Abstract
The biological effects of cytokines are mediated by STAT proteins, a family of dimeric transcription factors. In order to elicit transcriptional activity, the STATs require activation by phosphorylation of a single tyrosine residue. Our experiments revealed that fully tyrosine-phosphorylated STAT dimers polymerize via Tyr(P)-Src homology 2 domain interactions and assemble into paracrystalline arrays in the nucleus of cytokine-stimulated cells. Paracrystals are demonstrated to be dynamic reservoirs that protect STATs from dephosphorylation. Activated STAT3 forms such paracrystals in acute phase liver cells. Activated STAT1, in contrast, does not normally form paracrystals. By reversing the abilities of STAT1 and STAT3 to be sumoylated, we show that this is due to the unique ability of STAT1 among the STATs to conjugate to small ubiquitin-like modifier (SUMO). Sumoylation had one direct effect; it obstructed proximal tyrosine phosphorylation, which led to semiphosphorylated STAT dimers. These competed with their fully phosphorylated counterparts and interfered with their polymerization into paracrystals. Consequently, sumoylation, by preventing paracrystal formation, profoundly curtailed signal duration and reporter gene activation in response to cytokine stimulation of cells. The study thus identifies polymerization of activated STAT transcription factors as a positive regulatory mechanism in cytokine signaling. It provides a unifying explanation for the different subnuclear distributions of STAT transcription factors and reconciles the conflicting results as to the role of SUMO modification in STAT1 functioning. We present a generally applicable system in which protein solubility is maintained by a disproportionately small SUMO-modified fraction, whereby modification by SUMO partially prevents formation of polymerization interfaces, thus generating competitive polymerization inhibitors.
Keywords: Cytokine Action, Interferon, Phosphotyrosine, Phosphotyrosine Signaling, SH2 Domains, STAT Transcription Factor, Sumoylation
Introduction
STAT transcription factors comprise a structurally and functionally conserved family of proteins with indispensable roles in cytokine signaling (1). Cytokine binding to specific membrane receptors first activates receptor-associated JAK tyrosine kinases, which activate STAT proteins by phosphorylating a single C-terminal tyrosine (2). The STATs are nucleocytoplasmic shuttling homodimers that adopt antiparallel conformation involving N domain interactions (3, 4). Structural evidence indicates that such dimers are recruited to cytokine receptors for activation (4, 5). Upon activation, an additional dimer conformation (termed “parallel”) emerges, which is stabilized by mutual SH22 domain-phosphotyrosyl interactions. In this conformation, the STATs possess DNA binding activity and evoke transcriptional responses to cytokines (6). The activated dimers can enter the nucleus, but dephosphorylation is required for their export (7). The time required for dephosphorylation causes a transient accumulation of the activated dimers in the nucleus (3). Here, for previously unknown reasons, STAT3 localizes to distinct nuclear bodies or particles (8), whereas activated STAT1 distributes homogeneously (9).
The activity of STAT proteins is regulated both positively and negatively in a number of different ways (2). One rather poorly understood example is the covalent modification by SUMO, which among the STAT proteins is unique to STAT1. STAT1 harbors a functional sumoylation consensus sequence (ΨKXE, where Ψ represents a large hydrophobic residue) with the SUMO acceptor lysine 703 in position +2 relative to the Tyr701 phosphorylation site (10). The SUMO consensus is evolutionarily conserved in STAT1 but mutated in the other STAT family members (11). Inactivation of the SUMO consensus increased STAT1 activity according to some studies (12, 13), but the effects differed depending on the amino acid alteration, which led to contradictory results (11). This indicates restraints on the amino acid side chains in this region where SUMO consensus and phosphorylation site overlap. A further complication to biochemical analyses is posed by the highly efficient SUMO deconjugation reaction, which mirrors the situation with many SUMO targets, because it leaves only an apparently negligible STAT1 fraction SUMO-modified at the steady state (10–12). In order to overcome these obstacles, we introduced a modest charge-neutralizing mutation Glu705 → Gln at Tyr+4 of the SUMO consensus to minimize SUMO-independent effects on STAT1.
This approach has greatly facilitated the analyses of STAT1 sumoylation and its physiological consequences. In the course of these studies, we found that tyrosine-phosphorylated STAT dimers polymerize via SH2 domain interactions. Electron microscopy demonstrated that the polymers can align to form paracrystalline arrays in the nucleus of cytokine-stimulated cells, as exemplified by STAT3 in liver cells after peritoneal lipopolysaccharide (LPS) injection that mimics systemic bacterial infection. Activated STAT1, in contrast, does not normally form paracrystals, which we demonstrate is due to the unique ability of STAT1 among the STATs to SUMO-conjugate. We further describe how SUMO modification generates competitive polymerization inhibitors that interfere with the paracrystal assembly of STAT1, which in turn curtails STAT1 activation in the nucleus. These results establish cytokine-induced polymerization as a mechanism to sequester activated STATs and suggest a general mechanism for SUMO-mediated regulation of protein solubility.
EXPERIMENTAL PROCEDURES
Animal Experimentation
In compliance with United Kingdom Home Office guidelines, 0.2 ml of PBS was injected into the peritoneum of C57Bl/6 mice without or with LPS (7.35 μg/g body weight). Two hours later, the animals were euthanized, and livers were isolated. For light microscopy, one half of each liver was embedded in optimal cutting temperature formulation (OCT) and frozen in supercooled isopentane for subsequent thin sectioning (8–10 μm). The remainder was cut into ∼1-mm3 cubicles and stored in fixative for electron microscopy.
Cell Culture
Cell culture and transient transfections were done as described (15). For gene silencing, 5 × 106 HeLa cells were treated twice (24 h each) with 0.8 μg of siRNA using siPort reagent according to Ambion's instructions. To facilitate detection of paracrystals in the siRNA-treated cells, IFNγ (5 units/ml) was added subsequently and left on the cells for 16 h to boost STAT1 expression, followed by 1 h of restimulation after IFNγ withdrawal for 4 h. Cytokine stimulation of cells is detailed in the supplemental Experimental Procedures.
Reporter Gene Assay
Reporter gene assays in transiently transfected cells were done as described (16). STAT1-mediated gene induction in response to IFNγ was assayed with a reporter construct containing a triple STAT1 binding site (Ly6E). STAT3-mediated gene induction in response to erythropoietin was examined using a construct containing tandem STAT3 binding sites (APRE) (17).
Fluorescence Microscopy
GFP epifluorescence and indirect immunofluorescence detection were done after fixation of cells with a Zeiss Axioplan 2 microscope as described (16). Confocal microscopy of fixed cells and live cells (performed at 37 °C) was done using the Ar/ArKr laser (GFP excitation at 488 nm, Cy3 at 568 nm) and diode laser 405 (Hoechst dye excitation at 461 nm) of a Leica TCP-SP2 equipped with an automated shutter and motorized x, y, and z stack controller together with a Q-Imaging CCD camera with 12-bit gray scale resolution. Fluorescence signal intensities were obtained with Leica LCS Lite 2.61 software.
Electron Microscopy
Transmission electron microscopy was done essentially as described (18). In brief, transfected HeLa cells or mouse liver cubicles were fixed in 0.25% glutaraldehyde, 3% formaldehyde, followed by treatment with 2% OsO4 and dehydration in a graded ethanol series prior to embedding in epoxy resin. Sections (60-nm thickness) were cut and contrasted with 2% uranyl acetate and saturated lead citrate before analysis at 100 kV with a Zeiss 902 A, a Tecnai F20, or a JEOL 1010 electron microscope.
pSTAT1 Co-immunoprecipitation Assay
HEK293T cells expressing sumoylatable (WT) or SUMO-free (EQ) STAT1-UBC9 fusion proteins were treated with IFNγ for 1 h, followed by whole cell extraction. For each STAT1 construct, eight individual experiments (cell transfections and extractions) were performed. Consecutive immunoblotting experiments with anti-Tyr(P)701-STAT1- and anti-STAT1-specific (C24) antibodies were done with all 16 extracts to determine specific Tyr701 phosphorylation (termed input). Additionally, the extracts were used for immunoprecipitation experiments. Anti-unphosphorylated STAT1 (uSTAT1) antibody (C136) was used for six extracts each of WT STAT1 and SUMO-free STAT1, respectively, whereas the two remaining extracts of each STAT1 construct were immunoprecipitated with phosphorylation-indifferent anti-STAT1 antibody (C24). The immunoprecipitated materials were subsequently immunoblotted, and the specific Tyr701 phosphorylation of the precipitates was determined (termed IP). Quantitative immunoblotting data incorporated in Fig. 5D are given in supplemental Fig. S6C and supplemental Table S2, respectively.
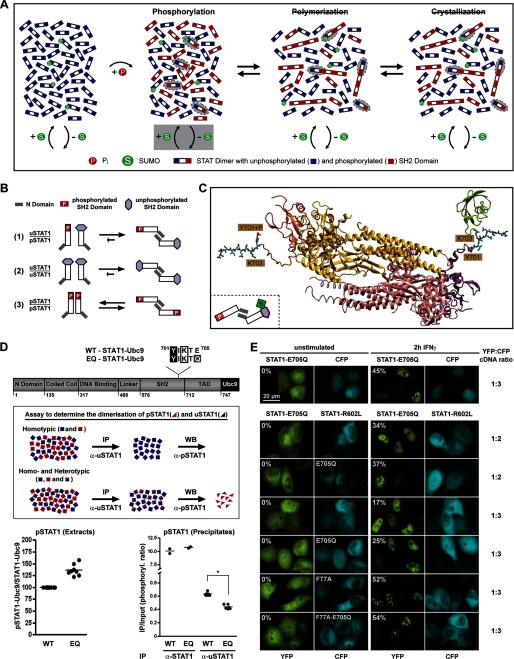
FIGURE 5.
