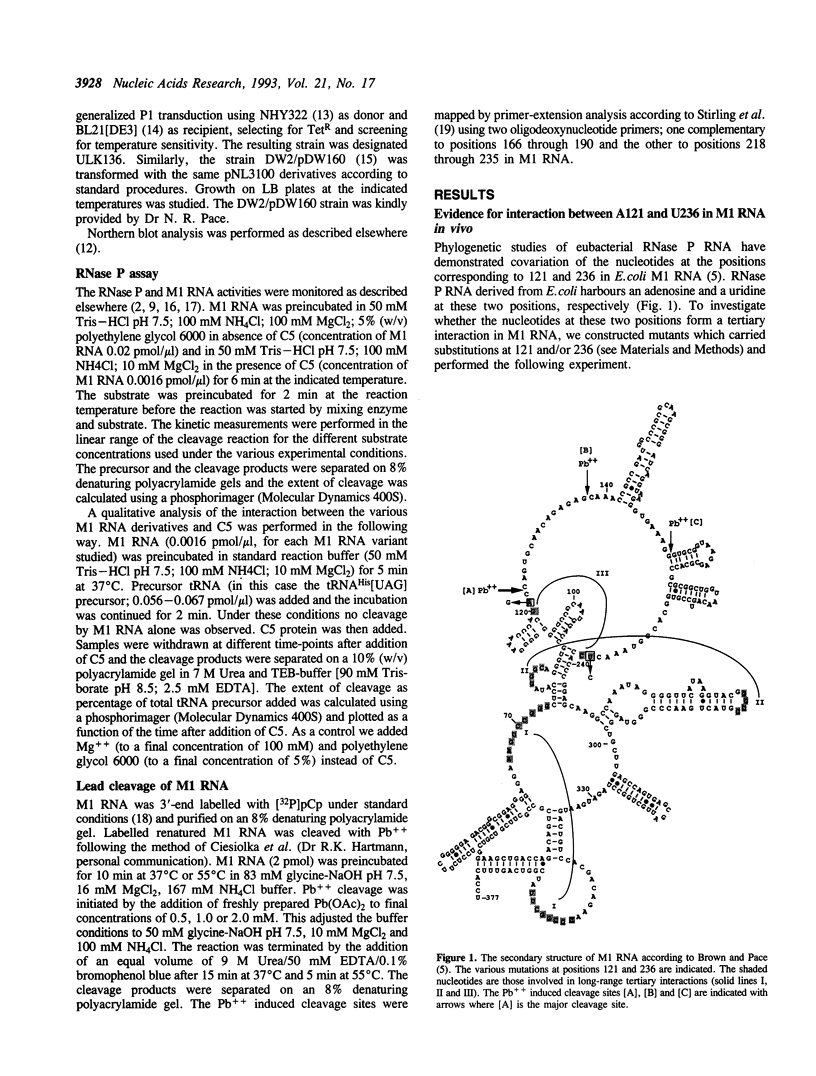
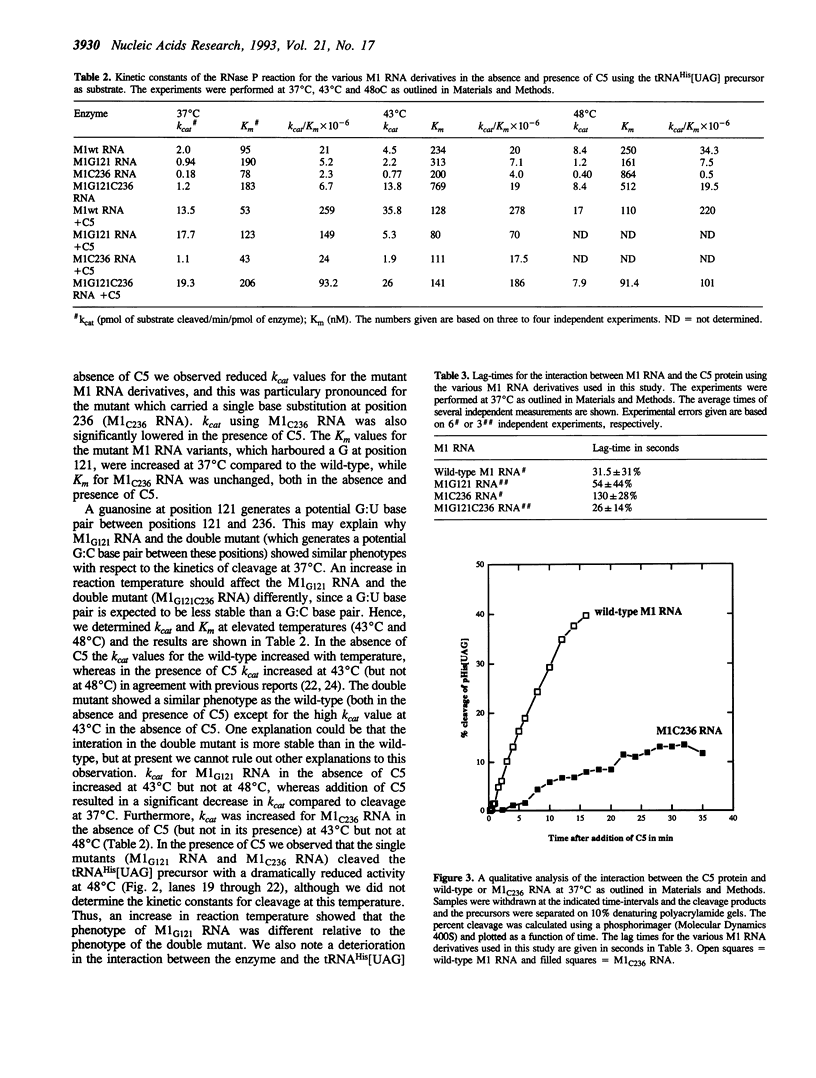
Phylogenetic covariation of the nucleotides corresponding to the bases at positions 121 and 236 in Escherichia coli RNase P RNA (M1 RNA) has been demonstrated in eubacterial RNase P RNAs. To investigate whether the nucleotides at these positions interact in M1 RNA we introduced base substitutions at either or at both of these positions. Single base substitutions at 121 or at 236 resulted in M1 RNA molecules which did not complement the temperature-sensitive phenotype associated with rnpA49 in vivo whereas wild-type M1 RNA or the double mutant M1 RNA, with restored base-pairing between 121 and 236, did. In addition, wild-type and the double mutant M1 RNA were efficiently cleaved by Pb++ between positions 122 and 123 whereas the rate of this cleavage was significantly reduced for the singly mutated M1 RNA variants. From these data we conclude that the nucleotides at positions 121 and 236 in M1 RNA establish a novel long-range tertiary interaction in M1 RNA. Our results also demonstrated that this interaction is not absolutely required for cleavage in vitro, however, a disruption resulted in a reduction in cleavage efficiency (kcat/Km), both in the absence and presence of C5.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altman S., Guerrier-Takada C. M1 RNA, the RNA subunit of Escherichia coli ribonuclease P, can undergo a pH-sensitive conformational change. Biochemistry. 1986 Mar 25;25(6):1205–1208. doi: 10.1021/bi00354a002. [DOI] [PubMed] [Google Scholar]
- Altman S., Kirsebom L., Talbot S. Recent studies of ribonuclease P. FASEB J. 1993 Jan;7(1):7–14. doi: 10.1096/fasebj.7.1.7916700. [DOI] [PubMed] [Google Scholar]
- Baer M. F., Wesolowski D., Altman S. Characterization in vitro of the defect in a temperature-sensitive mutant of the protein subunit of RNase P from Escherichia coli. J Bacteriol. 1989 Dec;171(12):6862–6866. doi: 10.1128/jb.171.12.6862-6866.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behlen L. S., Sampson J. R., DiRenzo A. B., Uhlenbeck O. C. Lead-catalyzed cleavage of yeast tRNAPhe mutants. Biochemistry. 1990 Mar 13;29(10):2515–2523. doi: 10.1021/bi00462a013. [DOI] [PubMed] [Google Scholar]
