Abstract
A bacterial xylanase gene, Nonomuraea flexuosa xyn11A, was expressed in the filamentous fungus Trichoderma reesei from the strong cellobiohydrolase 1 promoter as fusions to a variety of carrier polypeptides. By using single-copy isogenic transformants, it was shown that production of this xylanase was clearly increased (up to 820 mg/liter) when it was produced as a fusion protein with a carrier polypeptide having an intact domain structure compared to the production (150 to 300 mg/liter) of fusions to the signal sequence alone or to carriers having incomplete domain structures. The carriers tested were the T. reesei mannanase I (Man5A, or MANI) core-hinge and a fragment thereof and the cellulose binding domain of T. reesei cellobiohydrolase II (Cel6A, or CBHII) with and without the hinge region(s) and a fragment thereof. The flexible hinge region was shown to have a positive effect on both the production of Xyn11A and the efficiency of cleavage of the fusion polypeptide. The recombinant Xyn11A produced had properties similar to those of the native xylanase. It constituted 6 to 10% of the total proteins secreted by the transformants. About three times more of the Man5A core-hinge carrier polypeptide than of the recombinant Xyn11A was observed. Even in the best Xyn11A producers, the levels of the fusion mRNAs were only ∼10% of the level of cel7A (cbh1) mRNA in the untransformed host strain.
Nonomuraea flexuosa DSM43186 (previously Microtetraspora flexuosa or Actinomadura flexuosa [50]) is an actinomycete known to produce thermostable xylanases with alkaline optima (17). Xylanases with such properties are beneficial in many industrial processes, one of which is the enzymatic bleaching of kraft pulp. In enzyme-aided bleaching, pretreatment with xylanase removes xylans while preserving the cellulose content and decreases the amount of bleaching chemicals and/or increases the brightness of the paper (for a review, see reference 41).
Efficient and cost-effective industrial production of an enzyme having properties suitable for use at high process temperatures and pH is a challenge, because these enzymes originate mainly from relatively unstudied bacteria in which the production level is low. There may be little or no experience with cultivating these microbes in a fermentor, or they may otherwise be unsuitable for industrial scale production. Filamentous fungi, e.g., Aspergillus and Trichoderma, are used as producers of industrial enzymes, and genetically modified strains produce high levels of both homologous and heterologous fungal enzymes (for reviews, see references 27, 32, and 46). Thus, the filamentous fungi are possible choices as production organisms for bacterial enzymes. Unfortunately, the published yields of bacterial enzymes from filamentous fungi are low, not exceeding a few tens of milligrams per liter, and in many of the studies, the enzymes were detected only intracellularly (for reviews, see references 20 and 44). The only reported exception has been the Streptomyces hindustanus phleomycin-binding protein that was produced in Tolypocladium geodes at a high yield, 1,500 mg/liter (7).
Gene fusions have been successfully used in the filamentous fungi for increasing the yields of nonfungal heterologous proteins (for reviews, see references 12 and 32). For mammalian and plant proteins, increases in yield of 5- to >1,000-fold compared to the nonfused constructions have been obtained, resulting in protein levels of from 5 to >250 mg/liter. Fusions to both well-expressed full-length proteins and their catalytically active N-terminal domains (lacking the binding domains) have been successful. Positive effects of such gene fusions on the production of heterologous proteins have been observed at the (post)translational level and the transcriptional level (13, 21, 31). In T. reesei, fusions to the core-hinge domain of the major cellobiohydrolase I, Cel7A, have increased the production levels of calf chymosin (J. Uusitalo, unpublished data; referred to in reference 32), interleukin-6 (32; J. Demolder, X. Saelens, M. Penttilä, W. Fiers, and R. Contreras, Abstr. 2nd Eur. Conf. Fungal Genet., abstr. B38, 1994), murine-2-phenyloxazolone heavy Fd chain, and the antigen-binding Fab molecules after transformation of a heavy-chain construct to a strain producing light chain (30). An increase in yield could not be detected when the catalytic domain of the bacterial Dictyoglomus thermophilum xylanase was produced, after the synthetic xynB gene was fused to the cel7A core-hinge coding sequence (43).
In the present work, the gene fusion strategy was used to increase production of a bacterial enzyme from Trichoderma reesei. Previously, only low yields of the bacterial Thermomonospora fusca xylanase TfxA (19) were obtained by expressing its gene under a cel7A (cbh1) promoter in T. reesei (M. Paloheimo, unpublished data). The similarity between the TfxA and N. flexuosa Xyn11A proteins is 90%. Therefore, we considered that a carrier protein might be essential for efficient production of the Xyn11A xylanase in T. reesei. We investigated the required properties of the carrier polypeptide and the linker sequence between the fusion partners by using isogenic single-copy strains in which the fusion gene was expressed from a cel7A promoter and the constructed expression cassettes replaced the native cel7A locus. We used carriers known to be nondetrimental to the pulp-bleaching application. These were the core-hinge of T. reesei mannanase I (Man5A) (39) and the cellulose binding domain (CBD) of cellobiohydrolase II (42), belonging to the CBM1 family (information can be found at the website CAZy—Carbohydrate-Active enZYmes [http://afmb.cnrs-mrs.fr/CAZY/]). The Cel6A (CBHII) CBD was tested as a fusion partner for Xyn11A with and without the hinge (or double-hinge) region to analyze the effects of the flexible hinge on the production levels and on the efficiency of cleavage of the fusion polypeptide. The importance of the carrier having an intact domain structure was tested by fusing the xyn11A gene to sequences coding for fragments of the Man5A core or Cel6a CBD. As a control, an expression cassette was constructed in which the xyn11A gene was fused to the man5A signal sequence.
MATERIALS AND METHODS
Microbial strains, plasmids, growth media, and growth conditions.
Plasmids were propagated in Escherichia coli XL1-Blue (Stratagene, La Jolla, Calif.). The vector backbones used in the plasmid constructions were pUC18 (EMBL database accession no. L09136), pUC19 (L09137), and pBluescript SK(−) (Stratagene). All E. coli cultivations were performed overnight at 37°C in Luria-Bertani medium (37) to which ampicillin (50 μg/ml) had been added.
N. flexuosa DSM43186 (ATCC 35864) was cultivated in 1-liter fermentations (Biostat M; B. Braun, Melsungen AG, Melsungen, Germany) at 50°C using GPYB medium (14) supplemented with oat spelt xylan instead of glucose as described previously (17). Purified Xyn11A protein was used as a control in the Western blots.
The T. reesei strains ALKO3620 and ALKO4468 were used as parents for the transformations. T. reesei ALKO3620 is an endoglucanase II-negative strain. It was constructed from the low-protease mutant strain ALKO2221, derived from the strain VTT-D-79125 (3) by UV mutagenesis (A. Mäntylä, R. Saarelainen, R. Fagerström, P. Suominen, and H. Nevalainen, Abstr. 2nd Eur. Conf. Fungal Genet., abstr. B52, 1994) as follows. The endoglucanase 2 gene (cel5A or egl2; originally named egl3) (36) was replaced by the phleomycin resistance-encoding marker gene from S. hindustanus, Sh ble (9). The 3.3-kb BglII-XbaI fragment from the plasmid pAN8-1 (29), containing the ble gene flanked by the Aspergillus nidulans gpd promoter and trpC terminator, was used. The cel5A flanking sequences in the replacement cassette (the 5′ region was the 1.4-kb XhoI-SacI fragment ∼2.2 kb upstream from the cel5A gene, and the 3′ region was the 1.6-kb AvrII-SmaI fragment ∼0.2 kb from the end of the cel5A gene) were isolated from the egl3 λ clone (36). The strategy for the replacement was described previously (40). T. reesei ALKO4468 is an endoglucanase I- and II-negative strain. It was constructed from the strain ALKO3620 by further replacing the endoglucanase 1 gene, cel7B (egl1 [33]), with the E. coli hygromycin B phosphotransferase gene, hph (15), conferring resistance to hygromycin B. The 1.7-kb NotI-NsiI fragment from the plasmid pRLMEX30 (26) was used, in which the hph gene is expressed from the T. reesei pyruvate kinase (pki) promoter and transcription is terminated by using the cel6A terminator sequences. The plasmid pRLMEX30 was kindly provided by Christian P. Kubicek (Institut für Biochemische Technologie, Technische Universität Wien, Vienna, Austria). The cel7B flanking regions were as described previously (40). The single-copy replacements of the cel5A and cel7B genes by the marker genes in T. reesei ALKO3620 and ALKO4468 were verified by Southern blot analysis, as described previously (40).