SUMO-induced semiphosphorylated dimers inhibit STAT1 paracrystal assembly. A, a schematic representation of the direct influence of SUMO conjugation on the tyrosine 701 phosphorylation of STAT1. Semiphosphorylated dimers are highlighted with gray ellipsoids. B, model depicting predominantly antiparallel conformation of dimers of unphosphorylated and Tyr701-phosphorylated STAT1 as deduced from pull-down experiments (see supplemental Fig. S5A). Dimer conformer equilibria of unphosphorylated STAT1 and Tyr701-phosphorylated STAT1 are also shown. C, atomic model of an antiparallel STAT1 dimer consisting of a Tyr701-phosphorylated protomer (yellow with orange SH2 domain) and an unphosphorylated protomer (red with purple SH2 domain) with Lys703-conjugated SUMO1 (green). The tail segments (residues 701–710) and phosphoryl group are shown in a ball-and-stick representation. D, top, diagram of STAT1-Ubc9 fusion proteins and outline of the immunoprecipitation assay with anti-uSTAT1-specific antibody used to determine the abundance of semiphosphorylated dimers. Bottom, the left panel shows the specific Tyr701 phosphorylation of sumoylatable (WT; set as 100) and SUMO-free (EQ) STAT1-UBC9 fusion proteins present in the extracts (Input). The right panel depicts the Tyr701-phosphorylated STAT1 that co-precipitated with STAT1 (α-STAT1) or specifically with unphosphorylated STAT1 (α-uSTAT1) in the extracts containing sumoylatable or SUMO-free STAT1. Values represent the phosphorylation ratio (IP/Input) for each immunoprecipitation experiment. *, statistically significant differences using Student's t test (p < 0.0001). E, top row, fluorescence micrographs before and after IFNα treatment of HeLa cells co-expressing CFP and SUMO-free STAT1-E705Q (YFP-tagged). In the other rows, CFP was replaced by CFP-tagged non-phosphorylatable STAT1-R602L or the SUMO-free mutant thereof. Molar ratios of transfected cDNAs (YFP/CFP) and the percentages of paracrystal containing cells that express both fluorescent markers are given. WB, Western blot.
Quantitative Immunoblotting
Cell extraction using whole cell extraction buffer A (pH 7.4), SDS-PAGE, and immunoblotting were done as described (15). Primary antibodies were labeled with IRdye800-conjugated secondary immunoglobulin (Licor Biosciences) and detected and quantified with the Odyssey system (Licor Biosciences). Control readings were obtained with diluted cell extracts to confirm that signal intensities were within the linear range. Detection of pSTAT and total STAT was done consecutively on the same blot. Bound antibodies were stripped off the nitrocellulose membrane for 1 h at 65 °C in 25 mm glycine and 2% SDS (pH 2.0).
Molecular Modeling
The structural model of Fig. 3C was generated by superimposing one SH2 domain each of the crystal structures of two antiparallel STAT1 dimers (Protein Data Bank entry 1YVL) onto the SH2 domains of a STATa dimer (Protein Data Bank entry 1UUR). The STAT1 tail sequence (residues 700–710) was used in the conformation of phosphorylated STAT1 (Protein Data Bank entry 1BF5). The peptide segment that connects the tail sequence with the core region was model-built using the spdbv program (19). The structure was finally energy-minimized using the Amber9 package (20). The structural model of Fig. 5C is based on the structure of antiparallel STAT1 dimer (Protein Data Bank entry 1YVL). The Tyr701-phosphorylated STAT1 tail sequence in the conformation of phosphorylated STAT1 (Protein Data Bank entry 1BF5) was added to one protomer. To the other protomer the tail sequence was added in the same conformation without a phosphoryl group at Tyr701 but with Lys703-conjugated SUMO1 (Protein Data Bank entry 1WYW). Additional methods are described in the supplemental Experimental Procedures.
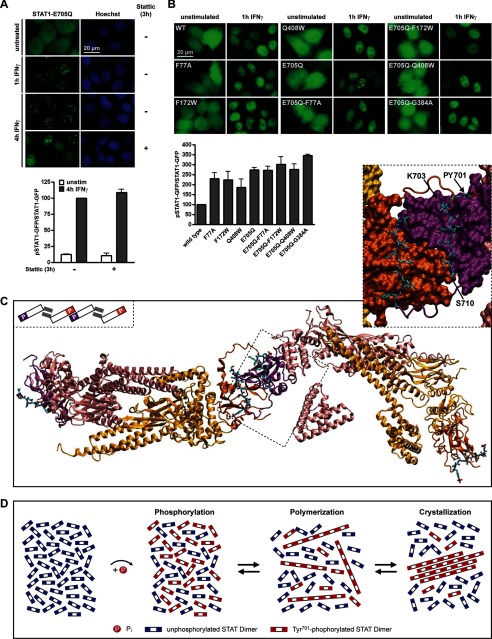
FIGURE 3.
Paracrystal assembly involves polymerization of antiparallel STAT1 dimers via SH2-Tyr(P) interactions. A, top, GFP fluorescence micrographs of HeLa cells expressing SUMO-free STAT1. The cells were treated as indicated with IFNγ and Stattic (50 μm). Bottom, bar diagram showing the specific Tyr701 phosphorylation of SUMO-free STAT1 in cells that were treated for 4 h with IFNγ (−) or that additionally received Stattic after 1 h. Results for IFNγ-treated cells were set as 100; data (mean ± S.D. (error bars)) were obtained from two independent immunoblotting experiments. B, top, IFNγ-induced relocalization of wild type STAT1 (WT), point mutants of STAT1 defective specifically in antiparallel dimerization (F77A, F172W, and Q408W), SUMO-free STAT1 (E705Q), and the respective SUMO-free dimerization mutants. Bottom, bar diagram showing the corresponding specific Tyr701 phosphorylation (wild type set as 100) after 1 h of IFNγ. Data display the mean ± S.D. obtained from three independent immunoblotting experiments. C, atomic model of two STAT1 antiparallel dimers (protomers colored red and yellow, respectively) that are linked by phosphopeptide-SH2 domain interactions (SH2 domains colored purple and orange). Additional stabilizing β-sheet interactions are circled. The inset shows details of the interdigitating SH2 domain surfaces with the phosphopeptide tail segments in a ball-and-stick display. The schematic model gives an overview highlighting the SH2 domains (colored) and N domains (gray bars). D, a schematic representation of events following the activation of SUMO-free STATs.
RESULTS
Cytokines Trigger the Localization of STAT3 and SUMO-free STAT1 to Discrete Nuclear Particles
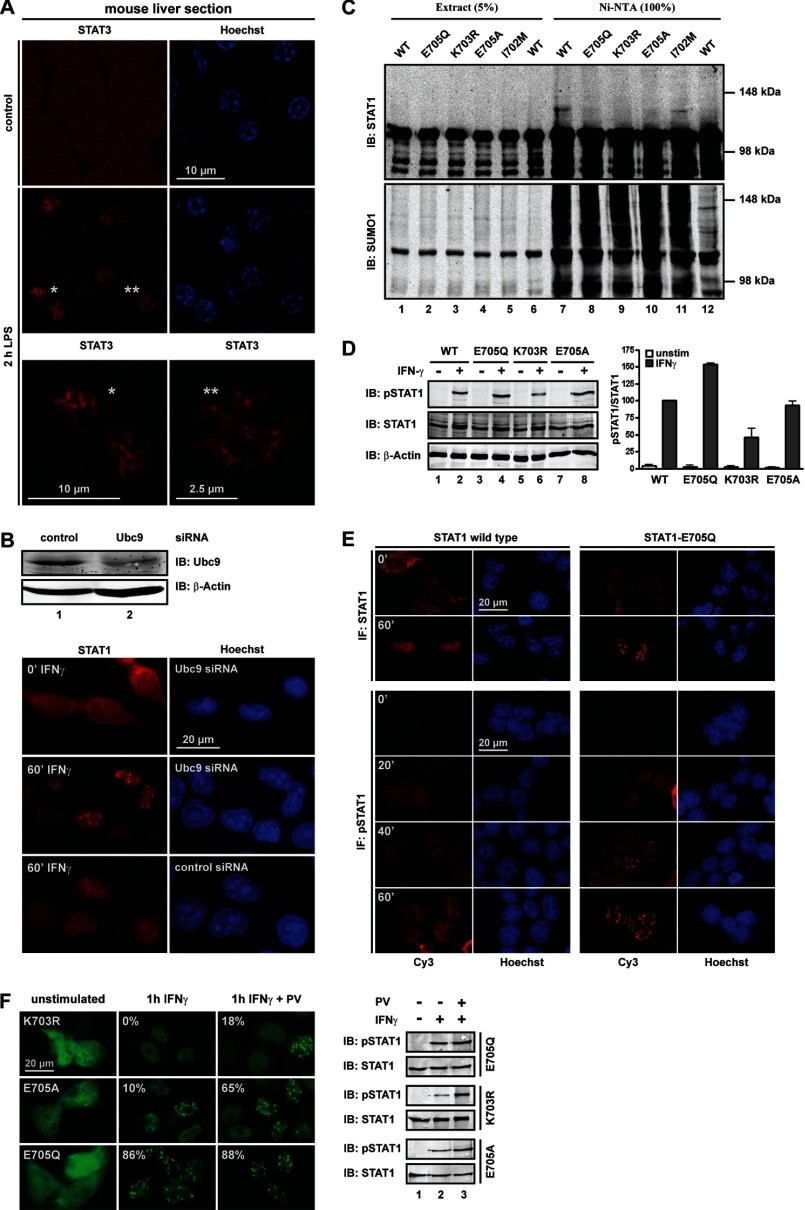
STAT1 and STAT3 distribute strikingly differently in the nucleus of cytokine-stimulated cells. Activation of STAT3 triggers its accumulation in nuclear particles. This has been observed with a number of different cell types from humans and mice (21–23). We extend these findings to primary human hepatocytes (supplemental Fig. S1A). A striking physiological in situ example of STAT3 particle assembly occurs during the liver immune response to systemic bacterial infection, which can be triggered in mice by intraperitoneal injection of bacterial LPS. The condition is associated with massive IL-6 release and activation of STAT3, which localized in abundant dotlike particles in virtually all hepatocytes 2 h after LPS injection (22) (Fig. 1A and supplemental Fig. S1B). Activated STAT1, in contrast, distributes homogeneously in the nucleus of cytokine-stimulated cells (Fig. 1B).