- Brown J. W., Pace N. R. Ribonuclease P RNA and protein subunits from bacteria. Nucleic Acids Res. 1992 Apr 11;20(7):1451–1456. doi: 10.1093/nar/20.7.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown R. S., Dewan J. C., Klug A. Crystallographic and biochemical investigation of the lead(II)-catalyzed hydrolysis of yeast phenylalanine tRNA. Biochemistry. 1985 Aug 27;24(18):4785–4801. doi: 10.1021/bi00339a012. [DOI] [PubMed] [Google Scholar]
- Dam E., Pleij K., Draper D. Structural and functional aspects of RNA pseudoknots. Biochemistry. 1992 Dec 1;31(47):11665–11676. doi: 10.1021/bi00162a001. [DOI] [PubMed] [Google Scholar]
- Deutscher M. P. Ribonucleases, tRNA nucleotidyltransferase, and the 3' processing of tRNA. Prog Nucleic Acid Res Mol Biol. 1990;39:209–240. doi: 10.1016/s0079-6603(08)60628-5. [DOI] [PubMed] [Google Scholar]
- England T. E., Bruce A. G., Uhlenbeck O. C. Specific labeling of 3' termini of RNA with T4 RNA ligase. Methods Enzymol. 1980;65(1):65–74. doi: 10.1016/s0076-6879(80)65011-3. [DOI] [PubMed] [Google Scholar]
- Guerrier-Takada C., Gardiner K., Marsh T., Pace N., Altman S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell. 1983 Dec;35(3 Pt 2):849–857. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- Guerrier-Takada C., van Belkum A., Pleij C. W., Altman S. Novel reactions of RNAase P with a tRNA-like structure in turnip yellow mosaic virus RNA. Cell. 1988 Apr 22;53(2):267–272. doi: 10.1016/0092-8674(88)90388-1. [DOI] [PubMed] [Google Scholar]
- Haas E. S., Morse D. P., Brown J. W., Schmidt F. J., Pace N. R. Long-range structure in ribonuclease P RNA. Science. 1991 Nov 8;254(5033):853–856. doi: 10.1126/science.1719634. [DOI] [PubMed] [Google Scholar]
- James B. D., Olsen G. J., Liu J. S., Pace N. R. The secondary structure of ribonuclease P RNA, the catalytic element of a ribonucleoprotein enzyme. Cell. 1988 Jan 15;52(1):19–26. doi: 10.1016/0092-8674(88)90527-2. [DOI] [PubMed] [Google Scholar]
- Kazakov S., Altman S. Site-specific cleavage by metal ion cofactors and inhibitors of M1 RNA, the catalytic subunit of RNase P from Escherichia coli. Proc Natl Acad Sci U S A. 1991 Oct 15;88(20):9193–9197. doi: 10.1073/pnas.88.20.9193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirsebom L. A., Altman S. Reaction in vitro of some mutants of RNase P with wild-type and temperature-sensitive substrates. J Mol Biol. 1989 Jun 20;207(4):837–840. doi: 10.1016/0022-2836(89)90250-7. [DOI] [PubMed] [Google Scholar]