T. reesei strain ALKO4332, used as a control in the cultivations, expresses the region coding for the Man5A core (from M1 to G373; man5A nucleotides 1 to 1248) from the cel7A promoter. The cel7A locus in this strain has been replaced by one copy of the expression cassette pALK1010, resulting in a transformant analogous to those constructed for this study (Paloheimo, unpublished). For the mannanase sequence, see reference 39.
T. reesei strains were sporulated on PD agar slants (potato dextrose broth; Difco, Detroit, Mich.). The transformants were selected on Trichoderma minimal medium containing acetamide as a nitrogen source (34). The fungal mycelia for DNA isolations were obtained after growing the strains for 2 days on Trichoderma minimal medium containing 2% proteose peptone (Difco). Complex lactose-based cellulase-inducing media (22) were used for enzyme production in shake flasks and fermentations. The transformants were screened using 50-ml cultivations, and the mycelium for the RNA isolations was collected from 200-ml cultivations. The shake flask cultivations were grown for 7 days at 30°C and 250 rpm. The laboratory scale fermentor cultivations were performed for 5 days in 1-liter Braun Biostat M fermentors.
DNA techniques.
Standard DNA methods (37) were used in constructing plasmids, transforming E. coli, and performing Southern blotting. Each enzyme and kit was used according to the instructions from the supplier. The enzymes for DNA modifications were purchased from Roche Diagnostics GmbH (Mannheim, Germany), New England Biolabs (Beverly, Mass.), and Finnzymes (Espoo, Finland). Qiagen (Hilden, Germany) columns or Magic Miniprep kits (Promega, Madison, Wis.) were used in the plasmid isolations. The ABI 381A DNA synthesizer and ABI 373A sequencer (Applied Biosystems, Foster City, Calif.) were used for synthesizing oligonucleotides and for analyzing sequencing reactions. PCRs were performed using a PTC-100 programmable thermal controller (MJ Research Inc., Watertown, Mass.). DNA fragments for subcloning and transformations were isolated from low-melting-point agarose gels (BioWhittaker Molecular Applications Inc., Rockland, Maine) by the freeze-thaw-phenol method (5), using β-agarase (New England Biolabs) or the Qiaex II gel extraction kit (Qiagen GmbH).
The genomic DNAs were isolated as described previously (35). Digoxigenin (Roche Diagnostics GmbH)-labeled expression cassettes were used as probes in the Southern blot hybridizations.
Construction of expression cassettes used in the study.
N. flexuosa xyn11A was expressed from the T. reesei cel7A promoter in all the cassettes constructed, pALK1118, pALK945, pALK948, pALK1021, pALK1022, pALK1264, and pALK1283 to -1286 (Fig. 1; also see Fig. 4). The promoter, transcription terminator, and 3′ flanking sequences were as described previously (23). The gene coding for acetamidase (amdS) was used as a marker in the transformations. The amdS gene was isolated from p3SR2 (24). A 3.1-kb SpeI-XbaI fragment was ligated between the cel7A terminator and the 3′ flanking region. The plasmid p3SR2 was kindly provided by M. Hynes (University of Melbourne, Melbourne, Australia).
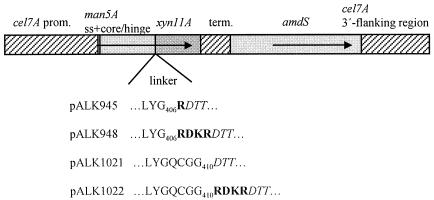
FIG. 1.
Expression cassettes constructed to study cleavage of Man5A-Xyn11A fusions. The gene fusions were expressed from the cel7A promoter (prom.), and termination of transcription was ensured by using a cel7A terminator (term.) sequence. The man5A signal (ss) and core-hinge sequences (M1 to G406 in pALK945 and pALK948; M1 to G410 in pALK1021 and pALK1022) were used to code for the carriers, the amdS gene was included as a transformation marker, and the cel7A 3′ flanking region, together with the cel7A promoter, was used to target the expression cassette into the cel7A locus by homologous recombination (for a more detailed description, see Materials and Methods). Synthetic linker sequences coding for an additional Arg in pALK945 and a Kex2-like protease cleavage signal, Lys-Arg (as RDKR), in pALK948 and pALK1022 were included to ensure cleavage of the fusion protein. The expression cassette pALK1021 does not contain an additional signal for proteolytic cleavage. The amino acids encoded by the man5A sequence are shown in regular type, those of xyn11A are in italics, and the synthetic amino acids for proteolytic cleavage are in boldface.
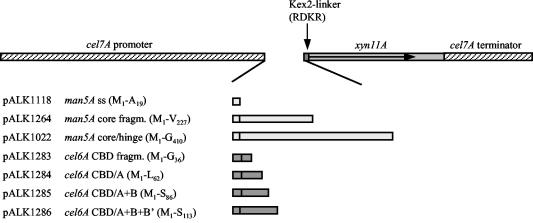
FIG. 4.
Expression cassettes constructed to study effect of carrier polypeptide on Xyn11A production. The overall structure of the expression cassettes was as described in the legend to Fig. 1. The amdS gene and the cel7A 3′ flanking region (not shown) were included as in the previous constructs. A sequence coding for the synthetic Kex2-like protein-processing signal RDKR, identical to that used in pALK948 and pALK1022, was included between the sequence coding for the carrier polypeptide and the xyn11A sequence, with the exceptions of pALK1118 and pALK1264. The expression cassette pALK1118 had the native man5A signal sequence (ss) cleaving signal and pALK1264 with an additional sequence coding for GQCGG preceding the Kex2 site. The constructs coding for the Man5A carrier (pALK1264 and pALK1022) had the man5A signal sequence, and the constructs coding for Cel6A CBD (pALK1283 to pALK1286) had the cel6A signal sequence. The numbers of native amino acids encoded by the carrier sequences, from M1 of the corresponding signal sequences, are shown. Cel6A CBD block A codes for the tail of the protein, B codes for the hinge, and B′ codes for the duplicated-hinge region. For a more detailed description, see Materials and Methods. fragm., fragment.
The N. flexuosa xyn11A gene (EMBL accession number AJ508952) was included in the expression cassettes as a 1.3-kb fragment ending at the MluI site ∼250 bp after the stop codon. The gene was linked from its N-terminal Asp44 to the man5A signal sequence (pALK1118 [man5A nucleotides 1 to 57]), to the man5A core-hinge coding sequences (pALK945 and -948 [nucleotides 1 to 1347] and pALK1021 and -1022 [nucleotides 1 to 1359]), or to a fragment of the man5A core (pALK1264 [nucleotides 1 to 681]). For the man5A sequence, see reference 39. The xyn11A fusions to the cel6A-derived carriers were to the cel6A CBD (block A in pALK1284 [cel6A nucleotides 1 to 288]), a fragment of the CBD (pALK1283 [nucleotides 1 to 156]), the CBD-hinge (blocks A and B in pALK1285 [nucleotides 1 to 306]), and the CDB-duplicated hinge (blocks A, B, and B′ in pALK1286 [nucleotides 1 to 387]). For the cel6A sequence, see reference 42. The sequence in the linker of pALK945 codes for an additional Arg that makes the amino acid sequence at the expected cleavage site identical to that of cel7A cleaved by an unknown protease (30). A synthetic sequence coding for the dipeptide Lys-Arg, a target of a Kex2-like protease (7), was included in the linkers of pALK948, pALK1022, and pALK1264 and all the Cel6A CBD constructs. In addition, the linker of pALK1264 was preceded by a sequence coding for the amino acids Gly-Gln-Cys-Gly-Gly. This additional sequence was included to increase the length of the linker between the nonintact carrier and Xyn11A. It derives from the beginning of the Man5A CBD. An identical sequence preceded the Xyn11A sequence in pALK1021 and pALK1022 (Fig. 1).
Exact fusions between the cel7A promoter and the signal sequences, carriers, linkers, and xyn11A were synthesized by PCR. An NruI recognition site (TCGCGA) was introduced into the Kex2 linker (encoded by the sequence CGCGACAAGCGC; the partial NruI sequence is underlined) to facilitate the construction of the fusions. The codon CGC was chosen for the arginines in the linker, and the third nucleotide of the native codon preceding the linker was changed to T when necessary. The modifications made did not change the amino acids encoded by the constructs.
Transformation of Trichoderma and analysis of the transformants.
