Abstract
Aberrant signaling of ErbB family members human epidermal growth factor 2 (HER2) and epidermal growth factor receptor (EGFR) is implicated in many human cancers, and HER2 expression is predictive of human disease recurrence and prognosis. Small molecule kinase inhibitors of EGFR and of both HER2 and EGFR have received approval for the treatment of cancer. We present the first high resolution crystal structure of the kinase domain of HER2 in complex with a selective inhibitor to understand protein activation, inhibition, and function at the molecular level. HER2 kinase domain crystallizes as a dimer and suggests evidence for an allosteric mechanism of activation comparable with previously reported activation mechanisms for EGFR and HER4. A unique Gly-rich region in HER2 following the α-helix C is responsible for increased conformational flexibility within the active site and could explain the low intrinsic catalytic activity previously reported for HER2. In addition, we solved the crystal structure of the kinase domain of EGFR in complex with a HER2/EGFR dual inhibitor (TAK-285). Comparison with previously reported inactive and active EGFR kinase domain structures gave insight into the mechanism of HER2 and EGFR inhibition and may help guide the design and development of new cancer drugs with improved potency and selectivity.
Keywords: Cancer Therapy, Crystal Structure, Protein-Drug Interactions, Receptor Tyrosine Kinase, X-ray Crystallography, EGFR, ErbB Family, HER2, Allosteric Activation
Introduction
Many solid tumors are characterized by aberrant signal transduction through activated receptor tyrosine kinases of the human ErbB protein family or epidermal growth factor receptor (EGFR)2 family (1). This family consists of EGFR (ErbB1), HER2 (ErbB2, HER2/neu), HER3 (ErbB3), and HER4 (ErbB4). The receptors play a role in the regulation of cell proliferation, differentiation, and migration (2). All receptors within the ErbB family contain a cytoplasmic tyrosine kinase region, and all except for HER2 bind to specific ligands through the extracellular domain (3, 4). In the case of HER3, the cytoplasmic kinase domain is generally thought of as a pseudokinase domain, although recent evidence suggests that there is low kinase activity. The catalytic activity is 1000-fold less than EGFR (5). Upon ligand binding to the extracellular domain, the receptors can make either homo- or heterodimers through which the cytoplasmic domains become catalytically active and undergo C terminus phosphorylation, which further initiates downstream signaling pathways (1, 6). HER2 appears to be the preferred binding partner of EGFR, and the HER2/EGFR heterodimer shows an increased signaling potency relative to EGFR homodimers (7). In contrast with most receptor tyrosine kinases, phosphorylation of the activation loop is not required for activation of the kinase, whereas the kinase is intrinsically autoinhibited in the cell. Crystallographic and mutagenesis studies recently provided evidence that EGFR and HER4 might be activated through an allosteric mechanism that occurs via dimer formation (8–10).
Several approaches have been taken in the therapeutic inhibition of EGF-stimulated signal transduction. Antibody-based drugs that compete with ligand binding to the extracellular domain of HER2 or EGFR as well as small molecule tyrosine kinase inhibitors that compete with ATP binding to the cytoplasmic tyrosine kinase domain of EGFR (OSI-774/erlotinib) or both HER2 and EGFR (GW572016/lapatinib) have received regulatory approval for the treatment of cancer. Clinical studies have shown that the antitumor effectiveness has improved for agents that target both receptors simultaneously when compared with selective agents (11, 12).
We report the first published crystal structure of the kinase domain of HER2, further referred to as HER2-KD. The HER2-KD structure is crystallized in complex with SYR127063 (13), a pyrrolo[3,2-d]pyrimidine-based potent and selective tyrosine kinase inhibitor of HER2. An allosteric mechanism of activation through dimer formation was observed for HER2, and the interaction of the monomers is comparable with previously reported activating dimers for EGFR and HER4 (8–10). Structural and sequence analysis also revealed a Gly-rich region unique to HER2 in the α-helix C-β4 loop immediately following the α-helix C. In the active kinase conformation of EGFR, Ser768 and Asp770 in this short loop form H-bond interactions with α-helix C, thereby stabilizing and positioning the α-helix in the correct orientation to enable catalysis. The Gly residues in HER2 prevent these interactions and could explain the low intrinsic catalytic activity that was previously reported for HER2 (14–16). In addition, we report the crystal structure of the complex of EGFR with TAK-285, a dual HER2/EGFR inhibitor with a pyrrolo[3,2-d]pyrimidine-based backbone (17). Structural comparison of HER2 and EGFR co-complex structures provides molecular insights into the basic inhibitor binding mode and enables the identification of key interactions to evaluate the potency and selectivity of ErbB inhibitors.
EXPERIMENTAL PROCEDURES
Chemical Synthesis
N-{2-[4-({3-Chloro-4-[3-(trifluoromethyl)phenoxy]phenyl}amino)-5H-pyrrolo[3,2-d]pyrimidin-5-yl]ethyl}-3-hydroxy-3-methylbutanamide (TAK-285) and 2-{2-[4-({5-chloro-6-[3-(trifluoromethyl)phenoxy]pyridin-3-yl}amino)-5H-pyrrolo[3,2-d]pyrimidin-5-yl]ethoxy}ethanol (SYR127063) were synthesized and purified according to methods described in the patents filed previously (13, 17).
Cloning, Expression, and Purification of HER2 and EGFR
For enzyme assays, DNA fragments encoding the human HER2 cytoplasmic domain (UniProtKB accession number P04626, amino acids 676–1255) and the human EGFR cytoplasmic domain (UniProtKB accession number P00533, amino acids 669–1210) with the N-terminal peptide (DYKDDDD) tag were subcloned into the pFastBac1 vector (Invitrogen). The constructs were transfected into Spodoptera frugiperda Sf9 cells, and the proteins were expressed using the Bac-to-Bac expression system. The expressed proteins were purified using anti-FLAG M2 affinity gel (Sigma-Aldrich). The human HER4 cytoplasmic domain with N-terminal hexahistidine tag was purchased from Upstate.
For structure determination of HER2, residues 703–1029 were amplified from cDNA by PCR and cloned into the pFastBac1 vector to acquire a C-terminal polyhistidine tag. Three N-terminal point mutations, M706A, Q711L, and M712L, were introduced into the HER2-KD. The three N-terminal mutations correspond to the equivalent residues in EGFR. Recombinant baculovirus incorporating the human HER2 kinase domain (residues 703–1029, M706A, Q711L, and M712L) was generated by transposition with the Bac-to-Bac system (Invitrogen), and high titer viral stocks were generated by infection of S. frugiperda Sf9 cells. Protein generated from this construct is further referred to as HER2-KD. Large scale production of recombinant protein was carried out in Sf9 cells utilizing 5-liter Wave Bioreactors (Wave Biotech). The human EGFR kinase domain (amino acids 696–1022) was expressed and purified as described previously (18) and is further referred to as the EGFR-KD. DNA encoding residues 696–1022 was amplified from full-length EGFR cDNA (UniProtKB accession number P00533) and cloned into the pFastBacHT vector (Invitrogen) to acquire the 6-histidine tag and a TEV protease cleavage site at the N terminus. The obtained recombinant transfer vector (Bac-to-Bac expression system, Invitrogen) was transfected into Sf9 cells to generate recombinant baculovirus. Large scale production of recombinant protein was carried out in Sf9 cells. Cells were harvested by centrifugation at 4000 × g and rapidly frozen for storage at −80 °C.