- Kirsebom L. A., Baer M. F., Altman S. Differential effects of mutations in the protein and RNA moieties of RNase P on the efficiency of suppression by various tRNA suppressors. J Mol Biol. 1988 Dec 20;204(4):879–888. doi: 10.1016/0022-2836(88)90048-4. [DOI] [PubMed] [Google Scholar]
- Kirsebom L. A., Svärd S. G. The kinetics and specificity of cleavage by RNase P is mainly dependent on the structure of the amino acid acceptor stem. Nucleic Acids Res. 1992 Feb 11;20(3):425–432. doi: 10.1093/nar/20.3.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzyzosiak W. J., Marciniec T., Wiewiorowski M., Romby P., Ebel J. P., Giegé R. Characterization of the lead(II)-induced cleavages in tRNAs in solution and effect of the Y-base removal in yeast tRNAPhe. Biochemistry. 1988 Jul 26;27(15):5771–5777. doi: 10.1021/bi00415a056. [DOI] [PubMed] [Google Scholar]
- Lawrence N. P., Altman S. Site-directed mutagenesis of M1 RNA, the RNA subunit of Escherichia coli ribonuclease P. The effects of an addition and small deletions on catalytic function. J Mol Biol. 1986 Sep 20;191(2):163–175. doi: 10.1016/0022-2836(86)90253-6. [DOI] [PubMed] [Google Scholar]
- Lumelsky N., Altman S. Selection and characterization of randomly produced mutants in the gene coding for M1 RNA. J Mol Biol. 1988 Aug 5;202(3):443–454. doi: 10.1016/0022-2836(88)90277-x. [DOI] [PubMed] [Google Scholar]
- Milligan J. F., Groebe D. R., Witherell G. W., Uhlenbeck O. C. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987 Nov 11;15(21):8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motamedi H., Lee K., Nichols L., Schmidt F. J. An RNA species involved in Escherichia coli ribonuclease P activity. Gene cloning and effect on transfer RnA synthesis in vivo. J Mol Biol. 1982 Dec 15;162(3):535–550. doi: 10.1016/0022-2836(82)90387-4. [DOI] [PubMed] [Google Scholar]
- Pan T., Gutell R. R., Uhlenbeck O. C. Folding of circularly permuted transfer RNAs. Science. 1991 Nov 29;254(5036):1361–1364. doi: 10.1126/science.1720569. [DOI] [PubMed] [Google Scholar]