T. reesei protoplasts were transformed with linear expression cassettes isolated from the vector backbones by EcoRI. The expression cassettes were transformed into T. reesei strain ALKO3620 (Cel5A−), with the exception of pALK1118 and pALK1264, which were transformed into T. reesei ALKO4468 (Cel5A− Cel7B−). The strains ALKO3620 and ALKO4468 produce similar amounts of heterologous xylanases when transformed with identical expression plasmid constructs (A. Mäntylä and M. Paloheimo, unpublished data). Transformations were performed as describecd previously (33) with the modifications described in reference 23. The transformants were purified on selection plates through single conidia prior to sporulating them on PD agar. Targeting to the cel7A locus was screened as a Cel7A-negative phenotype using a Minifold I-SRC 96 dot blotter (Schleicher & Schuell, Dassel, Germany). The monoclonal antibody CI-258 (1) was used in the detection of the Cel7A protein by the ProtoBlot Western Blot AP system (Promega). The genotypes of the chosen transformants were confirmed by using Southern blots in which several genomic digests were included, and the respective expression cassettes were used as probes. Strains containing a replacement of cel7A with one copy of the expression cassette were chosen for further studies.
Protein and enzyme assays and analysis of the mycelium dry mass.
Samples of the culture supernatants were run on 12% polyacrylamide slab gels containing 0.1% sodium dodecyl sulfate (SDS) on the Mini Protean II electrophoresis system (Bio-Rad, Hercules, Calif.). The proteins were stained with Coomassie brilliant blue R250. A polyclonal antibody raised against the native purified Xyn11A (Mäntylä and Paloheimo, unpublished) and the Protoblot AP System were used for the detection of the Xyn11A protein in the Western blots. The amounts of the proteins secreted into the culture supernatants were assayed after trichloroacetic acid precipitation by a method described previously (25). Bovine serum albumin (BSA) was used as a standard. Xylanase activity was assayed according to a method described previously (4) using 5-min reactions at pH 7 and 70°C in 50 mM McIlvaine's citrate-phosphate buffer. Birch xylan (catalog no. 7500; Roth, Karlsruhe, Germany) was used as a substrate. In the thermostability assays, 100 μg of BSA/ml was added to the reaction mixtures. Mannanase activity was assayed at 50°C and pH 5.3 using locust bean gum (G-0753; Sigma, Oslo, Norway) as a substrate, as described previously (39). The dry mass of mycelia was analyzed from 10-ml samples.
Isolation of RNA and analysis of transcripts.
Total RNA was isolated from mycelia grown in 200-ml shake flask cultivations for 3 and 4 days and from 1-liter fermentations after the strains were grown for 2 days. An RNeasy Plant Mini Kit (Qiagen GmbH) was used for the isolations. The Northern blot gels (1× MOPS [morpholinepropanesulfonic acid], 6% formaldehyde, 1.2% agarose) were run according to standard methods (37). A total of 2.0 μg of RNA was loaded per lane. An 81-mer oligonucleotide complementary to the transcribed region of the cel7A promoter (from +3 to −78) (18) was used to detect the mRNAs transcribed from the cel7A promoter. A 1.9-kb KpnI fragment containing the T. reesei actin gene (28) was used to normalize the signals obtained. The Trichoderma actin gene, cloned (49) and kindly donated by Christian P. Kubicek, was used. The oligonucleotide was labeled by using [γ-32P]ATP (Amersham Biosciences) according to standard methods (37). The actin probe was labeled by using [α-32P]dCTP and the Multiprime DNA labeling system RPN16004 (all from Amersham Biosciences). Hybridizations with both of the probes were performed at 42°C in 50% formamide- 6× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7])-5× Denhardt's solution-0.5% SDS- 100 μg of salmon sperm DNA/ml. The filters were washed with 2× SSC- 0.5% SDS for 5 min at room temperature and with 2× SSC- 0.1% SDS for 15 min at room temperature, followed by either one wash at 42°C for the filters probed with the oligonucleotide or two washes for the filters probed with actin. The filters probed with actin were again washed twice for 15 min at 42°C with 0.1× SSC- 0.5% SDS. The signals in the Northern blot filters were analyzed by using a Typhoon 8600 variable-mode imager and ImageQuant version 5.2 software (Molecular Dynamics, Sunnyvale, Calif.).
RESULTS
Production of the bacterial xylanase Xyn11A by using Man5A core-hinge carriers.
Four expression cassettes in which the sequence for the Man5A core-hinge was coded as a carrier polypeptide for Xyn11A were constructed (Fig. 1) (see Materials and Methods). These cassettes differed in the linker sequence inserted between the carrier and xyn11A. We tested two different proteolytic signals (pALK945 versus pALK948) and linkers of different lengths (pALK948 versus pALK1022) to analyze which type of linker would be the most efficiently cleaved. The fusion in the fourth cassette, pALK1021, did not include a synthetic protease cleavage signal, and we expected the transformants to produce an uncleaved fusion protein. The expression cassettes were isolated from the vector backbones and transformed into T. reesei ALKO3620. The transformants containing a single-copy replacement of the cel7A gene by the expression cassettes were screened for further studies. Several parallel single-copy strains from each construct were similar in protein and xylanase activity levels analyzed in the culture supernatants of shake flask cultivations. One single-copy representative from each construct was chosen to be cultivated in the fermentor. The culture supernatants from the fermentor cultivations were analyzed for the amounts of protein, mannanase, and xylanase activities (Table 1); cleavage of the fusion proteins (Fig. 2); and thermostability of the recombinant Xyn11A protein compared to the xylanase activity in the N. flexuosa culture supernatant (Fig. 3).
TABLE 1.
Production of recombinant Xyn11A protein from strains in which the Man5A core-hinge was used as a carrier for Xyn11Aa
| Strain | Expression cassette | Protein (mg/ml) | Xylanase (nkat/ml) | Mannanase (nkat/ml) | Xyn11A (mg/ml)b | Man5A (mg/ml)c | Xyn11A (nmol/ml)d | Man5A (nmol/ml)e |
|---|---|---|---|---|---|---|---|---|
| ALKO3620 | None | 15.3 | 150 | 310 | ND | 0.19 | 0 | 0.4 |
| ALKO4332 | pALK1010 | 11.7 | NDg | 7,800 | ND | 3.69 | 0 | 97.5 |
| ALKO4396 | pALK945 | 10.8 | 8,650 | 4,310 | 0.56 | 2.04 | 16.9 | 49.4 |
| ALKO4399 | pALK948 | 10.9 | 8,390 | 3,980 | 0.54 | 1.88 | 16.4 | 44.9 |
| ALKO4402 | pALK1021 | 10.0 | 8,610 | 4,020 | 0.55f | 1.90f | 16.8f | 46.1f |
| ALKO4405 | pALK1022 | 10.2 | 9,150 | 4,150 | 0.59 | 1.96 | 17.9 | 46.8 |
The results are from one fermentation each.
The specific activity determined for purified Xyn11A, 15,600 nkat/mg (Mäntylä and Paloheimo, unpublished), was used in the calculations.
The following specific activities were used: 1,645 nkat/mg for Man5A(ALKO3620) and the average of the two pI forms of purified mannanase (39) and 2,120 nkat/mg for the Man5A core and Man5A core-hinge (determined for the Man5A core protein) (M. Siika-aho, unpublished data).
The calculated molecular mass of 32.9 kDa was used for Xyn11A.
The following calculated molecular masses were used: Man5A, 44.3 kDa; Man5A core, 37.8 kDa; Man5A core-hinge in pALK945 and -948, 41.2 kDa, and in pALK1021 and -1022, 41.9 kDa.
The values were calculated from the activity values. An exact determination is not possible because of partial degradation of the fusion protein. Also, the activities of the partners in the uncleaved fusions are not known.
ND, not determined.
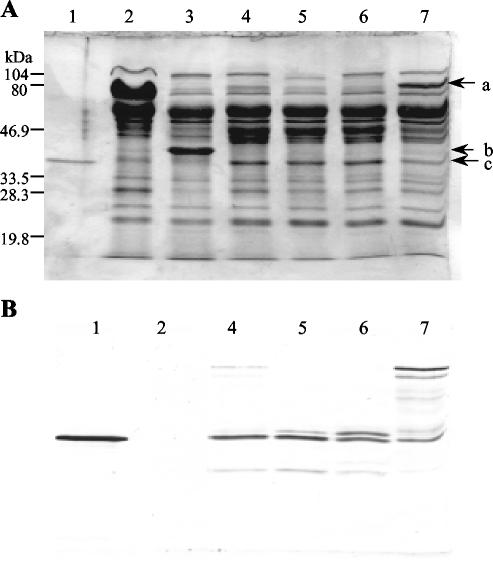
FIG. 2.