FIGURE 1.
STAT3 and SUMO-free STAT1 localize to nuclear particles. A, STAT3 immunofluorescence confocal microscopy of liver sections from control (PBS) and LPS-treated mice. Nuclei are Hoechst-stained. Asterisks denote cells shown in close-up view. B, immunoblot (IB) analysis of Ubc9 knockdown in HeLa cells (top) and corresponding STAT1 immunofluorescence microscopy. C, mutation of the SUMO consensus precludes SUMO conjugation of STAT1. Wild type or mutant STAT1 was co-expressed with Ubc9 (lanes 1–12) and His-tagged SUMO1 (lanes 1–11) in HEK293T cells. Results of immunoblotting experiments with anti-SUMO1-specific (bottom) and anti-STAT1-specific (top) antibody are shown for whole cell extracts and the corresponding bound fraction after affinity chromatography on nickel-NTA-agarose. D, U3A cells reconstituted with WT or SUMO-free STAT1 were treated for 60 min with IFNγ as indicated, followed by extraction and immunoblotting with anti-Tyr(P)701 STAT1-specific (top), anti-STAT1-specific (middle), and anti-β-actin-specific (bottom) antibody. The corresponding bar diagram shows the specific Tyr701 phosphorylation of mutants and wild type (set as 100) after 1 h IFNγ. Data display the mean ± S.D. (error bars) obtained from three independent immunoblotting experiments. E, top, STAT1 immunofluorescence confocal microscopy of U3A cells reconstituted with wild type or SUMO-free STAT1-E705Q. Bottom, confocal immunofluorescence microscopy time course showing the distribution of Tyr701-phosphorylated STAT1 in transfected HEK293T cells before and after IFNγ treatment. F, GFP fluorescence micrographs (left) and corresponding immunoblot analyses (right) of reconstituted U3A cells before and after treatment with IFNγ alone or IFNγ plus tyrosine phosphatase inhibitor pervanadate (PV). Pervanadate treatment started 30 min after the addition of IFNγ. The percentage of transfected cells with particles is denoted.
To find the cause of the differences in distribution, we explored the role of SUMO conjugation, an important regulator of protein association, because STAT1 is a SUMO target, whereas STAT3 is not. We therefore used siRNA to deplete cells by ∼70% (determined by quantitative Western blotting) of the essential SUMO ligase Ubc9 (Fig. 1B), which altered neither the intracellular distribution of STAT1 before cytokine stimulation nor its cytokine-induced nuclear accumulation. However, the nuclear accumulated STAT1 showed inhomogeneous punctuate distribution in Ubc9-depleted cells that resembled STAT3 nuclear particles (Fig. 1B). Identical results were obtained with GFP-tagged STAT1 (supplemental Fig. S1C). To determine whether particle suppression required the sumoylation of STAT1, we used published (K703R or E705A) or previously untested (E705Q) mutations to inactivate its SUMO consensus. In agreement with previous studies, we found that steady-state sumoylation of STAT1 was at the limit of detection (10–12). The STAT1 variants, fused to GFP to facilitate differentiation from endogenous STAT1, were therefore co-expressed in HEK293T cells with Ubc9 (Fig. 1C, lanes 1–12) and His-tagged SUMO1 (lanes 1–11) to boost sumoylation. His-tagged proteins were subsequently enriched by nickel-chelate affinity chromatography, as shown by Western blotting of respective cell extracts using SUMO1 antibody (Fig. 1C, bottom). Reprobing of the blot with STAT1 antibody (Fig. 1C, top) expectedly revealed the presence of STAT1 (∼110 kDa) in extracts and precipitates. In addition, SUMO-enriched fractions containing wild type STAT1 (lane 7) revealed an additional slower migrating band with STAT1 immunoreactivity (∼120 kDa). Because this band was absent when SUMO was absent (lane 12), we concluded that the ∼120 kDa band represented SUMO-conjugated STAT1. This band was absent too from cell extracts expressing STAT1 variants that deviated from the canonical SUMO consensus (K703R, E705A, or E705Q; lanes 8–10), but it was present if the SUMO consensus was maintained by exchanging Ile702 for Met, as present in some fish STAT1 orthologs (lane 11). Thus, lack of the canonical SUMO consensus resulted in SUMO-free STAT1, regardless of the actual side chain alteration. In contrast, the effects on STAT1 phosphorylation differed strongly with the mutation introduced (Fig. 1D), a finding reported previously (13). Of note, the structurally more disruptive changes (K703R and E705A) resulted in unaltered or even decreased STAT1 phosphorylation. This observation is difficult to reconcile with a role for these mutations solely in preventing SUMO conjugation of STAT1 because lack of sumoylation is bound to increase STAT1 phosphorylation due to the mutual exclusion of sumoylation and phosphorylation (24). Thus, the unaltered or even reduced phosphorylation of SUMO-free mutants K703R and E705A indicates additional SUMO-independent effects on STAT1 activation. On the contrary, when SUMO conjugation was precluded by the modest charge-neutralizing mutation E705Q, the expected outcome, namely increased STAT1 phosphorylation, was observed (Fig. 1D). However, STAT1 phosphorylation can be heightened too by its enhanced dimerization or DNA binding (3). We produced recombinant STAT1 (wild type and mutant E705Q) in insect cells and purified the Tyr701-phosphorylated variants to homogeneity (25) to test these possibilities by dynamic light scattering and EMSA analyses (supplemental Fig. S1, D and E). However, wild type and mutant STAT1 were indistinguishable, making gain-of-function mutation an unlikely cause of increased activation. We therefore inferred that E705Q mutation generated a superior reagent for studying the impact specifically of sumoylation on STAT1.
We then determined the subnuclear distribution of SUMO-free STAT1. Fig. 1E shows that it readily assembled elongated needle-shaped particles upon interferon stimulation. The time course at the bottom of Fig. 1E demonstrates that particles contained phosphorylated STAT1 and emerged 20–40 min after cytokine stimulation. Mutation of the STAT1 tyrosine phosphorylation site (Y701F) precluded particle assembly (not shown). Next, we confirmed that the different SUMO-free STAT1 variants share the ability to assemble particles (Fig. 1F, left). However, the poorly phosphorylated mutant K703R failed to form particles, whereas the better phosphorylated mutant E705A formed particles in about 10% of cells, compared with more than 80% for the highly activated E705Q variant. These results may explain why previous studies failed to report the STAT1 particles. Moreover, the positive correlation of particle prevalence and STAT1 phosphorylation suggested that differences in particle prevalence could be diminished if phosphorylation differences were evened out. We treated cells with tyrosine phosphatase inhibitor pervanadate, which increased phosphorylation of the K703R and E705A mutants, whereas the already well phosphorylated mutant E705Q was affected to a lesser degree (Fig. 1F, right). As expected, the increased phosphorylation enhanced particle formation of all SUMO-free STATs. Particles containing K703R or E705A STAT1 were present now in 18 or 65% of cells, respectively. In conclusion, these results demonstrated that particle formation is a shared quality of SUMO-free STAT1 variants; however, their differences in tyrosine phosphorylation bring about quantitative differences as to the prevalence of particles.
