Abstract
RcsF, a proposed auxiliary regulator of the regulation of capsule synthesis (rcs) phosphorelay system, is a key element for understanding the RcsC-D-A/B signaling cascade, which is responsible for the regulation of more than 100 genes and is involved in cell division, motility, biofilm formation, and virulence. The RcsC-D-A/B system is one of the most complex bacterial signal transduction pathways, consisting of several membrane-bound and soluble proteins. RcsF is a lipoprotein attached to the outer membrane and plays an important role in activating the RcsC-d-A/B pathway. The exact mechanism of activation of the rcs phosphorelay by RcsF, however, remains unknown. We have analyzed the sequence of RcsF and identified three structural elements: 1) an N-terminal membrane-anchored helix (residues 3–13), 2) a loop (residues 14–48), and 3) a C-terminal folded domain (residues 49–134). We have determined the structure of this C-terminal domain and started to investigate its interaction with potential partners. Important features of its structure are two disulfide bridges between Cys-74 and Cys-118 and between Cys-109 and Cys-124. To evaluate the importance of this RcsF disulfide bridge network in vivo, we have examined the ability of the full-length protein and of specific Cys mutants to initiate the rcs signaling cascade. The results indicate that the Cys-74/Cys-118 and the Cys-109/Cys-124 residues correlate pairwise with the activity of RcsF. Interaction studies showed a weak interaction with an RNA hairpin. However, no interaction could be detected with reagents that are believed to activate the rcs phosphorelay, such as lysozyme, glucose, or Zn2+ ions.
Keywords: Bacterial Signal Transduction, Histidine Kinases, Lipoprotein Structure, Metabolic Regulation, NMR, NMR Solution Structure, Capsule Synthesis, Disulphide Bridges, Outer Membrane Protein RcsF, Two-component System
Introduction
The regulation of capsule synthesis (rcs)2 system is a central regulatory network in enteric bacteria that is activated by external signals allowing rapid adjustment to the cellular environment. Received information is transmitted via the rcs multicomponent signaling cascade inside the cell, resulting in activation or deactivation of a variety of physiological pathways such as exopolysaccharide (EPS) biosynthesis, cell motility, antibiotic resistance, virulence, and many others (1–4).
The core of the rcs system is represented by the response regulator RcsB and the membrane-bound sensor kinase RcsC, both classical members of conventional bacterial two-component systems (5). RcsB is a DNA binding protein that can be activated via an N-terminal phosphoreceiver domain. The histidine kinase RcsC is supposed to form a complex with RcsD (formerly named YojN), a second membrane-bound sensor (6, 7). RcsC as well as RcsD have similar architectures and are composed of a periplasmic sensor domain, a transmembrane-spanning unit, a histidine kinase domain, and a C-terminal phosphorylation domain. However, the histidine kinase domain of RcsD appears to be inactive, as conserved residues in the active site are missing (6, 8, 9).
The multistep signaling mechanism of the rcs system is based on an unusual His-Asp-His-Asp phosphorelay, where the interaction between individual components is modulated by their state of phosphorylation. The sensor kinase RcsC is first autophosphorylated in response to environmental stimuli at a conserved His residue in its histidine kinase domain. The phosphoryl group is then transferred to the C-terminal phosphoreceiver domain of RcsC and further transmitted via the C-terminal histidine phosphotransferase domain of RcsD to the N-terminal phosphoreceiver domain of RcsB. Activated RcsB binds to the RcsB box, a conserved sequence motif present in a number of rcs-dependent promoters (10). rcs-dependent promoters involved in other biosynthetic pathways such as EPS biosynthesis contain the RcsAB box motif, which is recognized by a heterodimer composed of RcsB and the transcriptional coregulator RcsA (11).
The structural analysis of individual components of the rcs system helped to understand molecular details in its signaling mechanisms. Liquid-state NMR structures of the RcsB DNA binding domain, the RcsC phosphoreceiver domain, and the RcsD histidine phosphotransferase domain have been solved (12–14). In addition, a new structural motif, called ABL domain, (α-β-Loop domain) connecting the histidine kinase domain with the C-terminal phosphoreceiver domain or the histidine phosphotransferase domain, has been identified in RcsC and RcsD, respectively (14, 15). On the basis of the available NMR structures, protein interaction interfaces within the rcs signaling cascade could be identified.
The complex functional diversity of the rcs regulation mechanisms requires a number of accessory regulator proteins to expand the levels of control and to increase the number of checkpoints (16). Besides the coregulator RcsA and the cosensor RcsD, the outer membrane lipoprotein RcsF has come into focus as a further potential coregulator of the rcs system (17, 18). RcsF has been detected in many enteric bacteria, but its primary structure is somehow unique, and no homologues outside of the RcsF family have been found so far. RcsF is important for bacterial biofilm formation and pathogenicity, and its function is speculated to be connected to RcsC or RcsD, probably via direct or indirect interactions. RcsF most likely acts upstream of RcsC and does not affect the expression levels of the rcsC, rcsB, or rcsA genes (17). Because of being located in the periplasm, disulfide bridge formation is discussed to be crucial for RcsF activity (19, 20). RcsF may act as a signal acceptor, and it is part of the cellular response to treatment with antimicrobial peptides, lysozyme, Zn2+ ions, or antibiotics (20–22).
Although a variety of data on RcsF has been accumulated, its functional or structural properties are still not clearly understood. We have approached the structural evaluation of the Escherichia coli RcsF protein to understand the basic principles of its action within the rcs system. We could identify three motifs within RcsF, and we have determined the structure of the C-terminal RcsF domain by liquid-state NMR analysis. We could further identify and correlate the folding and oxidation state of RcsF to its function in the regulation of bacterial capsule biosynthesis.