SDS-PAGE (A) and Western blot (B) analyses of the fermentation supernatants of the transformants producing recombinant Xyn11A using Man5A core-hinge carriers. Lanes: 1, purified Xyn11A (1 μg in panel A and 200 ng in panel B); 2, T. reesei ALKO3620; 3, ALKO4332(pALK1010); 4, ALKO4396(pALK945); 5, ALKO4399(pALK948); 6, ALKO4405(pALK1022); 7, ALKO4402(pALK1021). A total of 2 μl of the undiluted and 2 μl of the 1:100 diluted culture supernatants was applied to the Coomassie blue-stained (A) and Western-blotted (B) SDS-polyacrylamide gels. A rabbit polyclonal antibody synthesized against the native Xyn11A was used to detect the recombinant Xyn11A protein. The positions of the Man5A core-hinge- Xyn11A fusion protein (a), Man5A core (b), and Xyn11A (c) are marked by arrows in panel A.
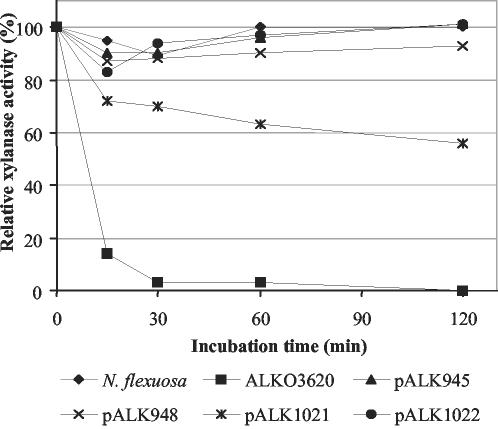
FIG. 3.
Thermostability of xylanase activity on culture supernatants deriving from strains producing recombinant Xyn11A. The thermostability of the xylanase activity was measured in the shake flask cultivation supernatants of the recombinant Xyn11A-producing T. reesei ALKO3620 transformants carrying the expression cassettes pALK945, pALK948, pALK1021, and pALK1022 and in the supernatant of the N. flexuosa fermentation cultivation. Samples to which BSA was added to a concentration of 100 μg/ml were incubated at pH 7 and 70°C, and the xylanase activity was measured at various time points from 0 to 120 min at pH 7 and 70°C using a reaction time of 5 min.
The largest amount of xylanase, 9,150 nkat/ml, representing 600 mg of Xyn11A protein, was measured in the culture supernatant of ALKO4405 carrying the expression cassette pALK1022. The T. reesei transformants ALKO4396(pALK945) and ALKO4399(pALK948) produced activities representing ∼550 mg of Xyn11A. The recombinant Xyn11A constituted 5 to 6% of the proteins secreted into the culture supernatants of the transformants. The mannanase activities were from ∼4,000 to 4,300 nkat/ml, representing ∼1.9 to 2 mg of the Man5A core protein/ml and constituting ∼20% of the proteins. The relative amount of Xyn11A was only one-third of the amount of the Man5A core-hinge carrier. Several single-copy transformants grown in 50-ml shake flask cultivations (data not shown) gave similar molar ratios for the Xyn11A and Man5A core-hinge carrier, from 2.5 to 3.2. The production levels in the shake flask cultivations were ∼70 to 80% of those obtained in the fermentor. The estimations for the amounts of Xyn11A calculated from the activity values correlate with the estimations from SDS-polyacrylamide gel electrophoresis (PAGE) gels and Western blots in which dilutions of the culture supernatant were run and the purified protein was used as a control (data not shown).
The mannanase and xylanase activities assayed from the culture supernatant of ALKO4402(pALK1021) were similar to those from the other three transformants. Estimations from SDS-PAGE gels suggest that the amount of Xyn11A (present in fused and unfused forms) was approximately the same as those from the transformants carrying the expression cassettes pALK945, pALK948, and pALK1022. However, it was not possible to make an exact estimate of the amount of Xyn11A produced by this strain. We do not know the activities of the partners in the full-length fusion protein, the fusion protein was partially cleaved, and significant proteolysis was observed (Fig. 2).
T. reesei strain ALKO4332 producing the Man5A core (pALK1010) produced about two times more Man5A core than the strains in which the Man5A core-hinge was used as a carrier of Xyn11A. The proportion of the Man5A core protein from the proteins secreted into the culture supernatant was also higher, ∼30%. The differences in the production levels of Xyn11A, Man5A core-hinge, and Man5A core can be seen on an SDS-PAGE gel (Fig. 2A).
The recombinant Xyn11A protein expressed from pALK945, pALK948, and pALK1022 had the native Xyn11A N terminus (data not shown). Thus, the cleavage of at least the major portion of the fusion products occurred after the dipeptide Lys-Arg (pALK948 and -1022) or after Arg (pALK945). The recombinant Xyn11A was detected as one or two major protein bands in the Western blot filters (Fig. 2B). The band with a molecular mass of ∼37 kDa migrates in parallel with the native purified Xyn11A. The other major band, with ∼2- to 3-kDa-higher molecular mass, was clearly detected in the supernatants of the strains carrying the expression cassettes pALK948 and pALK1022. This band represents a glycosylated form of Xyn11A, as proved by deglycosylation of the samples with endoglycosidase H (EndoHf) and peptide:N-glycosidase F (PNGase F)(data not shown). The strain carrying the expression cassette from pALK1021 produced an unprocessed full-length fusion protein of ∼80 kDa (the calculated molecular mass of the fusion protein is 74.8 kDa), but in addition, a band corresponding to the 37-kDa Xyn11A was detected. Also, several faint bands with molecular masses between 37 and 80 kDa were visible. These bands probably represent proteolytically cleaved forms of the fusion protein. The unprocessed fusion protein could also be faintly detected in the culture supernatants of the other transformants. The amounts were very small in the supernatants of the transformants containing the expression cassettes pALK948 and pALK1022, coding for the synthetic Kex2 protease-processing site. A polypeptide of ∼30 kDa was detected in all of the culture supernatants. This form of Xyn11A has the native N terminus (data not shown), suggesting that it has been cleaved from the C-terminal end.
The recombinant Xyn11A proteins had the same thermostability as the xylanase activity in the N. flexuosa cultivation supernatant (Fig. 3). The xylanase activities were stable for at least 2 h at 70°C and pH 7. The product from pALK1021 was an exception and lost nearly half of its activity during 2 h of incubation. The culture supernatants were also found to increase the brightness of pulp in the laboratory scale peroxide bleaching of kraft pulp at high temperature and pH in the same way as the culture supernatant from N. flexuosa (data not shown).
An intact domain structure of the carrier polypeptide is beneficial for high-yield production of Xyn11A.
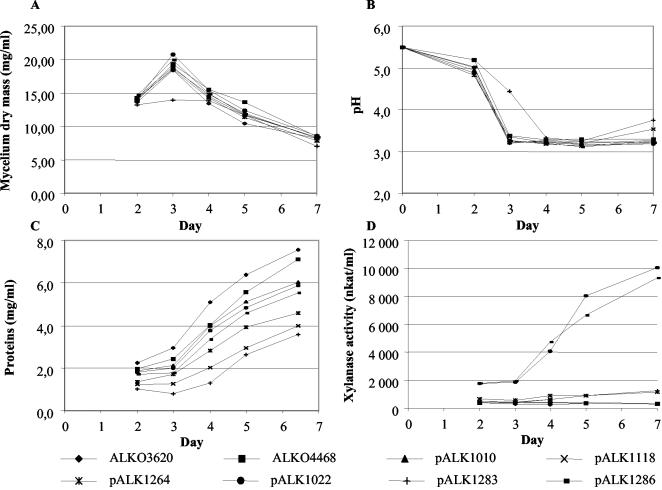
A variety of small polypeptides deriving from Cel6A and having different structures were tested as carriers and compared to the Man5A core-hinge-derived carrier from pALK1022 (Fig. 4). The Cel6a CBD (block A) (42) was encoded as a carrier in pALK1284. The coding regions for the CBD-hinge (blocks A and B) and CBD-duplicated hinge (blocks A, B, and B′) were used in pALK1285 and pALK1286 to evaluate the effects of the flexible linker region on the production and efficiency of cleavage of the fusion protein. Two incomplete domain structures, fragments from the Man5A core and Cel6A CBD, were included as carriers in pALK1264 and pALK1283 to analyze whether an intact domain structure is needed to achieve high production levels. A construct in which xyn11A was fused to the man5A signal sequence, pALK1118, was used as a control in the experiments. The expression cassettes were transformed into T. reesei host strains, single-copy integrants in which the expression cassettes replaced the cel7A locus were selected, and the chosen transformants were grown in shake flasks similarly to those with the previous Man5A core-hinge constructs. The production levels from several single-copy transformants were similar to each other. One transformant from each construct was grown in the fermentor and in 200-ml shake flask cultivations to analyze the growth, production levels, cleavage of Xyn11A from the carrier, and expression levels of the fusion mRNAs from the cel7A promoter (Fig. 5, 6, 7, and 8).