Cytokine-induced STAT Particles Are Dynamic Paracrystalline Assemblies
Next, transmission electron microscopy was used to compare the ultrastructure of particles formed by wild type STAT3 and SUMO-free STAT1 mutant. Acute phase liver sections obtained 2 h after the injection of mice with LPS were used to study STAT3 particles (Fig. 2A). Particles were recognized as well ordered entities consisting of a variable number (n = 6–36) of laterally aligned filaments with a diameter of 34.2 ± 2.8 nm and narrow variability in their center-to-center spacing (68.4 ± 7.8 nm). Such structures were absent from control liver. Particles of GFP-tagged STAT3 showed similar internal morphology with more compact lateral alignment of filaments (diameter 20.9 ± 2.8 nm) (supplemental Table S1). Similarly, examination of SUMO-free STAT1 in IFNγ-stimulated HeLa cells revealed particles consisting of a variable number (n = 11–275) of parallel filaments that stretch over the entire particle length of up to ∼2 μm (Fig. 2B). Inspection of micrographs taken before IFNγ stimulation failed to reveal such structures. Compared with STAT3 in liver, the STAT1 filaments are packed tighter and have a smaller diameter (25 ± 2.8 nm). Except for their generally higher filament numbers, particles of GFP-tagged STAT1 differed only minimally from their untagged counterparts (Fig. 2B and supplemental Table S1). We additionally used gold-conjugated antibodies to decorate particles, which established a direct connection between the structures observed in the EM and the localization of STAT1 or STAT3 therein (supplemental Fig. S2).
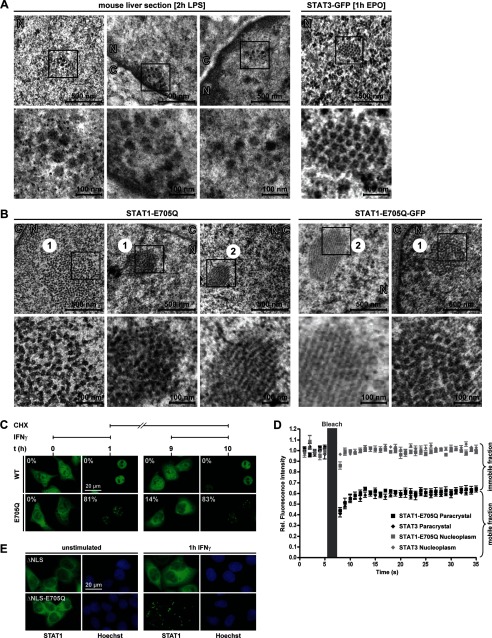
FIGURE 2.
Nuclear particles are reversible paracrystalline arrays. A, overview and close-up (boxed area) of electron micrographs of acute phase liver sections (left) and HeLa cells expressing GFP-tagged STAT3 stimulated with Epo (right). B, electron micrographs of IFNγ-stimulated HeLa cells expressing untagged (left) or GFP-tagged STAT1-E705Q (right). 1, cross section; 2, longitudinal section; N, nucleus; C, cytoplasm. C, HeLa cells expressing GFP-tagged WT or SUMO-free STAT1-E705Q were stimulated with IFNγ for 60 min. Subsequently, IFN was withdrawn, and incubation continued for 8 h in the presence of protein translation inhibitor cycloheximide (CHX), followed by restimulation with IFNγ for another 60 min. Representative images depicting the distribution of STAT1 are shown. The percentages of transfected cells with paracrystals are given. D, fluorescence recovery analyses after photobleaching of paracrystal-incorporated (labeled paracrystal) STAT3 and SUMO-free STAT1. Control measurements of soluble STATs were done in the nucleoplasm (labeled Nucleoplasm). Data are the normalized mean and S.D. (error bars) of six independent experiments carried out in IFNγ- or Epo-treated HeLa cells expressing GFP-tagged STAT1-E705Q or STAT3, respectively. E, GFP fluorescence microscopy of HeLa cells expressing nuclear import-deficient STAT1 (ΔNLS) and the SUMO-free variant thereof (ΔNLS-E705Q).
Particle dynamics and turnover was then researched. We expressed wild type or SUMO-free STAT1-GFP in HeLa cells, where IFNγ stimulation triggered particles in >80% of cells expressing mutant STAT1, whereas none where observed for wild type (Fig. 2C). Subsequent withdrawal of IFNγ and incubation with protein synthesis inhibitor cycloheximide for 8 h led to the reversal of nuclear accumulation and the disappearance of particles in almost all cells. However, upon restimulation with IFNγ, particles sprang up again in >80% of cells expressing SUMO-free STAT1. We thus concluded that STAT1 particles are fully reversible. In addition, GFP-tagged STAT3 and SUMO-free STAT1 were used in photobleaching (FRAP) experiments. To determine the exchange of STAT molecules in the particles with the nucleoplasm, we bleached a region of the particles (1-μm diameter) and subsequently followed the re-emergence of GFP fluorescence (Fig. 2D). We found that particles of STAT3 and SUMO-free STAT1 were essentially identical regarding their dynamic properties because both contained the same two kinetically distinct STAT fractions. The majority of molecules in the particles (∼60%) exchanged rapidly with soluble STATs in the surroundings (t50 < 2.5 s); the remainder, however, were essentially immobilized (t50 > 35 s). Control measurements taken outside of particles in the nucleoplasm confirmed highly mbile STAT molecules and the absence of an immobile fraction (Fig. 2D) (9).
Additionally, we asked whether paracrystals can form outside the nucleus (Fig. 2E). We inactivated the nuclear import signal of STAT1 (ΔNLS), which has no bearing on STAT1 activation but precludes import of phosphorylated STAT1 in the nucleus (15). Nonetheless, if import-deficient STAT1 remained SUMO-free (ΔNLS-E705Q), paracrystals readily formed, albeit in the cytoplasm. Thus, the requirements for paracrystal assembly and disassembly are not confined to the nuclear compartment. Taken together, the dynamic properties and regular ultrastructure of STAT particles resembled protein paracrystals. In further support of their crystalline nature, single STAT1 particles exhibited birefringence of ∼1 nm in fixed cells.3 We thus propose their designation as paracrystalline assemblies.
Activated STAT Dimers Polymerize via SH2-Tyr(P) Interactions
Paracrystals are protein self-assembly systems that involve the reversible polymerization of subunit molecules via non-covalent interactions (26). Our previous experiments demonstrated that STAT activation was indispensable for paracrystal assembly, suggesting that SH2-Tyr(P) interactions were involved. To test this assumption, we incubated cells with a chemical inhibitor of SH2-Tyr(P)-mediated dimerization both of STAT1 and of STAT3 (27, 28). When added 1 h after the cytokine-induced assembly of STAT1 paracrystals and left on the cells for 3 h, the compound did not measurably reduce STAT1 phosphorylation, yet it dissolved preformed paracrystals to a considerable extent, confirming the requirement for SH2-Tyr(P) interactions (Fig. 3A). Next, we destabilized the alternative, SH2 domain-independent dimer conformation by mutating key residues required for N domain-mediated (“antiparallel”) dimerization (F77A, F172W, G384A, and Q408W) (29). The mutations did not alter subcellular distribution or cytokine-induced nuclear accumulation but completely abrogated paracrystal formation of SUMO-free STAT1 (Fig. 3B). In agreement with previous studies (29), the mutants were highly phosphorylated, thus ruling out defective activation as a possible cause for defective paracrystal assembly (Fig. 3B).
These experiments strongly suggested the phosphorylated STAT dimer to be the polymer subunit required for paracrystal assembly. Because the interfaces of both dimer conformers were shown to participate, we propose two mechanisms as plausible explanations for the polymerization of STAT dimers. One posits parallel (Tyr(P)-SH2 domain-mediated) dimers, which polymerize via N domain-mediated (antiparallel) interactions; the other posits antiparallel dimers, which polymerize via Tyr(P)-SH2 domain interactions. To explore these possibilities, we used structural modeling of STAT1 based on analogy to STATa from Dictyostelium discoideum, which dimerizes by SH2-phosphopeptide binding yet adopts a fully extended arrangement (30). The arrangement of the SH2 domains in the D. discoideum dimer served as a template for modeling the interface between the SH2 domains that link two antiparallel STAT1 phosphodimers, which required only minor sterical adjustments. The flexible linker (residues 684–700) connecting the SH2 domain to Tyr(P)701 allowed phosphopeptide-SH2 interactions between two STAT1 dimers without causing any structural strain. Additionally, the two SH2 domains in the model interact through several van der Waals contacts and hydrogen bonds, in particular within two β hairpins corresponding to residues 652–662 (Fig. 3C). The proposed SH2-Tyr(P) linkage of antiparallel dimers thus forms a building block that allows further polymerization. The alternative model invoking polymerization of parallel dimers via antiparallel interactions was excluded due to steric overlap (supplemental Fig. S3). In conclusion, we describe polymerization of dimers as a mechanism for sequestering activated STATs and the formation of three-dimensional paracrystals, as diagrammed in Fig. 3D.