HER2-KD purification was carried out in which the cell pellet from a 5-liter Wave bag was suspended into lysis buffer consisting of 50 mm Tris-HCl (pH 7.9), 200 mm NaCl, 20 mm imidazole, 0.25 mm tris(2-carboxyethyl)phosphine hydrochloride, and protease inhibitor mixture (Complete EDTA-free, Roche Applied Science) and further lysed via Polytron for 2–4 min. The lysate was centrifuged at 4200 × g for 60 min, and clarified supernatant was batch-bound with 5 ml of ProBond nickel resin (Invitrogen). The resin slurry was washed with buffer containing 25 mm Tris-HCl (pH 7.9), 500 mm NaCl, 20 mm imidazole, and 2% glycerol, and then protein was eluted with buffer containing 200 mm NaCl and 200 mm imidazole. The sample was further purified by size exclusion chromatography utilizing an S3000 column equilibrated in 25 mm Tris-HCl (pH 7.9), 150 mm NaCl, and 2% glycerol. Collected fractions were then concentrated to 7–10 mg/ml utilizing YM10 Centricon (Millipore) and buffer-exchanged to the final buffer of 20 mm Tris-HCl (pH 7.9), 75 mm NaCl, 2 mm DTT, 2 mm benzamidine, and 2% glycerol.
EGFR-KD purification was performed through which frozen-thawed cells were resuspended in 200 ml of buffer (50 mm Tris-HCl (pH 8.0), 200 mm NaCl, 0.5 mm DTT, 10% glycerol, and protease inhibitor mixture (Complete EDTA-free, Roche Applied Science). The cells were homogenized using a Microfluidizer (M-110EH) at 15,000 p.s.i. (100 megapascals). The lysate was centrifuged at 10,000 × g for 30 min to remove insoluble material. The supernatant was batch-bound to 10 ml of nickel-nitrilotriacetic acid-agarose resin (Qiagen) for 2 h at 4 °C, and then the resin was packed into a column. The column was washed with 5 column volumes of a wash buffer (20 mm Tris-HCl (pH 8.0), 500 mm NaCl, and 10% glycerol) followed by the buffer containing 20 mm imidazole. The protein was eluted from the column with 10 × 1-column volume aliquots of an elution buffer (250 mm imidazole, 20 mm Tris-HCl (pH 8.0), 500 mm NaCl, and 10% glycerol). Fractions thought to contain the protein of interest were analyzed by SDS-PAGE, pooled according to purity, and concentrated to a volume of ∼10 ml by ultrafiltration. The concentrated solution was loaded onto a Superdex 200 gel filtration column (GE Healthcare) pre-equilibrated with 20 mm Tris-HCl (pH 8.0), 50 mm NaCl, 5 mm DTT, and 10% glycerol. Peak fractions were mixed with the AcTEV protease (Invitrogen) overnight at 4 °C to remove the N-terminal histidine tag and then loaded onto a Mono Q anion exchange column (GE Healthcare) pre-equilibrated in buffer containing 20 mm Tris-HCl (pH 8.0) and 5 mm DTT. The protein was eluted using a linear gradient from 50 to 300 mm NaCl containing 20 mm Tris-HCl (pH 8.0) and 5 mm DTT. Finally, the buffer was exchanged into a final buffer consisting of 20 mm Tris-HCl (pH 8.0) and 5 mm DTT using a HiPrep desalting column (GE Healthcare). The fractions containing the protein of interest were pooled, concentrated to 5–10 mg/ml by ultrafiltration, and flash frozen in liquid nitrogen for storage at −80 °C. The typical final yield was 5–10 mg for a 1-liter culture. Mass spectrometric analysis confirmed that the protein was non-phosphorylated protein.
EGFR, HER2, and HER4 Kinase Enzyme Assays
The EGFR, HER2, and HER4 kinase assays were performed using radiolabeled [γ-32P]ATP (GE Healthcare). The kinase reactions were performed in 50 mm Tris-HCl (pH 7.5), 5 mm MnCl2, 0.01% Tween 20, and 2 mm DTT containing 0.9 μCi of [γ-32P]ATP per reaction, 50 μm ATP, 5 μg/ml poly(Glu)-Tyr (4:1), and each purified cytoplasmic domain (0.25 μg/ml EGFR or 0.25 or 0.125 μg/ml of HER4) in a total volume of 50 μl. To measure the IC50 value for enzyme inhibition, the compounds were incubated with the enzyme for 5 min prior to the reaction at room temperature. The kinase reactions were initiated by adding ATP. After 10 min at room temperature, the reactions were stopped by the addition of 10% (final concentration) trichloroacetic acid. The γ-32P phosphorylated proteins were filtrated in a harvest plate (Millipore) with a cell harvester (PerkinElmer Life Sciences) and washed free of [γ-32P]ATP with 3% phosphoric acid. The plates were dried followed by the addition of 25 μl of MicroScint0 (PerkinElmer Life Sciences). The radioactivity was counted by a TopCount scintillation counter (PerkinElmer Life Sciences). IC50 values and 95% confidence intervals were calculated by nonlinear regression analysis of the percent inhibitions.
Crystallization and Structure Determination
The HER2-KD·SYR127063 complex was prepared by incubation of 7 mg/ml protein in the final buffer with 1 mm SYR127063 (in 100% DMSO) on ice for 1 h. Small crystals were obtained by sitting drop vapor diffusion against a precipitant of 20% PEG 3550, 200 mm diammonium tartrate, and 100 mm PIPES (pH 6.5) at 20 °C. From these initial crystals, seed stock was prepared using the Seed Bead kit (Hampton Research). Large crystals suitable for data collection were obtained by mixing 1 μl of protein to 1 μl of precipitant solution containing 15–20% PEG 3350, 150 mm sodium tartrate, 50 mm diammonium tartrate, and either 100 mm Bis-Tris or PIPES (pH 6.5–7.0). After equilibration of 1 day, microcrystals were seeded into the drop by addition of 0.1 μl of seed stock. For data collection, crystals were immersed in reservoir solution containing 25% ethylene glycol and flash frozen in liquid nitrogen. Diffraction data were collected from cryocooled crystals at the Advanced Light Source beamline 5.0.3. Data were reduced using the HKL2000 software package (19). The structure was determined by molecular replacement using the program MOLREP (20) of the CCP4 program suite utilizing EGFR (Protein Data Bank code 1M14) as a search model and refined with the program REFMAC (21). Several cycles of model building with XtalView (22) and refinement were performed for improving the quality of the model. Data reduction, refinement statistics, and quality of the structure are summarized in Table 1.