FIG. 5.
Results from 200-ml shake flask cultivations of T. reesei transformants producing Xyn11A. The strains were grown in parallel flasks for 7 days, and the growth was followed by determining the pH and mycelium dry mass. The masses of the lyophilized mycelia (A), the changes in pH (B), the amounts of the proteins secreted into the culture supernatants (C), and the xylanase activities (D) during cultivation are shown. The strains carrying the expression cassettes pALK1284 and pALK1285 were not included in the 200-ml cultivations. The values from duplicate flasks did not differ from each other on average by more than 2.2% (pH), 6.3% (biomass concentration), 6.7% (protein), and 7.0% (xylanase).
FIG. 6.
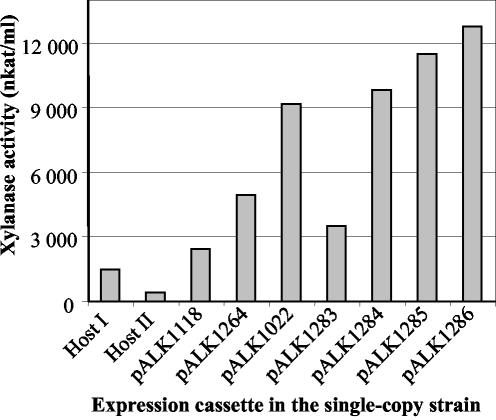
Production of recombinant Xyn11A in 1-liter fermentor cultivations using intact and incomplete domain structures as carrier polypeptides. The xylanase activities in the culture supernatants were analyzed after 5 days of fermentation; one fermentation was performed for each strain. The T. reesei strain ALKO4468 (Host II) was used as a host for the strains harboring pALK1118 and pALK1264 expression cassettes (ALKO4766 and ALKO4823), and ALKO3620 (Host I) was the host in the other transformations, resulting in ALKO4405(pALK1022), RF5007(pALK1283), RF5010(pALK1284), RF5013(pALK1285), and RF5017(pALK1286).
FIG. 7.
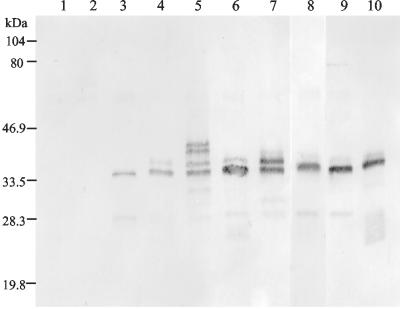
Western blot analysis of the fermentation supernatants of T. reesei transformants producing Xyn11A. Samples (3 μl of 1:100 dilutions of the culture supernatants) were run in SDS-PAGE, and the recombinant Xyn11A was detected from the Western blot filter by using a polyclonal antibody synthesized against the native Xyn11A. Lanes 1 and 2, T. reesei host strains ALKO3620 and ALKO4468; lanes 3 to 9, T. reesei transformed with the expression cassettes pALK1118, pALK1283, pALK1284, pALK1285, pALK1286, pALK1264, and pALK1022 (the same strains as in Fig. 6); lane 10 contains a 200-ng sample of the purified native Xyn11A protein.
FIG. 8.
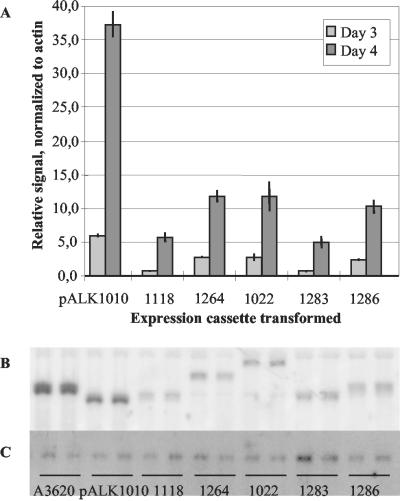
Northern blot results for fusion mRNAs expressed from the cel7A promoter. Samples of total RNA (2 μg), isolated from mycelia grown in shake flask cultivations for 3 and 4 days, were loaded in all lanes. The signals obtained by using a 32P-labeled oligonucleotide hybridizing to the untranslated region of the cel7A promoter were normalized using an actin probe. (A) Relative levels of mRNAs transcribed from the cel7A promoter. The mRNA levels shown are related to the level of the cel7A mRNA from T. reesei ALKO3620 mycelium analyzed on day 4, adjusted to 100. The bars indicate the averages from two parallel cultivations. The variation between the signals analyzed from the samples is shown by the vertical lines. (B and C) Northern blot filters of mycelia grown for 4 days and probed with the 32P-labeled oligonucleotide probe (B) and with the actin probe (C). The samples in the two lanes of each set of mycelia are from the two separate shake flask cultivations.
The strains grew similarly in the 200 ml cultivations, according to pH and mycelium dry mass (Fig. 5A and B), with the exception of RF5007 carrying the pALK1283 construct (the Cel6A CBD fragment). The decrease in pH in the culture supernatant of this strain was slower, and the mycelium dry mass did not reach the maximum levels of the others. The degrees of dependence of protein secretion into the culture medium on time were similar in all the cultivations (Fig. 5C). The highest levels of proteins (7.1 and 7.6 mg/ml) were produced by the host strains. The transformants could be divided into two groups according to the levels of proteins and xylanase activity they produced. The strains in which the carriers for Xyn11A had an intact domain structure (ALKO4405/pALK1022 and RF5017/pALK1286) produced higher levels of xylanase activity: 5.9 and 5.5 mg/ml of protein and ∼10,000 nkat/ml. Also, the strain producing the Man5A core (ALKO4332/pALK1010) belonged to this group (6.0 mg/ml of protein). The strains that had the signal sequence fusion to xyn11A or an incomplete domain structure as a carrier (ALKO4766/pALK1118, ALKO4823/pALK1264, and RF5007/pALK1283) produced lower levels of proteins and xylanase activity, 4.0, 4.6, and 3.6 mg/ml and only 1,100 to 1,200 nkat/ml. The background activity from the native Trichoderma xylanases was ∼300 nkat/ml under the conditions of the assay.
The results from the fermentor cultivations (Fig. 6) also showed that the best xylanase activities (12,780 and 11,500 nkat/ml, corresponding to 820 and 740 mg/liter) were obtained from the T. reesei strains in which the Cel6A CBD-hinge and CBD-duplicated hinge were used as carriers for Xyn11A (pALK1286 and pALK1285). When the Man5A core-hinge or Cel6A CBD without the hinge were used as carriers (pALK1022 and pALK1284), levels of 9,200 and 9,830 nkat/ml were reached (590 and 630 mg/liter). The xylanase activities from the strains without a carrier polypeptide (pALK1118/man5A signal sequence) or with an incomplete domain structure as a carrier (pALK1264 and pALK1283) were 2,460 to 4,940 nkat/ml (160 to 320 mg/liter). When the Cel6A CBD constructs were used as carriers, ∼10% of the proteins secreted consisted of Xyn11A. This higher relative level of Xyn11A (compared to 5 to 6% with the Man5A core-hinge) was due to both a higher level of xylanase activity in the cultivations and a lower level of proteins (6.8 to 8.1 mg/ml) produced by the strains with the Cel6A carriers compared to the strain with the Man5A core-hinge carrier (9.5 mg/ml).
The molecular mass of the recombinant Xyn11A was also similar to that of the native enzyme when Cel6A CBD was used as a carrier (Fig. 7). Thus, the fusion proteins appear to be correctly cleaved. Also, a glycosylated form of the protein with ∼2-kDa-higher molecular mass, as proved by deglycosylation of the samples by EndoHf and PNGaseF (data not shown), was detected in the culture supernatants. The relative amounts of the glycosylated form were different in the cultivations. The highest level of the glycosylated form was produced by the transformant containing the pALK1286 expression cassette. When Xyn11A was produced as a fusion to Cel6A CBD without the flexible linker (pALK1284), part of the product was detected as an uncleaved fusion protein with ∼7-kDa-higher molecular mass in the gel (the calculated mass of the Cel6A CBD is 4.6 kDa). Also, a glycosylated form of this fusion protein was visible (as proved by deglycosylation of the sample [data not shown]). Some degradation of Xyn11A was visible in all of the samples. In addition to the ∼30-kDa form representing Xyn11A cut from its C terminus, some polypeptides with lower molecular masses could be detected.