- Saiki R. K., Gelfand D. H., Stoffel S., Scharf S. J., Higuchi R., Horn G. T., Mullis K. B., Erlich H. A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988 Jan 29;239(4839):487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schedl P., Primakoff P. Mutants of Escherichia coli thermosensitive for the synthesis of transfer RNA. Proc Natl Acad Sci U S A. 1973 Jul;70(7):2091–2095. doi: 10.1073/pnas.70.7.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirling D. A., Hulton C. S., Waddell L., Park S. F., Stewart G. S., Booth I. R., Higgins C. F. Molecular characterization of the proU loci of Salmonella typhimurium and Escherichia coli encoding osmoregulated glycine betaine transport systems. Mol Microbiol. 1989 Aug;3(8):1025–1038. doi: 10.1111/j.1365-2958.1989.tb00253.x. [DOI] [PubMed] [Google Scholar]
- Streicher B., von Ahsen U., Schroeder R. Lead cleavage sites in the core structure of group I intron-RNA. Nucleic Acids Res. 1993 Jan 25;21(2):311–317. doi: 10.1093/nar/21.2.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier F. W., Moffatt B. A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986 May 5;189(1):113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- Svärd S. G., Kirsebom L. A. Several regions of a tRNA precursor determine the Escherichia coli RNase P cleavage site. J Mol Biol. 1992 Oct 20;227(4):1019–1031. doi: 10.1016/0022-2836(92)90518-o. [DOI] [PubMed] [Google Scholar]
- Tallsjö A., Kirsebom L. A. Product release is a rate-limiting step during cleavage by the catalytic RNA subunit of Escherichia coli RNase P. Nucleic Acids Res. 1993 Jan 11;21(1):51–57. doi: 10.1093/nar/21.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tranguch A. J., Engelke D. R. Comparative structural analysis of nuclear RNase P RNAs from yeast. J Biol Chem. 1993 Jul 5;268(19):14045–14055. [PubMed] [Google Scholar]
- Vioque A., Arnez J., Altman S. Protein-RNA interactions in the RNase P holoenzyme from Escherichia coli. J Mol Biol. 1988 Aug 20;202(4):835–848. doi: 10.1016/0022-2836(88)90562-1. [DOI] [PubMed] [Google Scholar]
- Waugh D. S., Pace N. R. Complementation of an RNase P RNA (rnpB) gene deletion in Escherichia coli by homologous genes from distantly related eubacteria. J Bacteriol. 1990 Nov;172(11):6316–6322. doi: 10.1128/jb.172.11.6316-6322.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]