TABLE 1.
X-ray data collection and refinement statistics
Values in parentheses are for the highest resolution shell. r.m.s., root mean square; ALS, Advanced Light Source; AU, asymmetric unit.
| HER2·SYR127063 | EGFR·TAK-285 | |
|---|---|---|
| Data collection | ||
| X-ray source | ALS 5.0.3 | SPring-8 BL41XU |
| Wavelength (Å) | 1.0 | 1.0 |
| Space group | P212121 | P212121 |
| Unit cell dimensions (Å) | a = 48.7, b = 79.0, c = 152.7 | a = 46.8, b = 68.9, c = 104.6 |
| Resolution (Å) | 2.25 | 1.50 |
| Unique reflections | 28,773 | 53,697 |
| Redundancy | 3.9 | 5.0 |
| Completeness (%) | 99.5 (96.6) | 97.8 (99.2) |
| I/σ(I) | 15.1 (1.8) | 14.0 (2.4) |
| Rsyma | 0.080 (0.553) | 0.048 (0.583) |
| Refinement | ||
| No. molecules/AU | 2 | 1 |
| Reflections used | 27,014 | 50,707 |
| r.m.s. bonds (Å) | 0.010 | 0.010 |
| r.m.s. angles (°) | 1.270 | 1.240 |
| Average B value (Å2) | 31.6 | 25.7 |
| R valueb | 0.185 | 0.219 |
| Ramachandranc | ||
| Favored (%) | 545 (95.5) | 273 (96.5) |
| Allowed (%) | 23 (4.0) | 8 (2.8) |
| Outliers (%) | 3 (0.5) | 2 (0.7) |
a Rsym = ΣhΣj| I(h) − I(h)j|/ΣhΣj I(h) where I(h) is the mean intensity of symmetry-related reflections.
b R value = Σ‖Fobs| − |Fcalc‖/Σ|Fobs|. Rfree for 5% of reflections was excluded from refinement.
c As reported by Coot/Dynarama.
The EGFR·TAK-285 complex was prepared by incubation of the kinase domain of EGFR (5–10 mg/ml) in the final buffer with a 3-fold molar excess of TAK-285 (in 100% DMSO) on ice for 2–3 h. A cluster of small crystals was grown by the sitting drop vapor diffusion method from the reservoir containing 0.1 m Bis-Tris (pH 6.5), 0.2 m lithium sulfate, and 25% PEG 3350 at 20 °C. These crystals were used as seed crystals with the Seed Bead kit (Hampton Research). Large crystals were typically grown by mixing 1 μl of protein with 0.5 μl of reservoir solution containing 0.1 m Bis-Tris (pH 6.5), 0.2 m lithium sulfate, 15–20% PEG 3350 at 20 °C, and 0.5 μl of seed stock. Crystals appeared within 1 week. For data collection, crystals were quickly dipped in reservoir solution containing 20% glycerol and flash frozen with liquid nitrogen.
Diffraction data were collected at 100 K at SPring-8 beam line BL41XU on the charge-coupled device detector Quantum 315r (Area Detector Systems Corp.) and processed with CrystalClear and d*TREK (Rigaku). The data processing statistics and structure quality are summarized in Table 1. The high resolution limit of data used in refinement was determined based on a signal-to-noise criterion (I/σ(I)). The structure was solved by molecular replacement with AMoRe of the CCP4 program suites (23) using the inactive EGFR structure (Protein Data Bank code 1XKK) as the starting model. The structure was rebuilt using Coot (24) and refined at 1.5-Å resolution using REFMAC (21). The refinement statistics and model quality are summarized in Table 1. All figures were prepared using PyMOL (Schrödinger) and ALINE (25).
Computational Chemistry
Homology models were constructed with the computer program MOE (Chemical Computing Group). Molecular dynamics was carried out with the AMBER software package (26). Ligand heats of formation were calculated at the 6-31G* theory level with the GAMESS software (27).
RESULTS AND DISCUSSION
Overall HER2 and EGFR Kinase Domain Crystal Structures
The HER2-KD crystal structure in complex with a selective HER2 inhibitor, SYR127063, was solved to 2.25-Å resolution. The crystals have one dimer in the asymmetric unit, and both monomers are occupied by the inhibitor. The two molecules are nearly identical with a root mean square deviation of 0.72 Å for 283 Cα positions as reported by Coot/SSM Superpose (24). The structure of EGFR-KD in complex with a dual EGFR/HER2 inhibitor, TAK-285, was determined at 1.5-Å resolution with one molecule in the asymmetric unit. The HER2-KD·SYR127063 and EGFR-KD·TAK-285 structures are highly similar with a root mean square deviation of 1.1 Å for 247 equivalent Cα positions as reported by Coot/SSM Superpose (24).
All structures encompass a shortened form of the human cytoplasmic tyrosine kinase domain of the respective kinases with part of the C-terminal tail segment. For HER2, this construct includes residues 703–1029 as well as three N-terminal mutations, M706A, Q711L, and M712L, which are to the equivalent residues in EGFR. These mutations were made to increase expression and foster crystal formation of the HER2·SYR127063 complex. Similar to HER2, a truncated form of the kinase domain of EGFR encompassing amino acids 696–1022 was used for crystallization. The mutations and shortened boundaries used for structure determination of HER2-KD and EGFR-KD did not significantly change the inhibitory activity (IC50) of SYR127063 and TAK-285 when compared with the full cytoplasmic domains of the wild-type proteins in eight-point kinase assays.3
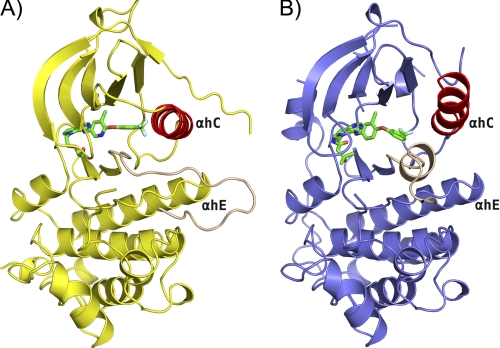
All structures adopt the typical kinase bilobed folding (Fig. 1). The N-terminal lobe (N-lobe) contains mostly β-strands and one α-helix, whereas the C-terminal lobe (C-lobe) is predominantly α-helical. The two lobes are connected by a flexible hinge region and separated by a deep cleft comprising the ATP binding site. The relative orientation of the two lobes has a distinct effect on the size of the ATP binding site, depending on the activation state of the kinase domain. Most of the residues associated with catalytic activity are located in the vicinity of the cleft, the glycine-rich nucleotide phosphate-binding loop (HER2: Leu726–Val734; EGFR: Leu718–Val726) and the α-helix C (αC; HER2: Pro761–Ala775; EGFR: Asn756–Ser768) of the N-lobe of the kinase; and the DFG motif (HER2: Asp863–Gly865; EGFR: Asp855–Gly857), the catalytic loop (HER2: Arg844–Asn850; EGFR: Arg836–Asn842), and the activation loop (A-loop; HER2: Asp863–Val884; EGFR: Asp855–Val876) of the C-lobe of the kinase. Polypeptide chain termini (HER2: Ser703–Ala705 and Glu1010–Gly1029; EGFR: Gly696–Asn700 and Ile1018–Gly1022) and several discrete regions of surface loops (HER2: Asp993–Ser998; EGFR: Glu734–Lys737, Arg748–Lys754, and Glu1004–Asp1009) are disordered and show poor electron density, and these residues are not included in the final model. The phosphate-binding loop (HER2: Gly729–Gly732; EGFR: Gly721–Gly724) is partially disordered, and the side chain of Phe731(HER2)/Phe723(EGFR) is solvent-exposed with conformational flexibility. A different conformation of Gly727–Ala730 in the monomers of the HER2/SYR127063 dimer was observed due to crystal packing. Part of the A-loop (HER2: Asp880–Gly881; EGFR: Glu868–Gly874) is also disordered in both the HER2·SYR127063 and EGFR·TAK-285 structures, respectively. In EGFR but not HER2, the N-terminal portion of the A-loop (Gly857–Gly863) forms a short α-helix and rotates the regulatory α-helix C by 45° away from the active site compared with the previously published active EGFR conformation (8, 28). In both EGFR and HER2, the salt bridge interaction between two highly conserved residues (HER2: Lys753 and Glu770; EGFR: Lys745 and Glu762), the canonical feature in the active conformation, is not observed. A part of the C-terminal tail segment (HER2: Pro999–Leu1009; EGFR: Asn992–Met1002) is traceable and forms a short α-helix adjacent to the hinge region with intermolecular contacts.