SUMO-mediated Suppression of Paracrystals Requires Blockage of Tyr701 Phosphorylation
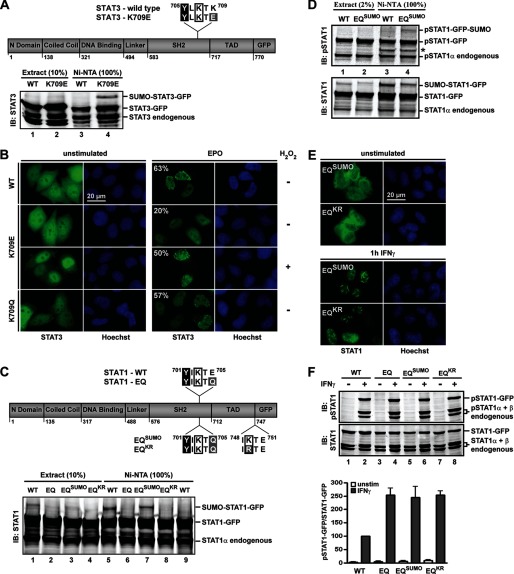
We then examined how SUMO suppresses the paracrystals of STAT1 and first asked whether its suppressive activity could be transferred to STAT3. This closely related protein harbors a potential SUMO acceptor Lys707 at Tyr+2, which is embedded in a SUMO consensus mutated at Tyr+4 (Fig. 4A, top). Therefore, the reverse experiment, reconstitution of the SUMO consensus (K709E) was done to examine the consequences of forced SUMO conjugation. Indeed, the STAT3 mutant was sumoylated at Tyr+2 with similar efficiency as STAT1 (Fig. 4A, bottom, lane 4). Moreover, although wild type STAT3 formed paracrystals in >60% of cells (Fig. 4B), the sumoylated variant generally adopted homogeneous subnuclear distribution similar to STAT1, with fewer and smaller paracrystals remaining only in 20% of cells (Fig. 4B). However, paracrystal formation was restored almost to the level of wild type when the cellular SUMO conjugation balance was tilted by H2O2 toward desumoylation (31) (Fig. 4B). The effectiveness of this treatment to reduce STAT3 sumoylation is shown in supplemental Fig. S4A. To additionally confirm that reduced crystal formation was consequential to SUMO conjugation rather than nonspecific structural perturbations of Tyr+4 mutants, we mutated Lys709 to Gln, which does not restore the SUMO consensus. Reassuringly this Tyr+4 mutant, like wild type STAT3, showed paracrystals in ∼60% of cells (Fig. 4B). It was thus concluded that the effects of SUMO on STAT3 distribution mirrored those on STAT1.
FIGURE 4.
Paracrystal suppression requires mutual exclusion of Lys703 sumoylation and Tyr701 phosphorylation. A, top, diagram depicting WT and SUMO consensus-reconstituted STAT3-K709E. The SUMO consensus-resembling amino acid sequence downstream of the Tyr705 phosphorylation site (black box), including the potential SUMO acceptor Lys707 (boxed) and the mutated residue 709 (gray box), is given. Bottom, immunoblotting (IB) results with anti-STAT3-specific antibody are shown for whole cell extracts and corresponding nickel-NTA-agarose-bound fractions. Cell extracts were prepared from COS7 cells co-expressing STAT3, Ubc9, and His-tagged SUMO1. B, PC3 cells reconstituted with wild type STAT3 or mutant K709E or K709Q. Treatment with Epo was for 1 h, and H2O2 (1 mm) was added where indicated after 30 min of Epo. The percentage of transfected cells with nuclear particles is given. C, top, diagram depicting WT and mutant STAT1 constructs, highlighting proximal and distal SUMO consensus sequences. Bottom, immunoblotting results with anti-STAT1-specific antibody are shown for whole cell extracts and nickel-NTA-agarose-bound fractions. Extracts were prepared from HEK293T cells co-expressing the indicated STAT1 variants, His-SUMO1, and Ubc9 (lanes 1–8). Cells analyzed in lane 9 did not co-express His-SUMO1. D, results of immunoblotting experiments with anti-Tyr(P)701-STAT1-specific (top) and anti-STAT1-specific (bottom) antibody are shown for whole cell extracts and nickel-NTA-agarose-bound fractions. Extracts were prepared from cells treated for 60 min with IFNγ as described in C. *, antibody cross-reactivity. E, GFP fluorescence micrographs of HeLa cells expressing STAT1 mutants. F, top, immunoblotting results of extracts from HeLa cells expressing STAT1 variants using anti-Tyr(P)701-STAT1-specific (top) and anti-STAT1-specific (bottom) antibody. Bottom, corresponding bar diagram showing the specific Tyr701 phosphorylation (wild type set as 100) after 1 h IFNγ. Data display the mean ± S.D. (error bars) of two independent immunoblotting experiments.
Next, we wondered whether the paracrystal-inhibiting activity of SUMO required conjugation specifically at Tyr+2 because it was demonstrated that sumoylation at this position and Tyr701 phosphorylation are mutually excluded (24). Thus, SUMO modifies exclusively the unphosphorylated STAT1, but all this modification appears to do is make sure that STAT1 remains unphosphorylated. How then can paracrystal assembly be inhibited, since the pool from which the paracrystals arise consists exclusively of unphosphorylated STAT molecules? To resolve this paradox, we devised a STAT1 variant that was SUMO-modified in the Tyr701-phosphorylated state. To this end, the native sumoylation site of STAT1 was inactivated by E705Q mutation (termed EQ), and another consensus sequence comprising IKTE (or the inactive variant IRTE) was placed distally at Tyr+48 (termed EQSUMO and EQKR, respectively (Fig. 4C, top)). The STAT1 variants were co-expressed with Ubc9 and His-tagged SUMO1, and sumoylation was detected by Western blotting after nickel-chelate affinity chromatography as described in the legend to Fig. 1C. This demonstrated comparable sumoylation of STAT1 at the native Tyr+2 site and the artificial distal position (Fig. 4C, bottom, compare lanes 5 and 7), whereas the mutants EQ and EQKR expectedly remained unmodified (Fig. 4C, bottom, lanes 6 and 8).
To see whether sumoylation at Tyr+48 no longer obstructed Tyr701 phosphorylation, we probed extracts containing proximally (wild type) or distally sumoylated STAT1 (EQSUMO) with an antibody reactive with Tyr701-phosphorylated STAT1, which is a reliable method to interrogate Tyr701 phosphorylation of sumoylated STAT1 (14). As shown in Fig. 4D, the Tyr+48 distally sumoylated STAT1 (EQSUMO) was indeed phosphorylated at Tyr701 (top, lane 4). In contrast, Tyr+2 proximally sumoylated wild type STAT1 remained unphosphorylated (Fig. 4D, top, lane 3). The mutual exclusion of proximal sumoylation and phosphorylation was further confirmed by in vitro sumoylation assays with purified recombinant STAT1 protein, which demonstrated SUMO conjugation of unphosphorylated STAT1, whereas the Tyr701-phosphorylated or E705Q-mutated variants remained unsumoylated (supplemental Fig. S4B).
We now asked whether the paracrystal inhibiting activity of SUMO required the blocking of Tyr701 phosphorylation. The answer was yes, because sumoylation at the distal position was without inhibitory effect on STAT1 paracrystals, which readily formed irrespective of SUMO conjugation at Tyr+48 (Fig. 4E). Thus, despite the presence of SUMO-conjugated STAT1 that was Tyr701-phosphorylated and hence potentially available for incorporation into paracrystals, SUMO was without adverse effects on crystal assembly. We therefore inferred that SUMO-mediated paracrystal dispersal occurred by an indirect mechanism that did not require the presence of SUMO in the crystals. This reasoning was supported by the unaltered paracrystal assembly of a fusion protein of SUMO1 and STAT1-E705Q (not shown).
It thus appeared that suppression of Tyr701 phosphorylation was pivotal. To corroborate this assumption, we used quantitative Western blotting and compared the consequences of distal and proximal sumoylation for STAT1 activation (Fig. 4F). This experiment clearly confirmed that proximal sumoylation reduced the phosphorylation of STAT1 (compare WT and mutant EQ), whereas distal sumoylation was without consequences for STAT1 phosphorylation (compare EQSUMO and EQKR). Because paracrystal-forming mutants all showed increased phosphorylation compared with WT, it could be argued that proximal sumoylation prevented paracrystals simply by suppressing STAT1 activation, such that heightening STAT1 activation could overcome the inhibitory effect of SUMO. Particle formation therefore was studied in STAT1-transfected fibroblasts deficient in the main STAT1 phosphatase TC45, which increases their IFNγ-induced STAT1 phosphorylation 2–3-fold (32), yet IFNγ stimulation did not induce paracrystals. However, paracrystals appeared when the cells received co-treatment with 1 mm H2O2, which led to diminished cellular SUMO conjugation without further increasing STAT1 phosphorylation (supplemental Fig. S4C). Thus, although blocking Tyr701 phosphorylation was sufficient for paracrystal inhibition by SUMO, we concluded that limiting the pool of activated STAT molecules was not an adequate explanation.
Sumoylation Promotes Semiphosphorylated STAT Dimers, Which Disperse Paracrystals
The preceding experiments indicated that SUMO, by preventing proximal Tyr phosphorylation, transformed unphosphorylated STAT1 from being inert to paracrystal growth to a potent inhibitor thereof. However, inhibition did not involve paracrystal incorporation of the SUMO-modified STAT1, and spurious amounts of it sufficed to keep STAT1 soluble. This conundrum can be resolved conclusively by a role for SUMO in generating a reversible inhibitor of STAT polymerization, namely semiphosphorylated dimers, because these could inhibit the elongation of pSTAT chains and hence antagonize paracrystals. Fig. 5A shows a schematic depicting this process. Semiphosphorylated dimers have been observed previously, but their structure and physiological role have remained unknown (33). We prepared unphosphorylated and Tyr701-phosphorylated full-length STAT1α recombinant protein, the C-terminally truncated STAT1 splice variant STAT1β (both unsumoylated and ∼20% SUMO-conjugated), and the experimental mutant STAT1tc, which is additionally lacking the N domain. These proteins were mixed pairwise, and their association was examined by Western blotting after an antibody pull-down assay. An extensive description of the controls and results can be found in supplemental Fig. S5. A summary of the results is shown in Fig. 5B. The assay confirmed the reported association of Tyr701-phosphorylated STAT1 and uSTAT1. However, their interaction was lost when the N domain, which is indispensable for antiparallel dimerization, was deleted from one of the partners. It was therefore concluded that semiphosphorylated dimers adopt antiparallel conformation because their single potential SH2-phosphotyrosyl interaction was insufficient to sustain stable parallel dimers. The assay further indicated that the sumoylation of unphosphorylated STAT1 did not preclude its interaction with phosphorylated STAT1. These experiments therefore supported a model whereby SUMO conjugation is compatible with stable semiphosphorylated STAT1 dimers, which adopt antiparallel conformation (see Fig. 5C for a model).