There is a block for production at the transcriptional level.
Total cellular RNAs were isolated from the mycelia cultivated for 3 and 4 days in the 200-ml shake flask cultivations (Fig. 5). The RNAs were probed using an oligonucleotide hybridizing to the untranslated region of the cel7A promoter (Fig. 8B), and the signals obtained were normalized using an actin probe (Fig. 8C).
The expression levels from the cel7A promoter were at their maximum in the samples from day 4, but the differences in the mRNA levels were already visible in the samples from day 3 (Fig. 8A). The highest relative levels of the fusion mRNA were detected in the strains producing the highest levels of the recombinant Xyn11A (Fig. 5D and 6), those in which the carriers had an intact domain structure (pALK1022 and pALK1286). The strains having fusions to the signal sequence or to the Cel6A CBD fragment (pALK1118 and pALK1283) accumulated only half of this amount of mRNA. Surprisingly, a high level of mRNA (equivalent to the amounts measured in the strains with pALK1022 and pALK1286) was also detected in the strain carrying the construct pALK1264 but producing a low level of Xyn11A.
Even the highest levels of the fusion mRNAs were only 10 to 12% of the level of the cel7A mRNA of the host and about one-third of the amount of the man5A core mRNA from ALKO4332(pALK1010). The mRNA level of ALKO4332 producing the Man5A core was 37% of the cel7A mRNA level of the host.
DISCUSSION
We produced the N. flexuosa 37-kDa xylanase, Xyn11A, in T. reesei using a variety of carrier polypeptides, including Man5A, Cel6A CBD, and fragments from both Man5A and Cel6A CBD. A carrier polypeptide was shown to be beneficial in obtaining high levels of recombinant Xyn11A. The transformants could be divided into two groups in their xylanase production, according to the type of carrier encoded by the expression cassette (Fig. 5D). Those having carriers with intact domain structures, Man5A core-hinge in pALK1022 and Cel6A CBD in pALK1286, produced xylanase activity of ∼10,000 nkat/ml in shake flask cultivations. The yields from the strains containing the incomplete carrier structures, fragments from the Man5A core (pALK1264) or Cel6A CBD (pALK1283), were eight- to ninefold less, 1,100 to 1,200 nkat/ml. In the fermentor cultivations, similar results were obtained even though the differences in yields between the two types of carriers were not so big (Fig. 6). The strain with the signal sequence fusion (pALK1118) produced the lowest yield in the fermentations. We suggest that the structure of the carrier has an important role in obtaining increased yields of the heterologous product, while the actual size of the carrier is not as meaningful.
The gene fusion constructs that have been successfully used to improve the yields of heterologous proteins in filamentous fungi have, in most of the reported cases, consisted of the whole secreted fungal protein or the core (and hinge) of such a protein, corresponding to the Man5A core-hinge construct we used. In addition to the high-molecular-mass carriers, sequences coding for shorter polypeptides, prosequences, and additional N-terminal amino acids of the well-secreted fungal proteins have been tested as carriers. With these, improvements in yields have usually not been obtained or they have been low compared to the yields with a long carrier polypeptide (45, 48). In Trichoderma an ∼1.4-fold increase in the production of chymosin was obtained when a sequence coding for 20 amino acids of the mature Cel7A (CBHI) was included prior to the chymosin sequence (16), but in this case the production level was also improved about fivefold with a fusion to the Cel7A core-hinge (Uusitalo, unpublished; referred to in reference 32). The Cel6A CBD carriers we used consisted of only 38 to 89 amino acids (8.5 to 19.9% of Cel6A), but still, levels of the recombinant Xyn11A similar to or even higher than those obtained using the Man5A core-hinge carriers (387 to 391 amino acids; 92.6 to 93.5% of Man5A) were achieved. We have also tested the 8-amino-acid-long Man5A prosequence as a short carrier polypeptide (pALK1116), but its use did not increase the yield of the recombinant Xyn11A, compared to the signal sequence fusion (pALK1118), in either the shake flask or fermentor cultivations (data not shown).
The recombinant Xyn11A was efficiently cleaved from the Man5A core-hinge when the Kex2 cleavage signal Lys-Arg was included in the construct. The result was as expected, since T. reesei has been shown to possess a Kex2-type dibasic endopeptidase activity (10; Demolder et al., Abstr. 2nd Eur. Conf. Fungal Genet.). We also tested another putative proteolytic site, an amino acid triplet, Tyr-Gly-Arg, in the construct pALK945. This triplet was analogous to that described previously (30) between the Cel7A core-hinge and the heavy Fd chain of murine anti-2-phenyloxazolone immunoglobulin G1 antibody. The Cel7A-Fd fusion was cleaved between Tyr and Gly by an uncharacterized T. reesei protease. The Man5A core-hinge- Xyn11A fusion protein was cleaved differently, after Arg. The most suitable cleavage site might finally depend on the structures of both fusion protein partners, not only on the amino acid sequence of the linker region, as has been suggested (e.g., 8, 38).
The hinge (linker) that naturally separates the catalytic and substrate-binding domains is thought to permit separate folding of the two independent domains. The hinge region, when included between the Cel6A CBD carrier and Xyn11A, was shown to have a positive effect on the production level (Fig. 6, compare pALK1285 and -1286, including the hinge- double-hinge regions, to pALK1284). Also, the cleavage of the fusion polypeptide was more efficient (Fig. 7). We presume that without the hinge the Kex2 cleavage site might be embedded in the fusion protein structure and thus not be efficiently recognized by the protease. Still, it seems that the three-dimensional structure of the polypeptides and fusion polypeptide has more effect on the efficiency of the cleavage than the hinge. Efficient cleavage was also obtained for the fusion products from the expression cassettes pALK1264 and pALK1283 (carriers of incomplete domain structures), in which no hinge region was included.
The Xyn11A sequence has five putative sites for N glycosylation. According to Western blot analysis (Fig. 2 and 7), the recombinant Xyn11A was produced as one or two major forms. The lower band in the gel had the same molecular mass as the native unglycosylated Xyn11A. The upper band was N glycosylated, as proved by using EndoH and PNGase treatment (data not shown). Also, a glycosylated form of the uncleaved fusion protein could be detected in the culture supernatants of the transformants carrying the expression cassettes pALK1021 and pALK1284. The relative amounts of the glycosylated form differed depending on the construct used and on the cultivation conditions (shake flask versus fermentor [data not shown]). N-glycans have a stabilizing effect on proteins (47), but whether there are differences in the properties of the two major forms of Xyn11A was not analyzed.
There was a correlation between the level of the mRNA and the level of the recombinant Xyn11A produced. The transformants producing the highest levels of Xyn11A also expressed the highest levels of mRNA from the cel7A promoter (Fig. 5D and 8), with the exception of the strain including the expression cassette pALK1264 (see below). The mRNA levels from the fusion constructs were higher than those from the fusion to the signal sequence. However, there appeared to be a restriction on production at the mRNA level. The levels of mRNA encoding the fusion protein even in the transformants producing the highest levels of Xyn11A were ∼12% of the amount of the cel7A mRNA from the host and about one-third of the level of mRNA expressed from pALK1010 in strain ALKO4332 producing the Man5A core. The lower level of mRNA from the constructs including the N. flexuosa xyn11A gene could be partly due to the difference in codon usage, which might result in either a lower transcription level or unstable mRNA. The lower mRNA level might also be due to some destabilizing motifs in the fusion transcript, or there might be some stabilizing regions included in the native cel7A transcript. Interestingly, the level of the Man5A core mRNA expressed using the cel7A promoter was also less, ∼37% of the level of the cel7A mRNA of the untransformed host. Why the level of the mRNA from a gene coding for a portion of a native Trichoderma protein, Man5A, is also lower than that from the cel7A gene is not known.