EXPERIMENTAL PROCEDURES
Strains, Plasmids, and DNA Manipulations
All cloning steps were performed following standard protocols (23). Polymerase chain reactions were carried out using Vent polymerase (New England Biolabs). The DNA fragments encoding the full-length RcsF protein (residues 1–134) and their fragments (RcsF-Δ16, RcsF-Δ30, and RcsF-Δ45) were amplified by using small synthetic oligonucleotides (Eurofins MWG Operon, Germany) as primers and chromosomal DNA of the E. coli strain XL1 (Stratagene) as a template. The DNA fragments were cloned with the restriction enzymes NcoI and BamHI into the expression vectors pET21d+, pET60m (Novagen), and pET60m_Ub, resulting in IPTG-inducible plasmids, including internal poly(His)6 tags (in the case of pET60m).
Protein Sample Preparation
For structural investigations, the corresponding constructs were transformed into the NEB T7 strain. Expression of uniformly 13C- and/or 15N-labeled proteins was achieved by growing the bacteria on M9 minimal medium containing 1 g/liter 15NH4Cl and 2.5 g/liter unlabeled or [U-13C]-d-glucose. Protein overproduction was induced with 1.0 mm IPTG. After 3 h of induction, the cells were harvested, resuspended in lysis buffer (50 mm Tris-HCl, 200 mm NaCl, 5% glycerol (pH 7.5)) and lysed. The proteins were loaded onto a Ni2+-chelate-Sepharose fast flow (Amersham Biosciences, GE Healthcare) column and eluted with a linear imidazole gradient (from 20 to 400 mm). Tobacco etch virus protease was added to the pure fused NusA- or Ub-RcsF-Δ30 protein at a ratio of 1:100 (w/w) in the presence of EDTA (2 mm) and β-mercaptoethanol (10 mm) to achieve ∼90% cleavage overnight at 18 °C. After cleavage, β-mercaptoethanol was removed by dialysis to facilitate the oxidation and disulfide bridge formation of RcsF-Δ30. Further purification and exchange into NMR buffer (50 mm Na2HPO4, 100 mm NaCl (pH 7.0)) was achieved by gel filtration with a Superdex75 column. Pure sample fractions were pooled and concentrated (0.2–0.7 mm). Usually, 15–20 mg of pure RcsF-Δ30 was obtained from 1 liter of culture. 5% 2H2O as a lock substance, 0.1 mm 4.4-dimethyl-4-silapentane-1-sulfonate as the internal proton chemical shift standard, and proteases inhibitor mixture (4.6 mm) were added to the samples. For titration experiments, the RNA hairpin G*G*CACUUCGGUGCC with phospho-thioester modification of the backbone phosphor (shown by asterisks) was used (representing a conserved protein interaction motif of the bacterial 16 S RNA).
NMR Spectroscopy and Structure Calculation
All NMR experiments necessary for resonance assignment and structure calculations were performed at 298 K on Bruker Avance spectrometers operating at 600, 700, 800, and 900 MHz proton frequencies. Proton chemical shifts were referenced relative to internal 4.4-dimethyl-4-silapentane-1-sulfonate. The 15N and 13C chemical shifts were referenced indirectly using the consensus ratios (24). Assignment of RcsF-Δ30 resonances was achieved following standard protocols (25). Proton-proton distances for structure calculations were derived from TROSY-based 15N- and 13C-edited three-dimensional NOESY spectra (mixing times 80 ms) and a 13C-edited 2D NOESY spectrum for aromatic ring protons (mixing time 120 ms). Prior to Fourier transformation, all time-domain data were zero-filled and multiplied by a 90°-shifted squared sine-bell window function in all dimensions. In vivo and post vivo experiments for overexpressed RcsF-Δ16, RcsF-Δ30, and RcsF-Δ45 constructs were performed at 308 K following published protocols (26, 27). Cells from 50 ml 15N-labeled M9 media were harvested by centrifugation at 2000 × g and resuspended in NMR buffer (50 mm Na2HPO4, 100 mm NaCl, 5% 2H2O (pH 7.5)). The NMR spectra were processed and analyzed using TOPSPIN 2.6 (Bruker BioSpin) and Sparky programs (28). The NOE-based distance restraints for RcsF-Δ30 were extracted from the 13C- and 15N-edited NOESY spectra using the CANDID module of the CYANA program (29), version 1.0.5. The final structure calculation was performed using a simulated annealing protocol with torsion angle dynamics (DYANA (30) version 1.5) on the basis of 1195 unambiguous upper-limit constraints generated by CANDID. 129 torsion angle restraints were included as predicted by the program TALOS on the basis of chemical shift values (31). In addition, 88 hydrogen bonds, deduced from the preliminary CANDID runs, were included as distance restraints (dH-O ≤ 2.1 Å and dN-O ≤ 3.1 Å) together with six upper and six lower distance restraints for two disulfide bonds (between Cys-74 and Cys-118, and Cys-109 and Cys-124). For the calculation of the final structural ensemble, 200 structures were calculated in 10,000 time steps per conformer. The 50 best DYANA conformers were refined in an explicit water shell (32) using the CNS software package (33), and 25 structures with the lowest energy (about −3500 kcal/mol) were selected for validation using PROCHECK-NMR 3.4 (34). The figures were prepared using the programs MOLMOL (35) and PyMOL (DeLano Scientific LLC). The atomic coordinates (PDB code 2L8Y) have been deposited in the Protein Data Bank.
The corresponding NMR assignments have been deposited in the Biological Magnetic Resonance Data Bank (accession number 17431).
EPS Quantification Anthrone Test
Cells were grown on sterile cellophane membranes (14-kDa cutoff) placed on lysogeny broth-agar plates. Cells were plated in five strokes per plate to provide sufficient nutrients for EPS synthesis. Prior to plating, 5 μl of a 100 mm IPTG solution was added to 100 μl of the cell suspension. The plates were incubated at 37° C for 12 h and after that at 20 °C for 24 h. Cells were harvested by suspension in 2 × 3 ml 0.85% NaCl. The cell suspensions were extensively vortexed for 5 min and centrifuged at 10,000 × g for 30 min. The supernatant was dialyzed against water (14-kDa cutoff). Samples of 100 μl were filled up with water to 1 ml volumes and added to 5 ml of anthrone reagent (200 mg anthrone in 100 ml concentrated H2SO4). The mixture was incubated at 100 °C for 10 min. The samples were chilled on ice, and the absorption at 625 nm was analyzed.