Next, we examined whether sumoylation regulated the abundance of semiphosphorylated STAT1 dimers in cells. STAT1 gene fusion with SUMO ligase Ubc9 made the experiment feasible by boosting STAT1 sumoylation as described (24). Mutation E705Q generated SUMO-free STAT1-Ubc9 (Fig. 5D and supplemental Fig. S6A). Interferon-γ-inducible nuclear accumulation and SUMO-sensitive particle formation indicated functionality of the Ubc9 fusions (supplemental Fig. S6B). We first determined the specific Tyr701 phosphorylation (pSTAT1/STAT1) in the cell extracts (termed Input), which confirmed increased phosphorylation of SUMO-free STAT1 (Fig. 5D, left). We then used an antibody with strongly reduced affinity toward activated STAT1 (34) to immunoprecipitate specifically unphosphorylated uSTAT1 from these extracts. The amount of co-precipitating phosphorylated pSTAT1 was determined by quantitative Western blotting, which served as a measure for the abundance of semiphosphorylated dimers. Control immunoprecipitations with an antibody that did not discriminate uSTAT1 and pSTAT1 were done in parallel. To normalize the different activation levels, data are presented as phosphorylation ratio, which is defined as the ratio of the specific STAT1 phosphorylation in the precipitates to the specific STAT1 phosphorylation in the corresponding extracts (IP/Input). The results are shown in Fig. 5D (right). The phosphorylation-indifferent control antibody (α-STAT1) generated nearly identical phosphorylation ratios for WT (10.0 ± 0.6) and SUMO-free STAT1 (10.7 ± 0.2) because the precipitation of pSTAT1 expectedly was proportional only to the abundance of pSTAT1 in the extracts. In contrast, the outcome was different if specifically unphosphorylated STAT1 was precipitated (α-uSTAT1). Now, the phosphorylation ratio of wild type (0.64 ± 0.03) was significantly higher compared with SUMO-free STAT1 (0.45 ± 0.03). Thus, although fewer pSTAT1 molecules were present in extracts where STAT1 was SUMO-conjugated (i.e. wild type), a significantly higher proportion of the pSTAT1 was associated with uSTAT1. We therefore concluded that SUMO conjugation increased the abundance of semiphosphorylated dimers.
Our model holds that the role of SUMO in paracrystal suppression is limited to the generation of semiphosphorylated dimers. Accordingly, non-phosphorylatable STAT1 too should have paracrystal-suppressing activity. Its inhibitory activity should be SUMO-independent, whereas the ability to associate with phosphorylatable STAT1 in order to assemble semiphosphorylated dimers should be indispensable. These predictions were tested in living cells with co-expression experiments of SUMO-free STAT1 and the non-phosphorylatable SH2 domain mutant R602L. The variants were expressed as fusions with different fluorescent proteins to allow their discrimination. The SH2 mutation prevents the docking to cytokine receptors, which not only leaves the protein permanently unphosphorylated at Tyr701 but also prevents it from interfering with the activation of co-expressed STAT1 (35). After confirming these previously demonstrated properties of the SH2 mutant (not shown), the co-expression experiments were done. In controls, IFNγ stimulation triggered paracrystals in ∼45% of cells expressing SUMO-free STAT1 (Fig. 5E). The presence of increasing concentrations of non-phosphorylatable STAT1, however, gradually reduced their prevalence to about 20%. Notably, the paracrystal-suppressing activity of the SH2 mutant was not reduced by its lack of sumoylation (E705Q) (Fig. 5E), which confirmed that the contribution of SUMO to paracrystal suppression was limited to preventing STAT phosphorylation. Participation of non-phosphorylatable STAT1 in antiparallel dimerization, in contrast, was indispensable because paracrystal inhibition was entirely lost when its N-domain interactions were weakened (F77A) (Fig. 5E). In strong support of this reasoning, co-expression of the isolated STAT1 N domain but not the F77A mutant sufficed for paracrystal suppression too (supplemental Fig. S6D). In summary, these experiments indicated that antiparallel dimerization with pSTAT1 bestowed crystal-inhibiting activity on uSTAT1. Moreover, the data agree with semiphosphorylated dimers being reversible inhibitors of STAT polymerization and paracrystal assembly.
SUMO-mediated Suppression of Paracrystals Accelerates STAT Dephosphorylation
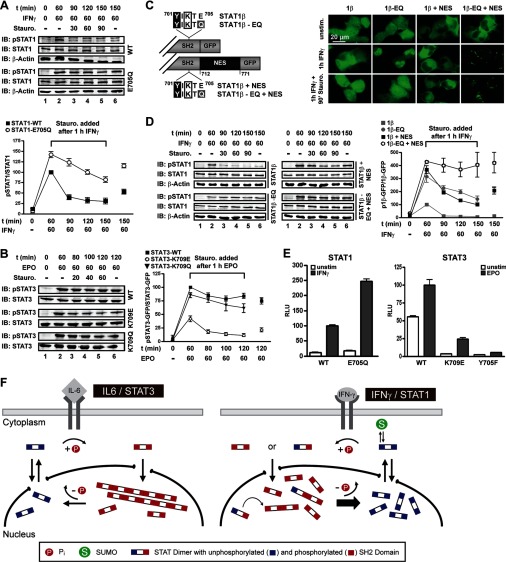
We then assessed the physiological impact of paracrystals and their solubilization by sumoylation. Because paracrystal formation was associated with elevated STAT Tyr phosphorylation, we probed the activation/inactivation kinetics using human fibrosarcoma cells stably reconstituted with wild type or SUMO-free STAT1. The cells were treated with IFNγ for 60 min followed by a pulse-chase with the kinase inhibitor staurosporine to block continued Tyr701 phosphorylation (7). Immunoblotting of cell extracts showed the expected elevated Tyr701 phosphorylation of the mutant STAT1 (Fig. 6A). However, the increase resulted not solely from enhanced activation because kinase inhibition revealed the markedly reduced dephosphorylation of SUMO-free STAT1 (Fig. 6A). These results were confirmed for STAT3. The naturally SUMO-free wild type STAT3 and the K709Q mutant were both more strongly activated and resistant to dephosphorylation compared with the K709E with reconstituted SUMO consensus and hence forced SUMO conjugation (Fig. 6B). Therefore, we asked whether reduced dephosphorylation of the SUMO-free STATs was consequential to the lack of sumoylation per se or rather to their reduced solubility. For these experiments, we took advantage of the STAT1 splice variant β (Fig. 6C), whose protracted nuclear export precludes efficient reactivation (36), as evident both in incomplete nuclear accumulation (Fig. 6C) and weak Tyr701 phosphorylation (Fig. 6D). Sumoylation deficiency (E705Q) increased Tyr701 phosphorylation of STAT1β (Fig. 6D), yet activation levels still did not sustain discernible paracrystals (Fig. 6C). We have previously shown that fusion of a nuclear export signal (+NES) to enhance nucleocytoplasmic shuttling causes increased Tyr701 phosphorylation of STAT1β in IFNγ-treated cells (36). In addition, the steady-state localization of STAT1β + NES is in the cytoplasm before and after cytokine stimulation due to its enhanced nuclear export (Fig. 6C). However, when mutation E705Q prevented the SUMO modification of STAT1β + NES, the protein readily formed paracrystals in IFNγ-stimulated cells, albeit in the cytoplasm (Fig. 6C). We then performed staurosporine pulse-chase experiments followed by immunoblotting of cell extracts to compare the inactivation kinetics of SUMO-free STAT1β in the absence and the presence of paracrystals. These experiments demonstrated efficient dephosphorylation of the non-paracrystal-forming variants, irrespective of NES fusion or mutation E705Q (Fig. 6D). In stark contrast, when enhanced export and lack of SUMO modification (i.e. STAT1β-EQ + NES) facilitated the assembly of paracrystals, dephosphorylation was considerably reduced (Fig. 6D). We therefore propose that incorporation into paracrystals protected activated STATs from dephosphorylation, probably by excluding phosphatase access. The persistence of paracrystals in cells treated with kinase inhibitor staurosporine, which rapidly depleted the pool of soluble pSTAT1, supported this conclusion (Fig. 6C). Finally, we used STAT1- or STAT3-dependent luciferase-reporter gene assays to examine cytokine-induced gene activation. As is shown in Fig. 6E, the reduced dephosphorylation associated with paracrystal formation of SUMO-free STATs (i.e. wild type STAT3 and STAT1 mutant E705Q) resulted in strongly enhanced gene activation in response to IFNγ or erythropoietin, respectively. Of note, as observed previously, low level constitutive activation of STAT3 resulted in considerable transcriptional activity already before cytokine stimulation of PC3 cells (37), which was absent for SUMO-conjugated STAT3. This indicated that the direct suppressive effect of sumoylation on STAT phosphorylation can affect transcription activation also in the absence of paracrystals. Together these results demonstrated that increased phosphorylation and reduced dephosphorylation of SUMO-free STAT transcription factors enhanced cytokine-inducible gene expression.