In addition to the restriction on production at the mRNA level, a posttranscriptional bottleneck(s) for the production of the bacterial xylanase in T. reesei can be presumed. The amount of the Man5A core-hinge deriving from the fusion protein was ∼1.9-fold less than the amount of the genetic Man5A core analyzed from an analogous T. reesei transformant, ALKO4332 (Table 1). This result is in quite good agreement with the threefold-lower level of mRNA from the fusion constructs. However, about three times more carrier polypeptide (Man5A core-hinge) than recombinant Xyn11A was detected in the culture supernatants of the strains containing the carrier constructs pALK945, pALK948, and pALK1022 (Table 1). Also, the transformant containing the expression cassette pALK1264 produced nearly 10-fold less Xyn11A, but the amount of mRNA was similar to those of the strains carrying pALK1022 and pALK1286. One of the main reasons for the lower yield of the heterologous proteins has been shown to be proteolysis, both in the growth medium (2) and inside the cell, in the last stages of the secretion pathway or by mycelium-bound proteases (11). Some proteolysis of the recombinant Xyn11A could be observed in the Western blots (Fig. 2 and 7), which at least partly explains the lower relative level of Xyn11A compared to the Man5A core-hinge. The proteolysis was enhanced in the culture supernatant by prolonged incubation and change of pH from 3 to 5 (not shown). The discrepancy in the relative amounts of the carrier and the heterologous protein from the fusions has been reported previously for several proteins, e.g., chymosin (48), Fab antibodies (31), and human interleukin-6 (13). Also, there are published examples in which such a discrepancy was not observed and the two protein partners were produced in equimolar amounts, e.g., interleukin-6, hen egg white lysozyme, and guar α-galactosidase expressed from a synthetic gene (8, 13, 21).
It has been shown that substantially higher levels of human interleukin-6 could be obtained from Aspergillus niger by using a fusion construct without the Kex2 site (6). We could not show any clear increase in the amount of Xyn11A from a pALK1021 fusion construct (Man5A core-hinge- Xyn11A), estimated from SDS-PAGE gels and Western blots. However, the estimation of the amount of the fusion protein was difficult because its specific activity is not known and the product was partially cleaved by proteases.
In conclusion, the bacterial Xyn11A produced in T. reesei appeared to be correctly cleaved and had properties (thermostability and bleaching properties) similar to those of the xylanase in N. flexuosa culture medium. It was shown that a higher production yield could be obtained by using carriers having an intact domain structure than with those having an incomplete domain structure. The recombinant Xyn11A constituted from 6% (Man5A carriers) to 10% (Cel6A CBD carriers) of the secreted proteins. The best yield of the recombinant Xyn11A from 1-liter fermentations was ∼820 mg/liter. This yield was obtained using the Cel6 CBD-double hinge as a carrier. Thus, the Cel6A CBD seems to have promise as a low-molecular-mass carrier for heterologous proteins in T. reesei. However, the yield of Xyn11A should be further increased to obtain industrially relevant production levels. In further strain constructions, several optimizations can be made, including changing the codons of the heterologous gene to better resemble the codon usage of the host, increasing the gene copy number, and mutagenization of the production strain. Also, an interesting area for further research will be to study how the different constructs induce the “unfolded protein response” pathway.
Acknowledgments
We thank Merja Helanterä, Jaana Oksanen, Outi Könönen, and Varpu Backman for skillful technical assistance. We acknowledge Elke Parkkinen, Sirpa Holm, Sirpa Okko, and Sanna Hiljanen-Berg for performing the laboratory-scale fermentations and Satu Hakola for constructing the plasmid pALK1118. We thank Tiina Pakula for her valuable help in analyzing the signals from the Northern blot filters and Nisse Kalkkinen for analyzing the N-terminal sequences of the recombinant Xyn11A proteins. We are grateful to Jari Vehmaanperä for useful comments, and John Londesborough is acknowledged for critically reading the manuscript and for correcting the language.
REFERENCES
- 1.Aho, S., V. Olkkonen, T. Jalava, M. Paloheimo, R. Bühler, M.-L. Niku-Paavola, E. H. Bamford, and M. Korhola. 1991. Monoclonal antibodies against core and cellulose-binding domains of Trichoderma reesei cellobiohydrolases I and II and endoglucanase I. Eur. J. Biochem. 200:643-649. [DOI] [PubMed] [Google Scholar]
- 2.Archer, D. B., D. A. MacKenzie, D. J. Jeenes, and I. N. Roberts. 1992. Proteolytic degradation of heterologous proteins expressed in Aspergillus niger. Biotechnol. Lett. 14:357-362. [Google Scholar]
- 3.Bailey, M. J., and K. M. H. Nevalainen. 1981. Induction, isolation and testing of stable Trichoderma reesei mutants with improved production of solubilizing cellulase. Enzyme Microb. Technol. 3:153-157. [Google Scholar]
- 4.Bailey, M. J., P. Biely, and K. Poutanen. 1992. Interlaboratory testing of methods for assay of xylanase activity. J. Biotechnol. 23:257-270. [Google Scholar]
- 5.Benson, S. A. 1984. A rapid procedure for isolation of DNA fragments from agarose gels. BioTechniques 2:66-68. [Google Scholar]
- 6.Broekhuijsen, M. P., I. E. Mattern, R. Contreras, J. R. Kinghorn, and C. A. M. J. J. Van den Hondel. 1993. Secretion of heterologous proteins by Aspergillus niger: production of active human interleukin-6 in a protease-deficient mutant by KEX2-like processing of a glucoamylase-hIL6 fusion protein. J. Biotechnol. 31:135-145. [DOI] [PubMed] [Google Scholar]
- 7.Calmels, T. P. G., F. Martin, H. Durand, and G. Tiraby. 1991. Proteolytic events in the processing of secreted proteins in fungi. J. Biotechnol. 17:51-66. [DOI] [PubMed] [Google Scholar]
- 8.Contreras, R., D. Carrez, J. R. Kinghorn, C. A. M. J. J. van den Hondel, and W. Fiers. 1991. Efficient Kex2-like processing of a glucoamylase-interleukin-6 fusion protein by Aspergillus nidulans and secretion of mature interleukin-6. Bio/Technology 9:378-381. [DOI] [PubMed] [Google Scholar]
- 9.Drocourt, D., T. Calmels, J.-P. Reynes, M. Baron, and G. Tiraby. 1990. Cassettes of the Streptoalloteichus hindustanus ble gene for transformation of lower and higher eukaryotes to phleomycin resistance. Nucleic Acids Res. 18:4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goller, S. P., D. Schoisswohl, M. Baron, M. Parriche, and C. P. Kubicek. 1998. Role of endoproteolytic dibasic proprotein processing in maturation of secretory proteins in Trichoderma reesei. Appl. Environ. Microbiol. 64:3203-3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gouka, R. J., P. J. Punt, J. G. M. Hessing, and C. A. M. J. J. van den Hondel. 1996. Analysis of heterologous protein production in defined recombinant Aspergillus awamori strains. Appl. Environ. Microbiol. 62:1951-1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gouka, R. J., P. J. Punt, and C. A. M. J. J. van den Hondel. 1997. Efficient production of secreted proteins by Aspergillus: progress, limitations and prospects. Appl. Microbiol. Biotechnol. 47:1-11. [DOI] [PubMed] [Google Scholar]
- 13.Gouka, R. J., P. Punt, and C. A. M. J. J. van den Hondel. 1997. Glucoamylase gene fusions alleviate limitations for protein production in Aspergillus awamori at the transcriptional and (post)transcriptional levels. Appl. Environ. Microbiol. 63:488-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greiner-Mai, E., R. M. Kroppenstedt, F. Korn-Wendisch, and H. J. Kutzner. 1987. Morphological and biochemical characterization and emended descriptions of thermophilic actinomycetes species. Syst. Appl. Microbiol. 9:97-109. [Google Scholar]
- 15.Gritz, L., and J. Davies. 1983. Plasmid-encoded hygromycin B resistance: the sequence of hygromycin B phosphotransferase gene and its expression in Escherichia coli and Saccharomyces cerevisiae. Gene 25:179-188. [DOI] [PubMed] [Google Scholar]
- 16.Harkki, A., J. Uusitalo, M. Bailey, M. Penttilä, and J. K. C. Knowles. 1989. A novel fungal expression system: secretion of active calf chymosin from the filamentous fungus Trichoderma reesei. Bio/Technology 7:596-603. [Google Scholar]
- 17.Holtz, C., H. Kaspari, and J.-H. Klemme. 1991. Production and properties of xylanases from thermophilic actinomycetes. Antonie Leeuwenhoek 59:1-7. [DOI] [PubMed] [Google Scholar]
- 18.Ilmén, M. 1997. Molecular mechanisms of glucose repression in the filamentous fungus Trichoderma reesei. VTT publication 315. VTT Offsetpaino, Espoo, Finland.