RESULTS
Sequence Analysis of RcsF and Secondary Structure Prediction
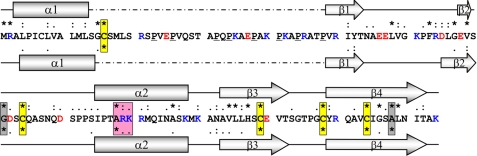
Alignment of all annotated RcsF sequences from UniProtKB revealed strong conservation of the protein within enterobacterial species (Fig. 1 and supplemental Fig. S1) and even outside of this bacterial family. By combining the observed homology pattern with secondary structure predictions (PsyPred server (36), we could identify three conserved motifs within RcsF. 1) The highly conserved N terminus-spanning amino acid positions 1 to 15 have a predicted α-helical structure and are proposed to form a membrane-anchored segment of RcsF. The helix is terminated by the conserved Cys-16 residue for which lipidation forming an N-acyl-diacylglycerylcysteine was suggested (18, 37). The proposed membrane-anchored helix has significant homology to other N-terminal helices in various periplasmic proteins, such as iron transporters (C6WPA8_ACTMD), proteins involved in thiamin biosynthesis (Q5LLK1_SILPO), and nitrite reductases (D5V9Q4_MORCR). There is no direct evidence for the transmembrane nature of this N-terminal helix which is, with 15 amino acids, too short for a normal transmembrane helix. However, it has been shown that this segment is necessary for periplasmatic localization and correct processing of the protein (18, 20). 2) The proline and positive charge-rich sequence from amino acid positions 17 to 48 is not conserved among RcsF sequences and is predicted to form an unstructured region. 3) The C-terminal region from Val-49 to Lys-134 is again conserved, and secondary structure prediction indicates the presence of a well folded domain.
FIGURE 1.
Sequence alignment and secondary structure prediction for E. coli RcsF. The amino acid sequence of E. coli RcsF (P69411) is shown (positively charged amino acids are shown in blue, negatively charged amino acids in red, and prolines are underlined). The top row of symbols represents conservation within Enterica sequences, and the bottom row represents total conservation between all annotated RcsF sequences. The secondary structure elements predicted by the PsiPred server for E. coli RcsF (P69411) and the most distant annotated RcsF sequence from Aeromonas hydrophilia (A0KMN2) are presented above and below the E. coli sequence, respectively. The boxes emphasize conserved residues (yellow, cysteines; red, positive charged cluster; gray, others).
Overall, the alignment identified only few absolutely conserved amino acids: the small amino acids Gly-71, Ala-88, and Ala-128 as well as Cys-16, Cys-74, Cys-109, Cys-118, and Cys-124. The only non-conserved cysteine, Cys-07, is located in the membrane-anchored helix and is absent in RcsF sequences of organisms more distantly related to Escherichia (supplemental Fig. S1).
Overexpression of RcsF Derivatives for Structural and Functional Studies
On the basis of this analysis of the sequence, several RcsF expression constructs were designed for structural and functional studies (supplemental Table S1). Full-length RcsF as well as its derivatives were expressed either alone or N-terminally fused to a modified ubiquitin (Ub-RcsF), cleavable by TEV protease. In addition, RcsF-Δ16 lacking the predicted membrane-anchored helix as well as RcsF-Δ30 and RcsF-Δ45 with partial or complete deletions of the proposed unstructured loop region were expressed (supplemental Fig. S2). Ub-RcsF as well as its mutants expressed mostly as inclusion bodies, although a fraction of approximately 15% stayed soluble (data not shown). However, the limited solubility of all full-length RcsF expression constructs combined with their tendency to aggregate prevented structural characterization of the complete protein.
Therefore, we focused on the structure of the periplasmic part of RcsF. The derivatives Ub-RcsF-Δ30 and NusA-RcsF-Δ30 were expressed with high efficiencies with yields of approximately 100–200 mg/liter culture. The proteins were soluble, and the Ub or NusA moieties could be released from the purified fusion proteins by TEV cleavage in batch reactions under reducing conditions. After cleavage, β-mercaptoethanol was removed from the cleavage reaction by dialysis to facilitate the oxidation and disulfide bridge formation of RcsF-Δ30. Subsequent purification of RcsF-Δ30 by size exclusion chromatography revealed that between 10 and 20% of the protein behaved like a monomeric and well folded protein, whereas the rest was strongly aggregated. Overall, 10–15 mg of folded and monomeric RcsF-Δ30 protein could be obtained from 1 liter of culture. The purified RcsF-Δ30 protein was highly stable and could be concentrated up to 1 mm.