FIGURE 6.
Localization in paracrystals protects STATs from dephosphorylation. A, STAT1 dephosphorylation kinetics in U3A cells reconstituted with WT or SUMO-free STAT1 revealed by treatment with IFNγ and tyrosine kinase inhibitor staurosporine. Top, representative example of immunoblotting (IB) experiments with Tyr(P)701-STAT1-, STAT1-, and β-actin-specific antibodies. Bottom, diagram showing the specific Tyr701 phosphorylation of SUMO-free and wild type STAT1 (value after 1 h of IFNγ was set as 100). Data display the mean ± S.D. obtained from three independent immunoblotting experiments. B, STAT3 dephosphorylation kinetics in Epo-stimulated PC3 cells reconstituted with STAT3, STAT3-K709Q, or sumoylatable STAT3-K709E. The experiment was done as described in A using anti-Tyr(P)705-STAT3- and anti-STAT3-specific antibodies. Specific Tyr705 phosphorylation was determined in two (K709Q) or six (WT and K709E) independent experiments. C, top, diagram depicting STAT1β variants used in C and D. Bottom, fluorescence micrographs of U3A cells reconstituted with the indicated STAT1β variants. D, corresponding dephosphorylation kinetics of STAT1β variant proteins. The experiment was done as described in A. For each STAT1β variant, specific Tyr701 phosphorylation depicted in the diagram was determined in three independent experiments. E, left, IFNγ-induced STAT1 reporter gene activity in U3A cells reconstituted with the indicated STAT1 variants. Right, Epo-induced STAT3 reporter gene activity in PC3 cells reconstituted with the indicated STAT3 variants. Non-phosphorylatable STAT3 (Y705F) was used as a negative control. Shown are normalized luciferase activities after 6 h of cytokine treatment (mean ± S.D. (error bars), n = 6–18). F, model of paracrystal formation of STAT3 during IL-6 signaling and the impact of SUMO conjugation on the intranuclear distribution and the tyrosine dephosphorylation of STAT1 during IFNγ signaling. For further details, see “Discussion.”
DISCUSSION
Herein we describe polymerization of STAT dimers as a mechanism for sequestering activated STATs in dynamic paracrystalline arrays. We propose that this discovery is of importance for many if not all STAT family transcription factors (Fig. 6F). We further describe how SUMO modification facilitates STAT1 inactivation and paracrystal dissolution by generating competitive polymerization inhibitors, which suggests a general mechanism for SUMO-mediated regulation of protein solubility (Fig. 6F).
Our experiments were facilitated by the use of a previously untested mutation to inactivate the SUMO consensus. We found that mutation of the SUMO consensus resulted in SUMO-free STAT1; however, in agreement with earlier studies, the mutants differed considerably in regard to their tyrosine phosphorylation (11, 12). Because the residues in question lie in a critical location of the STAT1 phosphorylation site and dimerization interface, we aimed to minimize the possible impact of SUMO consensus mutations on the structure of STAT1. Therefore, residue Glu705 in position Tyr+4, which is surface-exposed and not part of the dimerization interface (6), was altered in the structurally most conservative manner, namely Glu → Gln. We found no indication that this mutation altered the dimerization or DNA binding of STAT1. Moreover, the conservative side chain alteration resulted in increased STAT1 activation, as would be expected from exposing the proximal tyrosine 701 phosphorylation site. In contrast, with structurally more disruptive mutations, this SUMO-dependent effect was outweighed by their additional SUMO-independent constraints on STAT1 activation.
The use of this improved reagent has led to the discovery of STAT paracrystals. These structures are steady-state systems that are maintained by a constant flux of activated STAT dimers, suggesting that their formation is governed by the principle of self-organization (38). Paracrystalline protein associations are known to occur with hemoglobin S in sickle cells (26) and viral proteins during virion assembly (39); however, to our knowledge, this is the first report to implicate such structures in cellular signal processing. STAT paracrystal assembly involves mutual N domain and Tyr(P)-SH2 domain interactions that both are of very high affinity, with dissociation constants of ∼50 nm (28). Importantly, N and SH2 domains each separately sustain STAT1 dimerization at physiological concentrations, which affirms the model presented here. However, high affinity SH2-mediated interactions require Tyr phosphorylation, which drives polymerization and explains the cytokine dependence of STAT paracrystals. In addition to the findings reported here, nuclear particles have been observed also for activated STAT24 and STAT5 (40). Because the interaction surfaces required for dimerization and polymerization are conserved in the STAT family, we propose that paracrystals are common in cytokine signaling. STAT1, however, is the notable exception due to its SUMO conjugation. Although our data demonstrated that polymerization of activated STAT proteins is indispensable for paracrystal growth, they do not formally rule out the participation of additional proteins or components. However, aside from specific requirements concerning the structure and assembly of STAT dimers, there appears to be only one further need for paracrystal formation, i.e. a minimum threshold concentration of tyrosine-phosphorylated STATs. When these requirements were met, cytokine stimulation triggered paracrystals, whose location was not subcellular compartment-specific and whose size correlated with STAT expression levels, which makes the requirement for additional constituents rather improbable.
The experiments furthermore revealed how SUMO prevents STAT1 from assembling paracrystals. A characteristic of SUMO modification is that the biological consequences of conjugation do not appear proportionate to the small fraction of substrate that is modified. The global increase in STAT1 solubility therefore is a showcase example for what has been dubbed by Hay the “SUMO enigma” (41). Here we provide an explanation that is based on SUMO being a bulky obstacle that precludes phosphorylation of the proximal tyrosine 701 residue. Our data suggest that SUMO interferes in the very moment that STAT1 is phosphorylated at the membrane-bound cytokine receptors, thus generating a protein with different properties from a protein that has never been SUMO-modified, namely a semiphosphorylated STAT1 dimer. This reasoning agrees with SUMO being a cytoplasmic modifier (42) and with current structural ideas about STAT activation, which state that STATs approach cytokine receptors as dimers, such that the two protomers are Tyr-phosphorylated at the same time (4, 5). Importantly, high steady state sumoylation levels would defy the purpose because only semisumoylated dimers will generate semiphosphorylated ones. Thus, in order to prevent doubly sumoylated STAT1 dimers, which simply remain unphosphorylated, rapid SUMO turnover and hence very low steady-state concentrations of SUMO-modified STAT1 are mandatory. On the other hand, given their function as competitive polymerization inhibitors, low concentrations of semiphosphorylated STAT1 dimers suffice to keep STAT1 soluble. STAT1 dimers, in stark contrast to the exceedingly short lived SUMO conjugation, are persistent units with slow dissociation rates (t½ = 20–40 min), a prerequisite to minimize the exchange of activated and unphosphorylated protomers (28). Whether SUMO modification of STAT1 is subject to regulation is unknown, but it appears to occur constitutively in many cell types (10). It is noteworthy, therefore, that large assemblies of activated STAT2, STAT3, and STAT1 are found in the nucleus of mouse oocytes and early embryos (43).
From the work on STAT1, two simple generally applicable principles of SUMO-mediated solubility regulation can be deduced. First, we propose that proteins whose interactions involve polymerization of stable dimeric subunits are predisposed to regulation by SUMO. Second, SUMO is required to prevent the activation/inactivation of a polymerization interface of just one of the dimer subunits, thus generating a competitive polymerization inhibitor. The polymerization and assembly of cytoplasmic intermediate filaments might be another example where these principles apply; recent results suggest that their assembly is negatively regulated by a minimal SUMO-modified fraction of the cytoplasmic intermediate filament protein B-1A (44).
SUMO conjugation diminished the activity of STAT1 by two additive mechanisms, namely reducing STAT1 activation due to its direct interference with phosphorylation of the proximal tyrosine and enhanced inactivation due to the solubilization of paracrystals, which were identified as reservoirs for the phosphorylated STAT proteins. Thus, contrary to other transcription factors, the inhibition of STAT1 by SUMO does not appear to involve the modulation of interactions at gene promoters, i.e. recruitment of nuclear co-repressors, such as histone deacetylases (45), because the Tyr-phosphorylated and hence DNA binding-competent STAT1 remained SUMO-free. Nonetheless, curtailed activation resulted in diminished STAT1- or STAT3-dependent gene transcription in response to cytokine treatment of cells. Moreover, experiments with macrophages derived from knock-in mice expressing SUMO-free STAT1 demonstrated hyperresponsiveness to IFNγ, namely strongly increased expression of inducible nitric oxide (NO) synthase and release of NO.5 Further work is required to fully explore the importance of STAT1 paracrystal dispersion for IFNγ and cytokine signaling. One attractive possibility considers the well known but functionally elusive heterodimers of STAT1 (46), which in conjunction with SUMO may interfere with the polymerization of STAT family members. This, in turn, could provide a structural basis for the cross-regulation of multiple cytokine signaling pathways by IFNγ.