- 19.Irwin, D., E. D. Jung, and D. B. Wilson. 1994. Characterization and sequence of a Thermomonospora fusca xylanase. Appl. Environ. Microbiol. 60:763-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jeenes, D. J., D. A. MacKenzie, I. N. Roberts, and D. B. Archer. 1991. Heterologous protein production by filamentous fungi. Biotechnol. Genet. Eng. Rev. 9:327-367. [PubMed] [Google Scholar]
- 21.Jeenes, D. J., D. A. MacKenzie, and D. B. Archer. 1994. Transcriptional and post-transcriptional events affect the production of secreted hen egg white lysozyme by Aspergillus niger. Transgenic Res. 3:297-303. [DOI] [PubMed] [Google Scholar]
- 22.Joutsjoki, V. V., T. K. Torkkeli, and K. M. H. Nevalainen. 1993. Transformation of Trichoderma reesei with the Hormoconis resinae glucoamylase P (gamP) gene: production of a heterologous glucoamylase by Trichoderma reesei. Curr. Genet. 24:223-228. [DOI] [PubMed] [Google Scholar]
- 23.Karhunen, T., A. Mäntylä, K. M. H. Nevalainen, and P. L. Suominen. 1993. High frequency one-step gene replacement in Trichoderma reesei. I. Endoglucanase I overproduction. Mol. Gen. Genet. 241:515-522. [DOI] [PubMed] [Google Scholar]
- 24.Kelly, J. M., and M. J. Hynes. 1985. Transformation of Aspergillus niger by the amdS gene of Aspergillus nidulans. EMBO J. 4:475-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lowry, O. H., N. J. Roseborough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the folin phenol reagent. J. Biol. Chem. 193:265-275. [PubMed] [Google Scholar]
- 26.Mach, R. L., M. Schindler, and C. P. Kubicek. 1994. Transformation of Trichoderma reesei based on hygromycin B resistance using homologous expression signals. Curr. Genet. 25:567-570. [DOI] [PubMed] [Google Scholar]
- 27.Mäntylä, A., M. Paloheimo, and P. Suominen. 1998. Industrial mutants and recombinant strains of Trichoderma reesei, p. 291-309. In C. P. Kubicek and G. E. Harman (ed.), Trichoderma and Gliocladium, vol. 2. Taylor and Francis Ltd., London, United Kingdom.
- 28.Matheucci, E., F. Henrique-Silva, S. el-Gogary, C. H. Rossini, A. Leite, J. E. Vera, J. C. Urioste, O. Crivellaro, and H. el-Dorry. 1995. Structure, organization and promoter expression of the actin-encoding gene in Trichoderma reesei. Gene 8:103-106. [DOI] [PubMed] [Google Scholar]
- 29.Mattern, I. E., P. Punt, and C. A. M. J. J. van den Hondel. 1988. A vector for Aspergillus transformation conferring phleomycin resistance. Fungal Genet. Newsl. 35:25. [Google Scholar]
- 30.Nyyssönen, E., M. Penttilä, A. Harkki, A. Saloheimo, J. K. C. Knowles, and S. Keränen. 1993. Efficient production of antibody fragments by the filamentous fungus Trichoderma reesei. Bio/Technology 11:591-595. [DOI] [PubMed] [Google Scholar]
- 31.Nyyssönen, E., and S. Keränen. 1995. Multiple roles of the cellulase CBHI in enhancing production of fusion antibodies by the filamentous fungus Trichoderma reesei. Curr. Genet. 28:71-79. [DOI] [PubMed] [Google Scholar]
- 32.Penttilä, M. 1998. Heterologous protein production in Trichoderma, p. 365-382. In C. P. Kubicek and G. E. Harman (ed.), Trichoderma and Gliocladium, vol. 2. Taylor and Francis Ltd., London, United Kingdom.
- 33.Penttilä, M., P. Lehtovaara, H. Nevalainen, R. Bhikhabhai, and J. Knowles. 1986. Homology between cellulase genes of Trichoderma reesei: complete nucleotide sequence of the endoglucanase I gene. Gene 45:253-263. [DOI] [PubMed] [Google Scholar]
- 34.Penttilä, M., H. Nevalainen, M. Rättö, E. Salminen, and J. Knowles. 1987. A versatile transformation system for the cellulolytic filamentous fungus Trichoderma reesei. Gene 61:155-164. [DOI] [PubMed] [Google Scholar]
- 35.Raeder, U., and P. Broda. 1985. Rapid preparation of DNA from filamentous fungi. Lett. Appl. Microbiol. 1:17-20. [Google Scholar]
- 36.Saloheimo, M., P. Lehtovaara, M. Penttilä, T. T. Teeri, J. Stålberg, G. Johansson, G. Pettersson, M. Clayessens, P. Tomme, and J. K. C. Knowles. 1988. EGIII, a new endoglucanase from Trichoderma reesei: characterization of both the gene and enzyme. Gene 63:11-21. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 38.Spencer, J. A., D. J. Jeenes, D. A. MacKenzie, D. T. Haynie, and D. B. Archer. 1998. Determinants of the fidelity of processing glucoamylase-lysozyme fusions by Aspergillus niger. Eur. J. Biochem. 258:107-112. [DOI] [PubMed] [Google Scholar]
- 39.Stålbrand, H., A. Saloheimo, J. Vehmaanperä, B. Henrissat, and M. Penttilä. 1995. Cloning and expression in Saccharomyces cerevisiae of a Trichoderma reesei β-mannanase gene containing a cellulose binding domain. Appl. Environ. Microbiol. 61:1090-1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suominen, P. L., A. L. Mäntylä, T. Karhunen, S. Hakola, and H. Nevalainen. 1993. High frequency one-step gene replacement in Trichoderma reesei. II. Effect of deletions of individual cellulase genes. Mol. Gen. Genet. 241:523-530. [DOI] [PubMed] [Google Scholar]
- 41.Suurnäkki, A., M. Tenkanen, J. Buchert, and L. Viikari. 1997. Hemicellulases in the bleaching of chemical pulps. Adv. Biochem. Eng. Biotechnol. 57:261-287. [DOI] [PubMed] [Google Scholar]
- 42.Teeri, T., P. Lehtovaara, S. Kauppinen, I. Salovuori, and J. Knowles. 1987. Homologous domains in Trichoderma reesei cellulolytic enzymes: gene sequence and expression of cellobiohydrolase II. Gene 51:43-52. [DOI] [PubMed] [Google Scholar]
- 43.Te'o, V. S. J., A. E. Cziferszky, P. L. Bergquist, and K. M. H. Nevalainen. 2000. Codon optimization of xylanase gene xynB from the thermophilic bacterium Dictyoglomus thermophilum for expression in the filamentous fungus Trichoderma reesei. FEMS Microbiol. Lett. 190:13-19. [DOI] [PubMed] [Google Scholar]
- 44.van den Hondel, C. A. M. J. J., P. Punt, and R. F. M. van Gorcom. 1991. Heterologous gene expression in filamentous fungi, p. 241-250. In J. W. Bennet and L. L. Lasure (ed.), More gene manipulations in fungi. Academic Press, San Diego, Calif.
- 45.van den Hondel, C. A M. J. J., P. Punt, and R. F. M. van Gorcom. 1992. Production of extracellular proteins by the filamentous fungus Aspergillus. Antonie Leeuwenhoek 61:153-160. [DOI] [PubMed] [Google Scholar]
- 46.Verdoes, J. C., P. Punt., and C. A. M. J. J. van den Hondel. 1995. Molecular genetic strain improvement for the overproduction of fungal proteins by filamentous fungi. Appl. Microbiol. Biotechnol. 43:195-205. [Google Scholar]
- 47.Wang, C., M. Eufemi, C. Turano, and A. Giartosio. 1996. Influence of the carbohydrate moiety on the stability of glycoproteins. Biochemistry 35:7299-7307. [DOI] [PubMed] [Google Scholar]
- 48.Ward, M., L. J. Wilson, K. H. Kodama, M. W. Rey, and R. M. Berka. 1990. Improved production of chymosin in Aspergillus by expression as a glucoamylase-chymosin fusion. Bio/Technology 8:435-440. [DOI] [PubMed] [Google Scholar]
- 49.Zeilinger, S., R. L. Mach, and C. P. Kubicek. 1998. Two adjacent protein binding motifs in the cbh2 (dellobiohydrolase II-encoding) promoter of the fungusHypocrea jecorina (Trichoderma reesei) cooperate in the induction by cellulose. J. Biol. Chem. 273:34463-34471. [DOI] [PubMed] [Google Scholar]
- 50.Zhang, Z., Y. Wang, and J. Ruan. 1998. Reclassification of Thermomonospora and Microtetraspora. Int. J. Syst. Bacteriol. 48:411-422. [DOI] [PubMed] [Google Scholar]