FIGURE 1.
Overall structure of HER2·SYR127063 and EGFR·TAK-285. Comparison of the overall structure of HER2 inhibited by SYR127063 (A) and of EGFR inhibited by TAK-285 (B). For HER2·SYR127063, the kinase domain is represented as a yellow schematic. For EGFR·TAK-285, the kinase domain is represented as a slate blue schematic. For both structures, α-helix C and the A-loop are colored red and tan, respectively. SYR127063 and TAK-285 are shown in a green stick representation colored by atom type. HER2 represents the active-like form of the kinase with the A-loop in an extended conformation. EGFR represents the inactive form of the kinase with a short α-helix present in the N-portion of the A-loop that stabilizes the active-site outward conformation of the α-helix C. αh, α-helix.
Allosteric Mechanism of HER2 Activation
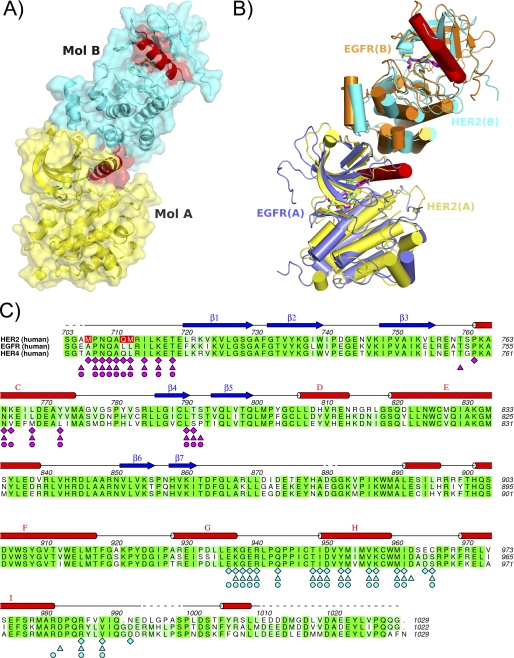
Recently, Zhang et al. (8, 9) and Qiu et al. (10) published evidence for an allosteric mechanism of activation of EGFR and HER4 that occurs through asymmetric dimer formation between two kinase domains (8–10). The interaction was originally identified through crystal packing interactions in structural studies and characterized by an extensive hydrophobic interface burying ∼2000 Å2 of surface area between the monomers. As a result of dimer formation, the conformation of the α-helix C of one monomer is restrained and correctly positioned to enable catalysis. Further evidence of the allosteric activation mechanism in EGFR and HER4 was obtained through mutagenesis studies on residues present at the dimer interface (8–10). The authors explained that the mechanism resembles a cyclin-like mechanism of kinase activation in which the engagement of α-helix C of the kinase through interaction with cyclin is crucial for activation (8). A similar dimer was observed in the crystal packing of the HER2 crystals. In the EGFR and HER4 activating dimers, one kinase domain in a pair is rotated by 120° about an axis relative to the other and translated by ∼45 Å with crystallographic symmetry. In the HER2 crystals, the monomers are related by a similar rotation and translation function in a non-crystallographic symmetry pair (Fig. 2). A hydrophobic interface burying 2054 Å2 of surface area was measured between the monomers (calculated by AREAIMOL of CCP4 (29)). Similar regions and residues within the monomers are involved in packing interactions in all ErbB activating dimers (Fig. 2C). This suggests that activation of HER2 might occur through an allosteric mechanism comparable with the one observed for EGFR and HER4. Mainly residues in the N terminus (Ala705–Thr718), α-helix C (Pro761–Ala775), and the loop between β-strand 4 and β-strand 5 (Leu790–Ser792) of one monomer interact with portions of α-helix G (Pro926–Lys937), the loop between α-helix G and α-helix H (Gly938–Pro945), α-helix H (Ile949–Trp959), and the loop between α-helix H and α-helix I (Met960–Cys965) of the other monomer. Several residues in the α-helix C (Lys765, Leu768, Asp769, and Tyr772) of one monomer (Fig. 2, A and B, Molecule A) make close, mainly hydrophobic, intermolecular interactions with the C-lobe of the kinase domain of the other HER2 monomer (Fig. 2, A and B, Molecule B). HER2 kinase dimerization and activation in vitro was recently reported using a nickel-chelating lipid-liposome system (30). In the EGFR and HER4 activating dimer crystal structures, both kinases were present in an active conformation (8–10). In HER2, both monomers are present in an active-like kinase conformation. Similar to previously published active EGFR structures and the activating EGFR dimer, the activation loop in HER2 is well ordered and does not form any secondary structure (8, 28). This is in contrast to previously published inactive EGFR structures where a short α-helix at the N-terminal end of the A-loop inserts into a hydrophobic pocket behind the α-helix C. However, in our HER2 structure, we call it an active-like kinase conformation because no salt bridge is formed between Lys753 in β-strand 3 and Glu770 in α-helix C, which is crucial for activation. This salt bridge is disrupted through the bulky 3-trifluoromethylphenyl of the inhibitor, which induces a shift of 20° in α-helix C away from the kinase active site.
FIGURE 2.