NMR Solution Structure of RcsF-Δ30
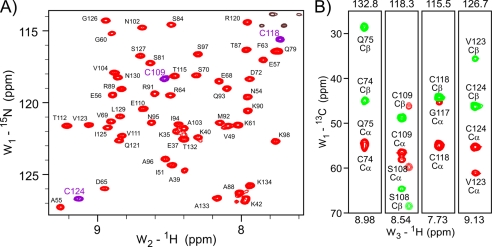
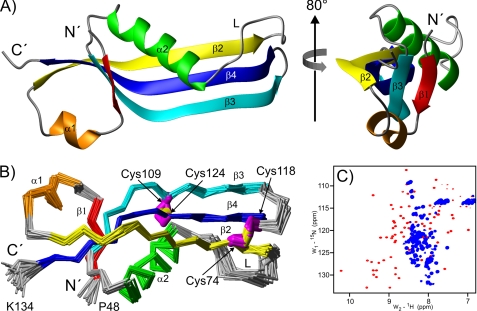
The [15N,1H]HSQC-TROSY spectrum of monomeric oxidized RcsF-Δ30 indicated that the protein is folded and exists in only one conformation (Fig. 2A). We determined the NMR solution structure based on 1195 NOEs and 129 torsion angle restraints obtained from chemical shifts as well as 88 hydrogen bonds. Analysis of the spectra revealed that the well structured part comprises residues Val-49 to Lys-134, whereas the N terminus (residues 30–47) shows characteristics of an unstructured segment. The structure of the folded core domain of RcsF-Δ30 (Fig. 3, structural statistics in supplemental Table S2) is formed by a central β-sheet surrounded by two α-helices, a smaller helix α1 (residues Ala-55 to Val-59) and a larger helix α2 (residues Ile-85 to Ser-97). The β-sheet consists of three major antiparallel β-strands, β2 (residues Phe-63 to Gln-75), β3 (residues Ala-103 to Thr-115) and β4 (residues Cys-118 to Ile-131) (Fig. 3A). Strand β1 (residues Arg-50 to Tyr-52), which is in parallel orientation relative to β3, is significantly shorter than β3 and forms hydrogen bonds with only ∼1/3 of the residues of β3. The two minor structural elements, strand β1 and helix α1, are somewhat distant from the core of the RcsF domain, forming a small N-terminal subdomain. The β-sheet core builds an inner surface contacted on one side by helix α2. Although helix α2 covers only approximately half of the length of the β-sheet, the long loop L (residues Ala-76 to Ser-84), which connects β2 and α2, is significantly ordered and covers the remaining inner surface of the β-sheet core.
FIGURE 2.
NMR spectra of RcsF-Δ30 oxidized monomeric form. A, representative area of the RcsF-Δ30 [15N,1H]HSQC-TROSY spectrum. The backbone HN resonances (generally red, HN resonances of cysteines, purple) are almost completely assigned. The non-assigned black contours in the upper right corner belong to Asn/Gln side chains. B, HNCACB strips demonstrating oxidation of all four Δ30 RcsF cysteines. The HN resonance of Cys-74 is not seen in the RcsF-Δ30 TROSY-HSQC spectrum. The positive Cα resonances are shown in red and the negative Cβ resonances in green. Values of >40 ppm for cysteine Cβ resonances indicate an oxidized state.
FIGURE 3.
Structure of the RcsF-Δ30 oxidized monomeric form. A, ribbon diagram representing the RcsF-Δ30 structure. The structure on the right was obtained by 80° rotation around the indicated axis. The unstructured residues 30–47 are not included. B, Cα traces of the final RcsF-Δ30 structural ensemble of 25 conformers. The indicated cysteine side-chains are marked in magenta and the -S–S- bridge in yellow. The structure was obtained from the one shown in A by 180° rotation around the x axis. C, RcsF-Δ30 [15N,1H]HSQC-TROSY spectrum under oxidizing conditions (red contours) and after reduction (blue contours).
An important feature of the RcsF structure is the disulfide bridge network (Fig. 3B). The pairwise contacts between Cys-74 and Cys-118 as well as between Cys-109 and Cys-124 were unambiguously identified via many NOE signals. Accordingly, the Cα and Cβ chemical shift values for all four cysteines indicated oxidized states (Fig. 2B). These bridges are critical for the stability of the RcsF-Δ30 structure. The structural consequences of disulfide bond reduction were monitored by time-dependent NMR spectroscopy of RcsF-Δ30 at 25 °C in the presence of 1 mm TCEP. Immediately after TCEP addition, disulfide bonds started to become reduced, followed by protein unfolding and resulting in a largely unstructured RcsF-Δ30 domain after 2 h. Unfolding was completed after 4 h (Fig. 3C).
As the RcsF primary structure does not show significant homologies to other protein families, we performed similarity searches using the DALI program (38). The best hits (Z = 8.5–9.9, RMSD = 2.2–3.2 Å) were represented by oligomeric proteins with five or more monomers but without functional annotations (PDB codes 1VR4, 2GTC, and 1Y2I). Further structural homologues (Z = 5.7–6.5, RMSD = 2.5–3.5 Å) were archaebacterial proteins with flavin-like ligands and pentameric selenium binding proteins (Z = 5.4–6.3, RMSD = 2.6–3.3 Å) (supplemental Figs. 3, A and B ). However, highly conserved residues important for their function within these homologous protein families are missing in RcsF. Structural homology was further identified in a number of proteins where the homologous proteins are part of larger complexes (Z = 2.0–4.7, RMSD = 2.5–5.0 Å), e.g. thiamine monophosphate kinases, acyl carrier protein synthases, or viral capsid proteins.
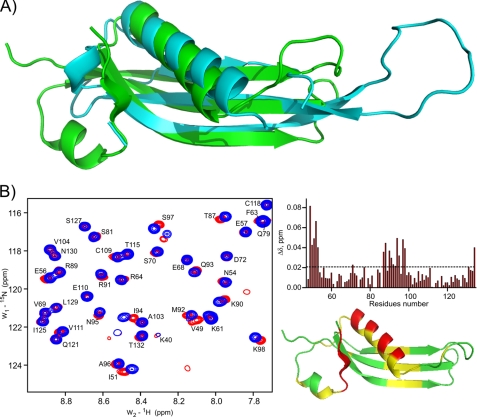
Interestingly, structural homology to RcsF-Δ30 was also detected in some of RNA binding proteins, in particular in ribosomal proteins such as L22 (1I4J-B) (Fig. 4A and supplemental Fig. S3C). This potential hint to a possible RNA or DNA binding function of RcsF was further analyzed by NMR titration experiments (Fig. 4B). Titration with small duplex DNA did not induce significant chemical shift perturbations (CSP) in standard conditions (50 mm Na2HPO4, 150 mm NaCl (pH 7.5)). tRNA or mononucleotides did not induce specific CSP either. Only some nonspecific binding of random DNA at lower pH or in cell lysates was observed, probably because of the strong electropositive surface of RcsF. However, a 14-meric RNA hairpin, representing a conserved protein interaction motif of the bacterial 16 S RNA, showed some degree of specific interactions (Fig. 4B). The RNA-induced CSP are weak, but their mapping on the RcsF 3D surface shows some clustering located near β1 and α2 (Fig. 4B).