Supplementary Material
Acknowledgments
We thank Dr. D. Lorenz, M. Ringling, F. Wolfram (Leibniz-Institut für Molekulare Pharmakologie, Berlin), Dr. S. Anderson (Nottingham University), and Dr. J. Skepper (Cambridge University) for electron microscopy advice and experiments; Dr. S. Diebold (King's College London) for animal experimentation; Drs. A. Bennett and E. Gottschalg (Nottingham University FRAME Laboratory) for providing human liver cells; and Drs. T. Hirano, F. Horn, F. Melchior, P. O'Hare, and M. Tremblay for sharing reagents. We thank Drs. J. E. Darnell and G. R. Stark for critical reading of the manuscript and many helpful suggestions and D. E. Vinkemeier also for text editing. Expert technical assistance was provided by C. Pelzel.
This work was supported by Deutsche Forschungsgemeinschaft (DFG;KFO 192) Grants VI 218/4 and 218/5 (to U. V.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S6, Tables S1 and S2, and Experimental Procedures.
U. Vinkemeier and R. Oldenbourg, unpublished observations.
M. Droescher and U. Vinkemeier, unpublished observations.
A. Begitt, M. Droescher, K. P. Knobeloch, and U. Vinkemeier, submitted for publication.
- SH2
- Src homology 2
- uSTAT
- unphosphorylated STAT1
- pSTAT
- phosphorylated STAT
- SUMO
- small ubiquitin-like modifier.
REFERENCES
- 1. Wang Y., Levy D. E. (2006) FASEB J. 20, 1641–1652 [DOI] [PubMed] [Google Scholar]
- 2. Levy D. E., Darnell J. E., Jr. (2002) Nat. Rev. Mol. Cell Biol. 3, 651–662 [DOI] [PubMed] [Google Scholar]
- 3. Meyer T., Marg A., Lemke P., Wiesner B., Vinkemeier U. (2003) Genes Dev. 17, 1992–2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mao X., Ren Z., Parker G. N., Sondermann H., Pastorello M. A., Wang W., McMurray J. S., Demeler B., Darnell J. E., Jr., Chen X. (2005) Mol. Cell 17, 761–771 [DOI] [PubMed] [Google Scholar]
- 5. Ota N., Brett T. J., Murphy T. L., Fremont D. H., Murphy K. M. (2004) Nat. Immunol. 5, 208–215 [DOI] [PubMed] [Google Scholar]
- 6. Chen X., Vinkemeier U., Zhao Y., Jeruzalmi D., Darnell J. E., Jr., Kuriyan J. (1998) Cell 93, 827–839 [DOI] [PubMed] [Google Scholar]
- 7. Haspel R. L., Salditt-Georgieff M., Darnell J. E., Jr. (1996) EMBO J. 15, 6262–6268 [PMC free article] [PubMed] [Google Scholar]
- 8. Herrmann A., Sommer U., Pranada A. L., Giese B., Küster A., Haan S., Becker W., Heinrich P. C., Müller-Newen G. (2004) J. Cell Sci. 117, 339–349 [DOI] [PubMed] [Google Scholar]
- 9. Lillemeier B. F., Köster M., Kerr I. M. (2001) EMBO J. 20, 2508–2517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rogers R. S., Horvath C. M., Matunis M. J. (2003) J. Biol. Chem. 278, 30091–30097 [DOI] [PubMed] [Google Scholar]
- 11. Song L., Bhattacharya S., Yunus A. A., Lima C. D., Schindler C. (2006) Blood 108, 3237–3244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ungureanu D., Vanhatupa S., Kotaja N., Yang J., Aittomaki S., Jänne O. A., Palvimo J. J., Silvennoinen O. (2003) Blood 102, 3311–3313 [DOI] [PubMed] [Google Scholar]
- 13. Ungureanu D., Vanhatupa S., Grönholm J., Palvimo J. J., Silvennoinen O. (2005) Blood 106, 224–226 [DOI] [PubMed] [Google Scholar]
- 14. Jakobs A., Koehnke J., Himstedt F., Funk M., Korn B., Gaestel M., Niedenthal R. (2007) Nat. Methods 4, 245–250 [DOI] [PubMed] [Google Scholar]
- 15. Meyer T., Begitt A., Lödige I., van Rossum M., Vinkemeier U. (2002) EMBO J. 21, 344–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Begitt A., Meyer T., van Rossum M., Vinkemeier U. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 10418–10423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Matsui T., Kinoshita T., Hirano T., Yokota T., Miyajima A. (2002) J. Biol. Chem. 277, 36167–36173 [DOI] [PubMed] [Google Scholar]
- 18. Lorenz D., Krylov A., Hahm D., Hagen V., Rosenthal W., Pohl P., Maric K. (2003) EMBO Rep. 4, 88–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Guex N., Peitsch M. C. (1997) Electrophoresis 18, 2714–2723 [DOI] [PubMed] [Google Scholar]
- 20. Case D. A., Cheatham T. E., 3rd, Darden T., Gohlke H., Luo R., Merz K. M., Jr., Onufriev A., Simmerling C., Wang B., Woods R. J. (2005) J. Comput. Chem. 26, 1668–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bartlett N. W., Dumoutier L., Renauld J. C., Kotenko S. V., McVey C. E., Lee H. J., Smith G. L. (2004) J. Gen. Virol. 85, 1401–1412 [DOI] [PubMed] [Google Scholar]
- 22. Ray S., Boldogh I., Brasier A. R. (2005) Gastroenterology 129, 1616–1632 [DOI] [PubMed] [Google Scholar]
- 23. Tonozuka Y., Minoshima Y., Bao Y. C., Moon Y., Tsubono Y., Hatori T., Nakajima H., Nosaka T., Kawashima T., Kitamura T. (2004) Blood 104, 3550–3557 [DOI] [PubMed] [Google Scholar]
- 24. Zimnik S., Gaestel M., Niedenthal R. (2009) Nucleic Acids Res. 37, e30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Vinkemeier U., Cohen S. L., Moarefi I., Chait B. T., Kuriyan J., Darnell J. E., Jr. (1996) EMBO J. 15, 5616–5626 [PMC free article] [PubMed] [Google Scholar]
- 26. Eaton W. A., Hofrichter J. (1990) Adv. Protein Chem. 40, 63–279 [DOI] [PubMed] [Google Scholar]
- 27. Schust J., Sperl B., Hollis A., Mayer T. U., Berg T. (2006) Chem. Biol. 13, 1235–1242 [DOI] [PubMed] [Google Scholar]
- 28. Wenta N., Strauss H., Meyer S., Vinkemeier U. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 9238–9243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mertens C., Zhong M., Krishnaraj R., Zou W., Chen X., Darnell J. E., Jr. (2006) Genes Dev. 20, 3372–3381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Soler-Lopez M., Petosa C., Fukuzawa M., Ravelli R., Williams J. G., Müller C. W. (2004) Mol. Cell 13, 791–804 [DOI] [PubMed] [Google Scholar]
- 31. Bossis G., Melchior F. (2006) Mol. Cell 21, 349–357 [DOI] [PubMed] [Google Scholar]
- 32. ten Hoeve J., de Jesus Ibarra-Sanchez M., Fu Y., Zhu W., Tremblay M., David M., Shuai K. (2002) Mol. Cell. Biol. 22, 5662–5668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gupta S., Yan H., Wong L. H., Ralph S., Krolewski J., Schindler C. (1996) EMBO J. 15, 1075–1084 [PMC free article] [PubMed] [Google Scholar]
- 34. Marg A., Meyer T., Vigneron M., Vinkemeier U. (2008) Cytometry A 73A, 1128–1140 [DOI] [PubMed] [Google Scholar]
- 35. Shuai K., Stark G. R., Kerr I. M., Darnell J. E., Jr. (1993) Science 261, 1744–1746 [DOI] [PubMed] [Google Scholar]
- 36. Lödige I., Marg A., Wiesner B., Malecová B., Oelgeschläger T., Vinkemeier U. (2005) J. Biol. Chem. 280, 43087–43099 [DOI] [PubMed] [Google Scholar]
- 37. Yuan Z. L., Guan Y. J., Chatterjee D., Chin Y. E. (2005) Science 307, 269–273 [DOI] [PubMed] [Google Scholar]
- 38. Misteli T. (2001) J. Cell Biol. 155, 181–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Roingeard P. (2008) Biol. Cell 100, 491–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Herrington J., Rui L., Luo G., Yu-Lee L. Y., Carter-Su C. (1999) J. Biol. Chem. 274, 5138–5145 [DOI] [PubMed] [Google Scholar]
- 41. Hay R. T. (2005) Mol. Cell 18, 1–12 [DOI] [PubMed] [Google Scholar]
- 42. Geiss-Friedlander R., Melchior F. (2007) Nat. Rev. Mol. Cell Biol. 8, 947–956 [DOI] [PubMed] [Google Scholar]
- 43. Truchet S., Chebrout M., Djediat C., Wietzerbin J., Debey P. (2004) Biol. Reprod. 71, 1330–1339 [DOI] [PubMed] [Google Scholar]
- 44. Kaminsky R., Denison C., Bening-Abu-Shach U., Chisholm A. D., Gygi S. P., Broday L. (2009) Dev. Cell 17, 724–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gill G. (2005) Curr. Opin. Genet. Dev. 15, 536–541 [DOI] [PubMed] [Google Scholar]
- 46. Platanias L. C. (2005) Nat. Rev. Immunol. 5, 375–386 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.