HER2 allosteric activating dimer. A, the HER2 dimer present in the asymmetric unit of the crystal is shown as a surface representation to depict the tight packing of the two monomers: Molecule (Mol.) A has a yellow surface, Molecule B has a cyan surface, and α-helix C is colored red in both molecules. The α-helix in Molecule A makes a close interaction with hydrophobic residues in the C-lobe of Molecule B. As a result of this interaction, the active conformation of the kinase is induced. B, superposition of HER2/SYR127063 dimer with EGFR/erlotinib dimer (Protein Data Bank code 1M17). The HER2/SYR127063 dimer is shown in yellow and cyan with SYR127063 as a magenta stick representation. The EGFR/erlotinib dimer is shown in slate blue and orange with erlotinib as a green stick representation. α-Helix C is shown in red for all molecules. C, sequence alignment between the kinase domains of HER2, EGFR, and HER4. Identical residues are highlighted in green boxes. Residues involved in close interactions at the dimer interface in HER2, EGFR, and HER4 derived from crystal structure information (Protein Data Bank codes 1M17 and 3BCE) are denoted by diamonds, stars, triangles, and circles, respectively, and are colored magenta for Molecule A and blue for Molecule B of the dimer. Mutations made in the HER2 construct are shown as red boxes. Secondary structure elements are assigned from the HER2·SYR127063 structure.
Origins of α-Helix C Conformational Flexibility in HER2
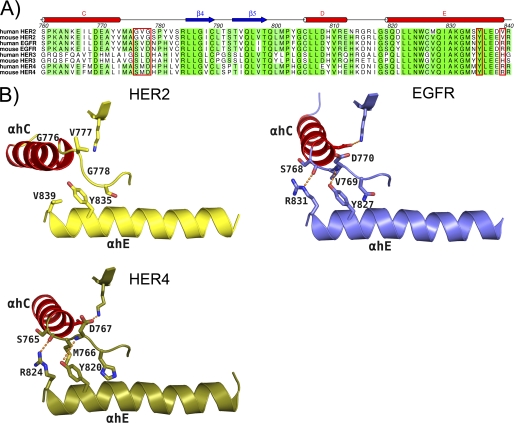
Superposition of all available crystal structures of the kinase domain of ErbB family members shows many unique positions of α-helix C, suggesting a clear conformational flexibility in α-helix C (data not shown). In this analysis, HER2 shows a particular high degree of flexibility in this helix compared with other family members. The position of α-helix C relative to the ATP binding site determines the activation state of the kinase. Comparison of the amino acid sequence between HER2 and other ErbB family members around the α-helix C and αC-β4-loop clearly reveals a non-conserved Gly-rich region in the αC-β4-loop of HER2 (Gly776–Ser779) (Fig. 3A). In our and previously published EGFR structures, we observed that the proper positioning of α-helix C for activation is highly controlled through the αC-β4-loop (768SVDN771) immediately following the α-helix C (30) (Fig. 3B). In the case of EGFR, α-helix C is rotated by ∼45° going from the inactive to active conformation through which the α-helix C is stabilized by hydrogen bond formation between the main-chain atoms of residues Ser768 and Asp770 of the αC-β4-loop and the side-chain atoms of Arg831 and Tyr827 of α-helix E in the C-lobe, respectively. Val769 sits in a hydrophobic pocket formed by Tyr827, Leu828, Arg831, Leu833, and Phe856 in the structure of EGFR. These hydrogen bonds are conserved in the structure of HER4 (10) (Fig. 3B). Gly776 and Gly778, which are unique to HER2, impart flexibility to the region so that α-helix C can adopt many positions of which a subset will be conducive to proper positioning of α-helix C for activation. The side chain of Tyr835 forms no hydrogen bond and is stacked to the main chain of Gly776 (Fig. 3B). The side chain of Val777 is exposed to solvent and totally disordered. These observations could explain the lower intrinsic kinase activity of HER2 when compared with EGFR (14–16). Fan et al. (16) confirmed the importance of both Gly776 and Gly778 in HER2 intrinsic kinase activity through mutagenesis studies. The authors showed that mutation of the two glycines to corresponding residues in EGFR resulted in a ∼10-fold increase in ATP binding, ∼7-fold increase in ATP turnover, and enhanced ligand-independent stimulation of downstream cell signaling. Moreover, genetic studies on cancer patients have shown increased HER2 catalytic activity as a result of mutations or insertion of hydrophobic residues (YVMA) before Gly776 (31–34). Presumably, this mutation would stabilize the loop region and therefore lead to a decrease in flexibility of α-helix C, but further studies need to be conducted to confirm this. Importantly, comparable mutations within this region have not been observed for EGFR. HER3 is unique among the other ErbB family members because it is generally considered as kinase-inactive (34). The recently published crystal structure of HER3 revealed a shortened α-helix C when compared with other family members and confirms the importance of this α-helix in the regulation of kinase activity (35, 36).
FIGURE 3.
Origins of α-helix C conformational flexibility in HER2. A, sequence alignment of the amino acids in the region between α-helix C and α-helix E of HER family members that contain a cytoplasmic tyrosine kinase region. A Gly-rich sequence unique to HER2 in a region immediately following the α-helix C was observed. Residues labeled in B are shown in the red boxes. B, the αC-β4-loop region plays an important role in the proper positioning of the α-helix C through interactions with α-helix E of the C-lobe of the respective kinase. HER2 is represented as a yellow schematic with important side chains rendered by atom type. EGFR (Protein Data Bank code 1M14) is represented as a slate blue schematic with important side chains rendered by atom type. HER4 (Protein Data Bank code 3BCE) is represented as an army green schematic with important side chains rendered by atom type. For all kinases, α-helix C is represented as a red schematic. In both EGFR and HER4, conserved interactions are observed, but in HER2, conserved interactions are not observed. The unique Gly-rich sequence in this region in HER2 imparts an increased flexibility of the α-helix C. αh, α-helix.
Tumor cells usually overexpress several ErbB family receptors with HER2 as a preferred partner during receptor dimerization (1). In particular, HER2 is the preferred partner of EGFR, and the HER2/EGFR heterodimer exhibits an increased rate of recycling, stability, and signaling potency compared with EGFR homodimers (7). Our structural data suggest that due to the increased flexibility in the α-helix C of HER2 and its less stable active conformation when compared with EGFR HER2 might predominantly function as a binding partner to activate and potentiate the activity of EGFR within the EGFR/HER2 heterodimer. This hypothesis supports the generally accepted theory that HER2 overexpression potentiates EGFR signaling (12, 37, 38). HER2 overexpression in EGFR-positive non-small cell lung cancer patients is associated with an increased response to gefitinib, a specific inhibitor of active EGFR but not of HER2 or inactive EGFR. Moreover, clinical studies have shown that the level of expression of only EGFR does not predict the sensitivity to gefitinib (39). This also explains why the most advanced and effective tyrosine kinase inhibitors in the clinic either target selectively active EGFR such as erlotinib and gefitinib or both EGFR and HER2 such as lapatinib but not selectively the kinase domain of HER2.