FIGURE 4.
Structural similarity of the RcsF-Δ30 oxidized monomeric form to DNA/RNA binding proteins and possible interaction with RNA. A, superimposition of the free L22 protein (cyan, PDB code 1I4J) and RcsF-Δ30 (green). The superimposition with L22 of the Thermus thermophilus 50 S ribosomal subunit (PDB code 2WRJ) is provided in supplemental Fig. 3. B, representative area of the RcsF-Δ30 [15N,1H]HQSC-TROSY spectrum before (red) and after (blue) addition of a 2-fold excess of a 14-mer RNA hairpin (left panel). Right panel, mapping of the observed CSP onto the sequence (top) and the structure (bottom). CSP values below 0.018 ppm (3 × statistical error) are shown in green, those between 0.018–0.036 ppm in yellow, and those with more than 0.036 ppm in red.
Role of RcsF Disulfide Bridges in Capsule Biosynthesis Regulation
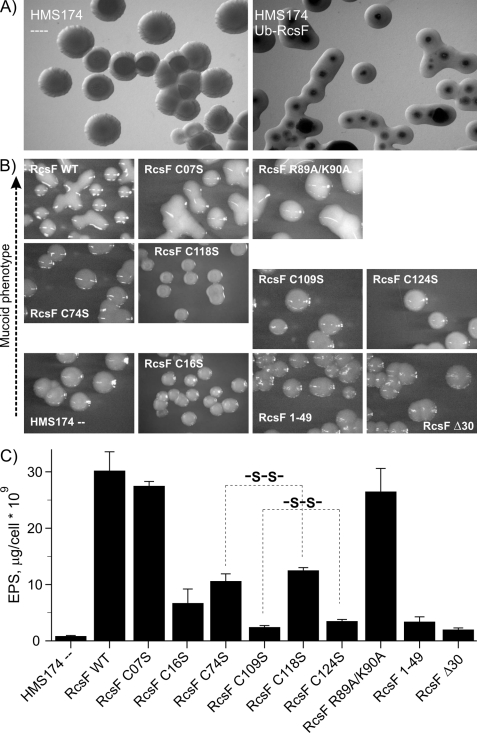
During the expression of the full-length Ub-RcsF protein in the E. coli strain HMS174 (Lys-12 derivative harboring the DE3 insert for T7RNAP expression), we noticed the induction of a well pronounced mucoid phenotype, which is characteristic for the activation of the RcsC pathway (Fig. 5A) as described previously (17, 18, 39). We have used this induction of a mucoid phenotype as a functional assay to evaluate the importance of the cysteine residues as well as residues comprising the positively charged surface of the β-sheet for the function of RcsF. The levels of synthesized EPS were quantified with the anthrone test for each mutant (40) Expression of Ub-RcsF induced a strong mucoid phenotype with EPS production of 30.2 ± 2.5 μg/cell·109, which was set to 100% of RcsF activity (Fig. 5). HMS174 cells not expressing Ub-RcsF do not show a mucoid phenotype and produce only 0.84 ± 0.12 μg/cell·109 of EPS (set to 0% RcsF activity). These data are in good agreement with previously reported values for rcs-dependent EPS biosynthesis upon RcsA overexpression (41).
FIGURE 5.
Role of the RcsF disulfide bridges in the RcsF-controlled EPS biosynthesis regulation. A, microphotographs of the empty (left panel) and transformed with pET60m_Ub_RcsF WT (right panel) HMS174 cells treated with IPTG and grown at 20 °C. B, microphotographs of HMS174 colonies after IPTG-induced expression of the various RcsF constructs. The pictures are placed in the four rows according to the level of mucoidity of the colonies. The RcsF constructs used in these experiments are indicated in each picture. C, quantification of the polysaccharides production in the HMS174 cells from B. The results of three independent anthrone tests are averaged and expressed in μg of total polysaccharides per 109 cells. Correlated effects of Cys-to-Ser substitutions in the RcsF C-terminal domain are highlighted with dashed lines.
The first non-conserved Cys-07 is located in the membrane-anchored helix of RcsF, and our mutagenesis analysis showed that it is not relevant for EPS biosynthesis. The mucoid phenotype (Fig. 5B) is similar to that of Ub-RcsF, and the levels of EPS synthesis are comparable (27.5 ± 0.8 μg/cell·109, 91% of RcsF activity) (Fig. 5C). In contrast, mutation of Cys-16 in RcsF (RcsF-C16S), which was shown to be important for anchoring RcsF in the outer membrane (18, 37), strongly decreases EPS biosynthesis (6.7 ± 2.5 μg/cell·109, 20% of RcsF activity). This result indicates an important but not absolutely essential role of this cysteine for RcsF activity. The remaining four cysteines are located in the folded RcsF domain and are involved in the formation of two disulfide bonds. These disulfide bonds are, as it was shown in the previous section, critical for the structural stabilization of RcsF. However, the impact of the two disulfide bridges on the function of RcsF could be different. Quantitative analysis of EPS production with bacteria expressing the RcsF-C74S (10.6 ± 1.3 μg/cell·109, 32% of RcsF activity) and RcsF-C118S (12.5 ± 0.5 μg/cell·109, 39.5% of RcsF activity) mutants revealed a significant reduction in EPS production down to ∼35% in both cases (Fig. 5C). An even stronger effect was observed in the case of RcsF-C109S (2.4 ± 0.3 μg/cell·109, 5.5% of RcsF activity) and RcsF-C124S (3.5 ± 0.3 μg/cell·109, 9.0% of RcsF activity), resulting in an EPS reduction down to <10%. The pairwise correlation of these four cysteine mutants in their effect on the RcsF activity supports further our assignment of the cysteine pairs in the disulfide bonds (Cys-74 to Cys-118 and Cys-109 to Cys-124) and indicates that reduction of the Cys-109 to Cys-124 bond has a stronger effect than reduction of the Cys-74 to Cys-118 bond.