Binding Mode of SYR127063 and TAK-285 in HER2 and EGFR
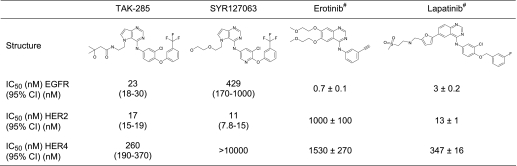
The enzyme inhibitory activity of the cytoplasmic domain of ErbB kinases was evaluated by phosphorylation of a peptide substrate (Table 2). TAK-285 and SYR127063 are low nanomolar pyrrolo[3,2-d]pyrimidine-based inhibitors of both HER2 and EGFR (Table 2). The in vitro potency of TAK-285 against HER2 and EGFR is similar. In ATP competition assays, both compounds were shown to bind to the respective kinase in an ATP-competitive manner.3 Consistent with these data, the crystal structures of both HER2·SYR127063 and EGFR·TAK-285 show that the inhibitors bind to the ATP site in a competitive manner (Fig. 4, A and B, and supplemental Fig. S1). Crystallographic data show that TAK-285 binds to the inactive conformation of EGFR (Fig. 4B) with a DFG-in and α-helix C-out conformation. One of the pyrrolo[3,2-d]pyrimidine ring nitrogen atoms makes a direct hydrogen bond with the main-chain nitrogen of Met793(EGFR) of the hinge region between the N- and C-lobes (supplemental Fig. S1B). The other ring nitrogen atom makes a water-mediated hydrogen bond to Thr854 (supplemental Fig. S1B). The bulky 3-trifluoromethylphenyl group occupies a pocket formed by Met766, Cys775, Leu777, Leu788, Thr790, Thr854, and Phe856 (Fig. 4A and supplemental Fig. S1B). The hydroxymethylbutanamide moiety on the other end of the molecule is exposed to the solvent (supplemental Fig. S1B), and its electron density is partly disordered. The amide oxygen does not make any direct hydrogen bond interactions with the protein.
TABLE 2.
In vitro enzymatic IC50 values of ErbB inhibitors
IC50 values and 95% confidence intervals (95% CI) were calculated by nonlinear regression analysis.
# Ki(app) (nm); previously reported values (40).
FIGURE 4.
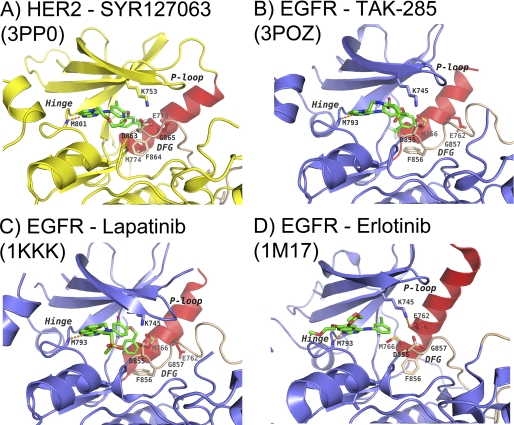
HER2 and EGFR mechanism of inhibition. Binding modes of SYR127063 in HER2 (A), TAK-285 in EGFR (B), lapatinib in EGFR (Protein Data Bank code 1XKK) (C), and erlotinib in EGFR (Protein Data Bank code 1M17) (D) are shown. The protein backbone in HER2 is represented as a yellow schematic, whereas in EGFR, it is represented as a slate blue schematic. The DFG motif and part of the activation loop are colored tan, and the α-helix C is colored red for all proteins. Key protein side chains are shown as sticks and colored according to atom type. Inhibitors SYR127063, TAK-285, lapatinib, and erlotinib are shown as green sticks and colored according to atom type. Hydrogen bonds are indicated as orange dashed lines. The α-helix C position and the A-loop conformation of the EGFR·TAK-285 complex remarkably resemble those of the EGFR·lapatinib complex. The binding mode of SYR127063 in HER2 differs from TAK-285 in EGFR through the different substituent appended to the pyrrolo[3,2-d]pyrimidine N5 atom and the conformation of the 3-trifluoromethylphenyl group. The A-loop conformation of the HER2·SYR127063 complex is similar to that of the active EGFR·erlotinib complex. P-loop, phosphate-binding loop.
In the HER2·SYR127063 structure, the compound binds to the active-like conformation of HER2. Similar interactions between the central region of SYR127063 and the hinge were observed as described for EGFR·TAK-285. These include both of the hydrogen bonds between the central pyrrolo[3,2-d]pyrimidine ring and Met801 and Thr862 as well as the hydrophobic interactions in the adenine site (Fig. 4, A and B, and supplemental Fig. S1A). The terminal trifluoromethylphenyl group is rotated ∼150° from its position in the EGFR·TAK-285 complex and packs against α-helix C, making hydrophobic contact with the side chains of Thr862, Glu770, Met774, Ser783, Leu785, Leu790, Leu796, and Phe864 (supplemental Fig. S1A).
Comparison of SYR127063 and TAK-285 Binding Mode with Erlotinib and Lapatinib
We compared our HER2 and EGFR crystal structures with previously published structures of EGFR in complex with lapatinib (Protein Data Bank code 1XKK) (Fig. 4C) and erlotinib (Protein Data Bank code 1M17) (Fig. 4D) to understand the molecular origins of inhibitor specificity for either HER2 or EGFR. Lapatinib (Tykerb/Tyverb) is a dual HER2/EGFR inhibitor and has been approved as front line therapy in triple positive breast cancer and as an adjuvant therapy in patients with HER2-overexpressing breast cancer (40). Erlotinib (Tarceva) is a selective inhibitor of EGFR and has been approved to treat non-small cell lung cancer, pancreatic cancer, and several other types of cancer (41, 42). TAK-285 and lapatinib, both dual EGFR/HER2 kinase inhibitors, bind to the inactive conformation of EGFR and show a similar binding mode in the active site. The activation loop forms a short α-helix that displaces the α-helix C from the active site. The 3-trifluoromethylphenoxy group of TAK-285 occupies roughly the same space as the 3-fluorobenzyloxy group of lapatinib (Fig. 4). SYR127063 binds to HER2 in an active-like conformation, and erlotinib binds to the active conformation of EGFR. In both structures, the activation loop is well ordered and shows no secondary structure at the N-terminal end. Both active and inactive EGFR pockets are larger than the pocket observed in active HER2. However, the active and inactive EGFR pockets have regions where one is narrower than the other. In the HER2·SYR127063 structure, the α-helix C is slightly rotated outward (∼20° compared with active EGFR) due to the bulky trifluoromethyl group of the compound. It is unclear whether EGFR can adopt an active conformation similar to that seen in our HER2 structure. The fluorophenyl of lapatinib is smaller than the trifluoromethylphenyl and so may be able to bind active-like HER2 in the same conformation as it does in EGFR. The pocket of HER2 in this region is smaller due to the position of Ser783 toward the trifluoromethylphenyl, and this may impact its binding to HER2 relative to EGFR (hence 4-fold weaker inhibitory activity but still very potent).
In the active EGFR·erlotinib structure, Met742 and Glu738 occupy the area that the trifluoromethylphenyl group of SYR127063 occupies in the HER2 structure. Given the very close sequence identity in the ATP site between HER2 and EGFR, it is interesting to note the selectivity profiles of TAK-285 and SYR127063: the former is a potent and non-selective inhibitor of HER2/EGFR, and the latter is a potent inhibitor of HER2 with 40-fold less potency for EGFR. We believe the explanation lies in how their trifluoromethylphenyl groups are oriented with respect to the Ser783(HER2)/Cys775(EGFR) and the low energy conformers of both compounds (Fig. 4, A and B, and supplemental Fig. S1). The key difference is the presence of the central pyridine ring nitrogen in SYR127063. TAK-285 can adopt the trifluoromethylphenyl conformation observed in the HER2·SYR127063 complex as it represents an energy minimum. The 3-trifluoromethylphenyl group of SYR127063 is flipped by ∼150° relative to the orientation of the same substituent in the EGFR·TAK-285 structure (Fig. 4B). One possible structural explanation for this change in ligand conformation is the presence of Ser783(HER2) compared with Cys775(EGFR): the side chain of Ser783 is hydrogen-bonded to Thr860, which precludes the EGFR·TAK-285 binding mode in HER2. Because Cys775 cannot hydrogen bond to the threonine, the active site of EGFR is larger, and the trifluoromethylphenyl of TAK-285 can fit into this region. However, SYR127063 cannot easily adopt the trifluoromethylphenyl conformation observed with TAK-285 in EGFR and suffers a penalty in excess of 20 kcal/mol to do so. Hence, because TAK-285 can adopt both conformations, it is a dual inhibitor, whereas SYR127063 suffers a 40-fold loss against EGFR due to the conformational penalty.
Interestingly, the HER2 T798I mutation (T790M in EGFR) has been shown to confer lapatinib resistance in a Ba/F3 cell-based assay (43) and is also associated with clinical drug resistance. A study done by Trowe et al. (43) suggested that lapatinib resistance in HER2 arose from replacing a smaller polar Thr with a bulkier Ile residue, thus causing steric clashes with lapatinib. To examine this and to determine the relevance to SYR127063 binding in HER2, Thr798 was manually mutated to Ile in the model building program Coot (24). A number of most favored side-chain rotamers were examined for steric clashes with SYR127063. Certain rotamers caused a steric clash, whereas others did not. Additional experiments are necessary to determine whether the mutant will confer resistance to the inhibitor.
In conclusion, we report the first three-dimensional crystal structure of HER2 in complex with a selective HER2 inhibitor, SYR127063. In addition, we report the crystal structure of EGFR in complex with TAK-285. TAK-285 is a potent dual inhibitor of EGFR and HER2 and is currently in Phase Ib clinical development for cancer. The HER2 structure gave insight into an allosteric mechanism of activation and gave molecular details on the determinants for selective inhibition. In addition, the structure gave structural evidence for the unique low intrinsic catalytic activity of HER2. Structural comparisons with other known EGFR kinase structures provided molecular details of the mechanism of activation and inhibition of HER2 and EGFR that are essential to design and develop new cancer drugs with improved potency and specificity profiles.
Supplementary Material
Acknowledgments
The authors thank Keith Wilson, Paul Steed, and David Weitz for critical review of this manuscript. We also thank Advanced Light Source, which is supported by the Director, Office of Science, Office of Basic Energy Sciences, Materials Sciences Division of the United States Department of Energy under Contract DE-AC03-76SF00098 at Lawrence Berkeley National Laboratory. The authors thank the office staff of Pharmaceutical Consortium for Protein Structure Analysis and the technical staff of the BL41XU at SPring-8 for facilitating synchrotron data collection. The authors also thank the TAK-285 project team of Millennium Pharmaceuticals and the senior management member of Pharmaceutical Research Division for consultation and research support.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
The atomic coordinates and structure factors (codes 3PP0 and 3POZ) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
K. Aertgeerts, R. Skene, J. Yano, B.-C. Sang, H. Zou, G. Snell, A. Jennings, K. Iwamoto, N. Habuka, A. Hirokawa, T. Ishikawa, T. Tanaka, H. Miki, Y. Ohta, and S. Sogabe, unpublished data.
- EGFR
- epidermal growth factor receptor
- KD
- kinase domain
- TEV
- tobacco etch virus
- Bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- N-lobe
- N-terminal lobe
- C-lobe
- C-terminal lobe
- A-loop
- activation loop
- αC
- α-helix C
- HER2
- human epidermal growth factor 2.
REFERENCES
- 1. Yarden Y., Sliwkowski M. X. (2001) Nat. Rev. Mol. Cell Biol. 2, 127–137 [DOI] [PubMed] [Google Scholar]
- 2. Schlessinger J. (2004) Science 306, 1506–1507 [DOI] [PubMed] [Google Scholar]
- 3. Carter P., Presta L., Gorman C. M., Ridgway J. B., Henner D., Wong W. L., Rowland A. M., Kotts C., Carver M. E., Shepard H. M. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 4285–4289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Slamon D. J., Godolphin W., Jones L. A., Holt J. A., Wong S. G., Keith D. E., Levin W. J., Stuart S. G., Udove J., Ullrich A., Press M. F. (1989) Science 244, 707–712 [DOI] [PubMed] [Google Scholar]
- 5. Shi F., Telesco S. E., Liu Y., Radhakrishnan R., Lemmon M. A. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 7692–7697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Qian X., LeVea C. M., Freeman J. K., Dougall W. C., Greene M. I. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 1500–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lenferink A. E., Pinkas-Kramarski R., van de Poll M. L., van Vugt M. J., Klapper L. N., Tzahar E., Waterman H., Sela M., van Zoelen E. J., Yarden Y. (1998) EMBO J. 17, 3385–3397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhang X., Gureasko J., Shen K., Cole P. A., Kuriyan J. (2006) Cell 125, 1137–1149 [DOI] [PubMed] [Google Scholar]