Mutating Arg-89 and Lys-90 in the positively charged surface of RcsF to alanine did not affect the EPS biosynthesis significantly (26.5 ± 4.1 μg/cell·109, 87.5% of RcsF activity), indicating that the potential DNA/RNA binding activity may not be relevant for the RcsF function in EPS biosynthesis. Control experiments with the N- and C-terminally truncated derivatives RcsF-Δ30 (2.0 ± 0.3 μg/cell·109, 4.0% of RcsF activity) and RcsF-1–49 (3.4 ± 0.9 μg/cell·109, 9.0% of RcsF activity) demonstrated that RcsF is only active after proper periplasmic localization of the protein and that its C-terminal domain is essential for activity.
RcsF Expresses as an Unfolded Protein and Needs to be Correctly Folded in the E. coli Periplasm
The high expression levels of the N-terminally truncated RcsF constructs RcsF-Δ16, RcsF-Δ30, and RcsF-Δ45 allowed us to monitor their folding stage by solution state NMR either directly in vivo in E. coli cells or immediately after cell lysis. [15N,1H]HSQC-TROSY experiments were used to investigate whether these constructs were able to adopt a specific structural fold upon expression in E. coli cells by in-cell NMR (26, 27). The results indicate that all expressed constructs were not able to form stable structures neither in-cell nor post vivo under normal cellular (e.g. reducing) conditions (supplemental Fig. S4). The HSQC-TROSY spectra showed mostly poorly dispersed signals around 8 ppm with few resolved signals, which is typical for unfolded proteins. These spectra are very similar to that of TCEP-treated RcsF-Δ30 (Fig. 3C), indicating similar states of the protein in the different conditions. In addition, the clear detection of the RcsF-Δ30 signals in the NMR spectra of intact cells or crude lysates indicated that this protein does not efficiently interact either with compounds of the cytoplasm nor with any other soluble compound of cell extracts.
Potential Interactions of RcsF
The titration experiments with the 14-mer RNA have revealed that RcsF can, in principle, interact with RNA hairpins. However, the physiological interaction partner of RcsF remains unknown. It was speculated that RcsF senses Zn2+, glucose, peptidoglycan, or damages of its layer. Recently, the physical interaction between RcsF and periplasmic domains of RcsC, RcsD, and/or YrfF/IgaA were proposed to be important for the RcsF function (20, 42). To investigate the interaction of RcsF with potential interaction partners, we have performed NMR titration experiments with monomeric oxidized RcsF-Δ30. Potential interaction partners were dissolved or transferred into the same buffer and added to the RcsF sample in several steps until a molar ratio > 1:1 was achieved. Besides the small 14-mer RNA, we did not observe any significant CSP for any of the components tested. Supplemental Table S3 summarizes these experiments. The widely accepted idea that RcsF directly participates in glucose or Zn2+ sensing was not confirmed experimentally in these titration experiments, indicating that RcsF might not be the sensor but rather an auxiliary protein.
DISCUSSION
The outer membrane lipoprotein RcsF, shown to transduce a distinct set of signals via RcsC and RcsD to the response regulator RcsB, remains a key element for a global understanding of molecular mechanisms modulating the rcs signaling chain. Analysis of the RcsF primary sequence revealed a structural architecture of three domains. Hydrophobic residues are significantly less populated in the part spanning residues 14–134, indicating that this whole area may be periplasmic. The highest similarity between RcsF proteins is found in the C-terminal domain, whereas the N-terminal membrane-anchored domain is less conserved. This N-terminal helix does most likely not participate in the RcsF folding and function but rather facilitates the periplasmic localization of RcsF. In our experiments, the proline-rich sequence of RcsF (residues 17–48) did not exhibit any structure. However, being exposed to other conditions, e.g. localized on or near membranes or involved in molecular interactions, this loop might adopt certain structures.
The structural fold indicates RcsF as a prototype of small bacterial lipoproteins with highly conserved secondary structural pattern but with low homology in the primary structure. Functional homologous of RcsF might therefore extend far beyond the Enterobacteriaceae, despite only little sequence homologies. According to the DALI search, the RcsF structure is most similar to a number of oligomeric proteins such as dodecines, small docameric flavoproteins, and selenium binding proteins. We did not observe any tendency of oxidized RcsF to aggregate or to oligomerize. In contrast, the oxidized monomeric RcsF could be highly concentrated, and its NMR properties were comparable with that of ubiquitin, remaining stable for weeks. However, we cannot exclude that RcsF can built some high-molecular-weight complexes under certain conditions during activation or upon its oxidation pathway in the periplasm.
Two disulfide bridges (Cys-74 to Cys-118 and Cys-109 to Cys-124) in the oxidized monomeric form of RcsF were identified based on NOE information, and both were shown to be essential for RcsF folding. The fact that RcsF was not able to obtain its structure without extensive oxidation indicates that the final RcsF structure might depend on how it is expressed, transported into the periplasm, and subsequently oxidized.
RcsF was found to be a substrate of DsbA, a major periplasmic chaperone involved in disulfide bridge formation in proteins and in disulfide bridge shuffling (19). However, the role of potential disulfide bridge formation in the function of RcsF remained unknown. In this work, we could directly correlate the redox state of RcsF to its structural folding. After synthesis in the cytoplasm, the reduced RcsF remains unfolded. After transport into the periplasm or into the outer membrane by yet unknown mechanisms, the protein becomes folded concomitant with the formation of disulfide bridges that obviously are essential to stabilize the structural fold. DsbA might play a key role in chaperoning the disulfide bridge formation; however, the oxidation and correct disulfide bridge formation can obviously also be initiated, at least to some extent, by cell disruption.
RcsF contains six cysteines, from which the first, Cys-07, is removed by processing after transport of RcsF into the outer membrane (18). This residue appears not to be necessary for correct transport, localization, or function, as a RcsFC7S substitution was not impaired in the induction of EPS biosynthesis. Cys-16 acts as a lipid anchor and is modified by N-acyl-diacylglycerol (18, 37). It is consequently not available for disulfide bridge formation, which is in agreement with our results. However, the residue is still important for RcsF activity, indicating that the membrane localization of RcsF is crucial for its activity.
For the remaining four cysteines, we have identified intrachain disulfide bridge formation between Cys-74 and Cys-118 and between Cys-109 and Cys-124, respectively. In a new approach using a combined chemical cross-linking and mass spectrometry strategy, evidence for intrachain bonds between Cys-74 and Cys-124 and between Cys-109 and Cys-118 were reported (43). These findings disagree with our data and might be based on the fact that rcsC-deficient strains were used for RcsF expression in that study. In our activity assays, we observed a good correlation between mutations of the identified cysteine pairs and induction of EPS biosynthesis.
RcsF may play a critical role in signal transduction form the cell surface to RcsC or RcsD (17). However, RcsF appears to be an optional player in the rcs system, as several responses are RcsF-independent (3, 18). RcsC is necessary to transduce the signals received via RcsF (18). Important is that RcsF activation is not affecting the level of RcsC expression in the cell; therefore, the mechanism of activation should rather be based on physical interactions between RcsF and RcsC. RcsF was initially characterized as a cytoplasmic protein (44). Later on, the periplasmic localization of RcsF was proposed and experimentally demonstrated by several studies (17–20), indicating that the potential interactions are initiated by damages of the cellular envelope and by destruction/perturbation of the peptidoglycan layer. However, we did not observe significant interactions of RcsF with Zn2+, glucose, lysozyme, AMP, and other components that are shown to be activators of the rcs phosphorelay. Direct interaction studies with the periplasmic domain of RcsC also showed no interaction. However, because of the membrane-bound nature of this domain, it might not have been in the correct conformation under the specific conditions of the titration experiment.
Conclusions
Our structural data provide new details and evidence for the function of RcsF. The structural homology to several multisubunit complexes of diverse function could be taken as indication of an oligomerization potential of RcsF, probably induced under particular conditions. In-cell NMR experiments combined with the molecular analysis of disulfide bridge formation clearly document the periplasmic localization of RcsF and therefore support previous reports based on biochemical data. A number of proposed ligands could be ruled out as RcsF interaction partners, whereas the presented NMR analysis will serve as basis for further studies of RcsF complex formation, in particular with the periplasmic domains of RcsC and RcsD.
Supplementary Material
Acknowledgments
We thank Christian Ross for providing competent HMS174 cells and Robert Hänsel for providing RNA samples for this work. We also thank Thomas Köhler for help in the initial part of this project.
This work was supported by Deutsche Forschungsgemeinschaft Grant BE 1911/4-1, the Center for Biomolecular Magnetic Resonance (BMRZ), and the Cluster of Excellence Frankfurt Macromolecular Complexes (CEF).
The atomic coordinates and structure factors (code 2L8Y) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
The NMR assignments have been deposited in the Biological Magnetic Resonance Data Bank (accession no. 17431).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4 and Tables S1–S3.
- rcs
- regulation of capsule synthesis
- EPS
- exopolysaccharide
- IPTG
- isopropyl 1-thio-β-d-galactopyranoside
- Ub
- ubiquitin
- CSP
- chemical shift perturbation(s)
- HSQC
- heteronuclear single-quantum correlation
- TROSY
- transverse relaxation optimized spectroscopy
- TCEP
- tris(2-carboxyethyl)phosphine.
REFERENCES
- 1. Majdalani N., Gottesman S. (2005) Annu. Rev. Microbiol. 59, 379–405 [DOI] [PubMed] [Google Scholar]
- 2. Gottesman S., Trisler P., Torres-Cabassa A. (1985) J. Bacteriol. 162, 1111–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Andresen L., Kõiv V., Alamäe T., Mäe A. (2007) FEMS Microbiol. Lett. 273, 229–238 [DOI] [PubMed] [Google Scholar]
- 4. Francez-Charlot A., Laugel B., Van Gemert A., Dubarry N., Wiorowski F., Castanié-Cornet M. P., Gutierrez C., Cam K. (2003) Mol. Microbiol. 49, 823–832 [DOI] [PubMed] [Google Scholar]
- 5. West A. H., Stock A. M. (2001) Trends Biochem. Sci. 26, 369–376 [DOI] [PubMed] [Google Scholar]
- 6. Takeda S., Fujisawa Y., Matsubara M., Aiba H., Mizuno T. (2001) Mol. Microbiol. 40, 440–450 [DOI] [PubMed] [Google Scholar]