- 9. Zhang X., Pickin K. A., Bose R., Jura N., Cole P. A., Kuriyan J. (2007) Nature 450, 741–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Qiu C., Tarrant M. K., Choi S. H., Sathyamurthy A., Bose R., Banjade S., Pal A., Bornmann W. G., Lemmon M. A., Cole P. A., Leahy D. J. (2008) Structure 16, 460–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wada T., Myers J. N., Kokai Y., Brown V. I., Hamuro J., LeVea C. M., Greene M. I. (1990) Oncogene 5, 489–495 [PubMed] [Google Scholar]
- 12. Ye D., Mendelsohn J., Fan Z. (1999) Oncogene 18, 731–738 [DOI] [PubMed] [Google Scholar]
- 13. Ishikawa T., Miwa K., Seto M., Banno H., Kawakita Y. (June 7, 2007) World Intellectual Property Organization Patent WO/2007/064045
- 14. Cohen B. D., Kiener P. A., Green J. M., Foy L., Fell H. P., Zhang K. (1996) J. Biol. Chem. 271, 30897–30903 [DOI] [PubMed] [Google Scholar]
- 15. Brignola P. S., Lackey K., Kadwell S. H., Hoffman C., Horne E., Carter H. L., Stuart J. D., Blackburn K., Moyer M. B., Alligood K. J., Knight W. B., Wood E. R. (2002) J. Biol. Chem. 277, 1576–1585 [DOI] [PubMed] [Google Scholar]
- 16. Fan Y. X., Wong L., Ding J., Spiridonov N. A., Johnson R. C., Johnson G. R. (2008) J. Biol. Chem. 283, 1588–1596 [DOI] [PubMed] [Google Scholar]
- 17. Ishikawa T., Taniguchi T., Banno H., Seto M. (December 15, 2005) World Intellectual Property Organization Patent WO/2005/118588
- 18. Stamos J., Sliwkowski M. X., Eigenbrot C. (2002) J. Biol. Chem. 277, 46265–46272 [DOI] [PubMed] [Google Scholar]
- 19. Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 20. Vagin A., Teplyakov A. (1997) J. Appl. Crystallogr. 30, 1022–1025 [Google Scholar]
- 21. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 22. McRee D. E. (1999) J. Struct. Biol. 125, 156–165 [DOI] [PubMed] [Google Scholar]
- 23. Navaza J. (1994) Acta Crystallogr. A 50, 157–163 [Google Scholar]
- 24. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bond C. S., Schüttelkopf A. W. (2009) Acta Crystallogr. D Biol. Crystallogr. 65, 510–512 [DOI] [PubMed] [Google Scholar]
- 26. Case D. A., Darden T. A., Cheatham T. E, III, Simmerling C. L., Wang J., Duke R. E., Luo R., Walker R. C., Zhang W., Merz K. M., Roberts B., Wang B., Hayik S., Roitberg A., Seabra G., Kolossváry I., Wong K. F., Paesani F., Vanicek J., Liu J., Brozell S. R., Steinbrecher T., Cai Q., Ye X., Cui G., Roe D. R., Seetin M. G., Sagui C., Luchko T., Gusarov S., Kollman P. A. (2010) AMBER 11, University of California, San Francisco [Google Scholar]
- 27. Schmidt M. W., Baldridge K. K., Boatz J. A., Elbert S. T., Gordon M. S., Jensen J. H., Koseki S., Matsunaga N., Nguyen K. A., Su S. J., Windus T. L., Dupuis M., Montgomery J. A. (1993) J. Comput. Chem. 14, 1347–1363 [Google Scholar]
- 28. Yun C. H., Boggon T. J., Li Y., Woo M. S., Greulich H., Meyerson M., Eck M. J. (2007) Cancer Cell 11, 217–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Collaborative Computational Project, Number 4 (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–76315299374 [Google Scholar]
- 30. Monsey J., Shen W., Schlesinger P., Bose R. (2010) J. Biol. Chem. 285, 7035–7044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Stephens P., Hunter C., Bignell G., Edkins S., Davies H., Teague J., Stevens C., O'Meara S., Smith R., Parker A., Barthorpe A., Blow M., Brackenbury L., Butler A., Clarke O., Cole J., Dicks E., Dike A., Drozd A., Edwards K., Forbes S., Foster R., Gray K., Greenman C., Halliday K., Hills K., Kosmidou V., Lugg R., Menzies A., Perry J., Petty R., Raine K., Ratford L., Shepherd R., Small A., Stephens Y., Tofts C., Varian J., West S., Widaa S., Yates A., Brasseur F., Cooper C. S., Flanagan A. M., Knowles M., Leung S. Y., Louis D. N., Looijenga L. H., Malkowicz B., Pierotti M. A., Teh B., Chenevix-Trench G., Weber B. L., Yuen S. T., Harris G., Goldstraw P., Nicholson A. G., Futreal P. A., Wooster R., Stratton M. R. (2004) Nature 431, 525–526 [DOI] [PubMed] [Google Scholar]
- 32. Wang S. E., Narasanna A., Perez-Torres M., Xiang B., Wu F. Y., Yang S., Carpenter G., Gazdar A. F., Muthuswamy S. K., Arteaga C. L. (2006) Cancer Cell 10, 25–38 [DOI] [PubMed] [Google Scholar]
- 33. Shigematsu H., Takahashi T., Nomura M., Majmudar K., Suzuki M., Lee H., Wistuba I. I., Fong K. M., Toyooka S., Shimizu N., Fujisawa T., Minna J. D., Gazdar A. F. (2005) Cancer Res. 65, 1642–1646 [DOI] [PubMed] [Google Scholar]
- 34. Lee J. W., Soung Y. H., Seo S. H., Kim S. Y., Park C. H., Wang Y. P., Park K., Nam S. W., Park W. S., Kim S. H., Lee J. Y., Yoo N. J., Lee S. H. (2006) Clin. Cancer Res. 12, 57–61 [DOI] [PubMed] [Google Scholar]
- 35. Citri A., Skaria K. B., Yarden Y. (2003) Exp. Cell Res. 284, 54–65 [DOI] [PubMed] [Google Scholar]
- 36. Jura N., Shan Y., Cao X., Shaw D. E., Kuriyan J. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 21608–21613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Worthylake R., Opresko L. K., Wiley H. S. (1999) J. Biol. Chem. 274, 8865–8874 [DOI] [PubMed] [Google Scholar]
- 38. Emlet D. R., Schwartz R., Brown K. A., Pollice A. A., Smith C. A., Shackney S. E. (2006) Br. J. Cancer 94, 1144–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cappuzzo F., Varella-Garcia M., Shigematsu H., Domenichini I., Bartolini S., Ceresoli G. L., Rossi E., Ludovini V., Gregorc V., Toschi L., Franklin W. A., Crino L., Gazdar A. F., Bunn P. A., Jr., Hirsch F. R. (2005) J. Clin. Oncol. 23, 5007–5018 [DOI] [PubMed] [Google Scholar]
- 40. Higa G. M., Abraham J. (2007) Expert Rev. Anticancer Ther. 7, 1183–1192 [DOI] [PubMed] [Google Scholar]
- 41. Schultz R. M. (2005) Advances in Targeted Cancer Therapy. Progress in Drug Research, Binkhäuser, Basel, Switzerland [Google Scholar]
- 42. Shepherd F. A., Rodrigues Pereira J., Ciuleanu T., Tan E. H., Hirsh V., Thongprasert S., Campos D., Maoleekoonpiroj S., Smylie M., Martins R., van Kooten M., Dediu M., Findlay B., Tu D., Johnston D., Bezjak A., Clark G., Santabárbara P., Seymour L. (2005) N. Engl. J. Med. 353, 123–132 [DOI] [PubMed] [Google Scholar]
- 43. Trowe T., Boukouvala S., Calkins K., Cutler R. E., Jr., Fong R., Funke R., Gendreau S. B., Kim Y. D., Miller N., Woolfrey J. R., Vysotskaia V., Yang J. P., Gerritsen M. E., Matthews D. J., Lamb P., Heuer T. S. (2008) Clin. Cancer Res. 14, 2465–2475 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.