- 7. Chen M. H., Takeda S., Yamada H., Ishii Y., Yamashino T., Mizuno T. (2001) Biosci. Biotechnol. Biochem. 65, 2364–2367 [DOI] [PubMed] [Google Scholar]
- 8. Gottesman S., Stout V. (1991) Mol. Microbiol. 5, 1599–1606 [DOI] [PubMed] [Google Scholar]
- 9. Clarke D. J., Joyce S. A., Toutain C. M., Jacq A., Holland I. B. (2002) J. Bacteriol. 184, 1204–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wehland M., Kiecker C., Coplin D. L., Kelm O., Saenger W., Bernhard F. (1999) J. Biol. Chem. 274, 3300–3307 [DOI] [PubMed] [Google Scholar]
- 11. Wehland M., Bernhard F. (2000) J. Biol. Chem. 275, 7013–7020 [DOI] [PubMed] [Google Scholar]
- 12. Pristovsek P., Sengupta K., Löhr F., Schäfer B., von Trebra M. W., Rüterjans H., Bernhard F. (2003) J. Biol. Chem. 278, 17752–17759 [DOI] [PubMed] [Google Scholar]
- 13. Rogov V. V., Bernhard F., Löhr F., Dötsch V. (2004) J. Mol. Biol. 343, 1035–1048 [DOI] [PubMed] [Google Scholar]
- 14. Rogov V. V., Rogova N. Y., Bernhard F., Koglin A., Löhr F., Dötsch V. (2006) J. Mol. Biol. 364, 68–79 [DOI] [PubMed] [Google Scholar]
- 15. Schmöe K., Rogov V. V., Rogova N. Yu, Löhr F., Güntert P., Bernhard F., Dötsch V. (2011) Structure 19, 577–587 [DOI] [PubMed] [Google Scholar]
- 16. Zhang W., Shi L. (2005) Microbiology 151, 2159–2173 [DOI] [PubMed] [Google Scholar]
- 17. Majdalani N., Heck M., Stout V., Gottesman S. (2005) J. Bacteriol. 187, 6770–6778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Castanie-Cornet M. P., Cam K., Jacq A. (2006) J. Bacteriol. 188, 4264–4270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kadokura H., Tian H., Zander T., Bardwell J. C., Beckwith J. (2004) Science 303, 534–537 [DOI] [PubMed] [Google Scholar]
- 20. Farris C., Sanowar S., Bader M. W., Pfuetzner R., Miller S. I. (2010) J. Bacteriol. 192, 4894–4903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Callewaert L., Vanoirbeek K. G., Lurquin I., Michiels C. W., Aertsen A. (2009) J. Bacteriol. 191, 1979–1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Laubacher M. E., Ades S. E. (2008) J. Bacteriol. 190, 2065–2074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sambrock J., Fritsch E. F., Maniatis T. (1989) in Molecular Cloning: A Laboratory Manual (Ford N., Nolan C., Ferguson M. eds) 2nd Ed., Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 24. Wishart D. S., Bigam C. G., Yao J., Abildgaard F., Dyson H. J., Oldfield E., Markley J. L., Sykes B. D. (1995) J. Biomol. NMR 6, 135–140 [DOI] [PubMed] [Google Scholar]
- 25. Rogov V. V., Löhr F., Rogova N. Y., Klammt C., Koglin A., Bernhard F., Dötsch V. (2007) J. Biomol. NMR 38, 165. [DOI] [PubMed] [Google Scholar]
- 26. Serber Z., Selenko P., Hänsel R., Reckel S., Löhr F., Ferrell J. E., Jr., Wagner G., Dötsch V. (2006) Nat. Protoc. 1, 2701–2709 [DOI] [PubMed] [Google Scholar]
- 27. Serber Z., Corsini L., Durst F., Dötsch V. (2005) Methods Enzymol. 394, 17–41 [DOI] [PubMed] [Google Scholar]
- 28. Goddard T. D., Kneller D. G. SPARKY 3, University of California, San Francisco [Google Scholar]
- 29. Herrmann T., Güntert P., Wüthrich K. (2002) J. Mol. Biol. 319, 209–227 [DOI] [PubMed] [Google Scholar]
- 30. Güntert P., Mumenthaler C., Wüthrich K. (1997) J. Mol. Biol. 273, 283–298 [DOI] [PubMed] [Google Scholar]
- 31. Cornilescu G., Delaglio F., Bax A. (1999) J. Biomol. NMR 13, 289–302 [DOI] [PubMed] [Google Scholar]
- 32. Linge J. P., Williams M. A., Spronk C. A., Bonvin A. M., Nilges M. (2003) Proteins 50, 496–506 [DOI] [PubMed] [Google Scholar]
- 33. Brunger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice R. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 34. Laskowski R. A., Rullmannn J. A., MacArthur M. W., Kaptein R., Thornton J. M. (1996) J. Biomol. NMR 8, 477–486 [DOI] [PubMed] [Google Scholar]
- 35. Koradi R., Billeter M., Wüthrich K. (1996) J. Mol. Graph. 14, 51–55 [DOI] [PubMed] [Google Scholar]
- 36. Bryson K., McGuffin L. J., Marsden R. L., Ward J. J., Sodhi J. S., Jones D. T. (2005) Nucleic Acids Res. 33, W36–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Seydel A., Gounon P., Pugsley A. P. (1999) Mol. Microbiol. 34, 810–821 [DOI] [PubMed] [Google Scholar]
- 38. Holm L., Sander C. (1996) Science 273, 595–603 [DOI] [PubMed] [Google Scholar]
- 39. Hagiwara D., Sugiura M., Oshima T., Mori H., Aiba H., Yamashino T., Mizuno T. (2003) J. Bacteriol. 185, 5735–5746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Dubois M., Gilles K. A., Hamilton J. K., Rebers P. A., Smith F. (1956) Anal. Chem. 28, 350–356 [Google Scholar]
- 41. Bernhard F., Poetter K., Geider K., Coplin D. L. (1990) Mol. Plant-Microbe Interact. 3, 429–437 [DOI] [PubMed] [Google Scholar]
- 42. Clarke D. J. (2010) Future Microbiol. 5, 1173–1184 [DOI] [PubMed] [Google Scholar]
- 43. Singh P., Shaffer S. A., Scherl A., Holman C., Pfuetzner R. A., Larson Freeman T. J., Miller S. I., Hernandez P., Appel R. D., Goodlett D. R. (2008) Anal. Chem. 80, 8799–8806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gervais F. G., Phoenix P., Drapeau G. R. (1992) J. Bacteriol. 174, 3964–3971 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.