Abstract
Previously, we reported that mitogen-activated protein kinase kinase 1 (MEK1) activated in the mid-stage of skeletal muscle differentiation promotes myogenic differentiation. To elucidate the molecular mechanism, we investigated an activity of MEK1 for MyoD. Activated MEK1 associates with MyoD in the nucleus of differentiating myoblasts. In vitro kinase assay using active MEK1, a 32P-labeled protein band corresponding to GST-MyoD was observed but not to mutant GST-MyoD-Y156F. Tyrosine phosphorylation of endogenous MyoD was detected with a specific anti-pMyoD-Y156 antibody; however, this response was blocked by PD184352, a MEK-specific inhibitor. These results indicate that activated MEK1 phosphorylates the MyoD-Y156 residue directly. Interestingly, the protein level of mutant MyoD-Y156F decreased compared with that of wild type but was recovered in the presence of lactacystin, a proteasome inhibitor. The protein level of MyoD-Y156E, which mimics phosphorylation at Tyr-156, was above that of wild type, indicating that the phosphorylation protects MyoD from the ubiquitin proteasome-mediated degradation. In addition, the low protein level of MyoD-Y156F was recovered over that of wild type by an additional mutation at Leu-164, a critical binding residue of MAFbx/AT-1, a Skp, Cullin, F-box (SCF) E3-ubiquitin ligase. The amount of MyoD co-precipitated with MAFbx/AT-1 also was reduced in the presence of active MEK1. Thus, these results suggested that the phosphorylation probably interrupts the binding of MAFbx/AT-1 to MyoD and thereby increases its stability. Collectively, our results suggest that MEK1 activated in differentiating myoblasts stimulates muscle differentiation by phosphorylating MyoD-Y156, which results in MyoD stabilization.
Keywords: Cell Differentiation, Proteasome, Protein Degradation, Protein Phosphorylation, Signal Transduction, Skeletal Muscle, Ubiquitin, MAFbx/AT-1, MEK1, MyoD
Introduction
The terminal differentiation process of skeletal muscle is orchestrated by the family of myogenic regulatory factors (MRFs),2 a class of basic helix-loop-helix (HLH) transcription factors, including MyoD, Myf5, myogenin, and MRF4 (1, 2). These basic HLH transcriptional factors activate muscle-specific gene transcription by binding E-box sequences (CANNTG) as heterodimers with ubiquitous E proteins (e.g. E12, E47, and HeLa E-box binding protein), in cooperation with myocyte enhancer factor 2 family of MADS-box proteins (3). Among MRFs, MyoD is usually considered as “a determination factor” because it induces the withdrawal from the cell cycle as well as the activation of muscle-specific genes expression crucial for skeletal muscle differentiation (4). Thus, to elucidate the mechanism regulating stability as well as transcriptional activity of MyoD is critical in understanding skeletal muscle development and regeneration.
MyoD phosphorylation plays pivotal roles in regulating its stability as well as transcriptional activity. For example, MyoD phosphorylation at Ser-200 by Cdk1/2/cyclin E destabilizes MyoD through the ubiquitin/proteasome pathway (5, 6), which is blocked in the presence of p57kip2, a Cdk inhibitor (7). c-Abl1 activated by genotoxic stress phosphorylates MyoD at Tyr-30 directly, resulting in repression of its transcriptional activity (8). Mutation of MyoD at Thr-115, a putative PKC phosphorylation site, enhances transcriptional activity, suggesting that PKC-mediated MyoD phosphorylation at Thr-115 negatively regulates its function (9). By contrast, Mos, an upstream kinase of mitogen-activated protein kinase kinase (MEK)1/2 expressed in adult skeletal muscle (10), increases MyoD heterodimerization with E12 via direct phosphorylation of Ser-237, thus promoting myogenic differentiation (11, 12).
MyoD is also degraded via the ubiquitin-proteasome pathway (13, 14). Differential expression screening has identified two genes whose expression is significantly increased in atrophied skeletal muscles, muscle atrophy F-box/Astrogin-1 (MAFbx/AT-1) and muscle ring finger 1 (MuRF1) (15, 16), which encode E3-ubiquitin ligases (15). When knocked out, MAFbx/AT-1(−/−) and MuRF1(−/−) mice appeared resistant to the effects of denervation-induced muscle atrophy, with a 56 and 36% respective sparing of muscle loss, compared with littermate controls (15). Then, MAFbx/AT-1 and MuRF1 may be, in part, responsible for the ubiquitin proteasome-mediated muscle protein degradation observed in muscle atrophy conditions (17, 18). Recently, it was demonstrated that MyoD is a direct substrate of MAFbx/AT-1, and the LXXLL conserved motif sequence (residues 160–164) in the helix 2 region of the MyoD HLH domain is essential for MAFbx/AT-1 binding (19).
The ERK signaling cascade (Raf/MEK/ERK) appears to regulate numerous cellular events, including cell growth, migration, survival, and other biological processes (20). However, the physiological role of the ERK pathway in myogenic differentiation has been controversial (21–25). In the recent study employing the Tet-Off system expressing constitutively active nuclear MEK1 under the control of the Tet-response element, myogenic differentiation was inhibited if MEK1 activation was induced within 24 h of the change to differentiation medium (DM), but was stimulated after that point (26). These data suggest that the activation of MEK1 plays contrary stage-specific roles in skeletal myogenesis and that the activated MEK1 is a positive regulator in the mid-stage of skeletal myogenesis.
In an effort to understand the stimulatory role of MEK1 in the mid-stage of myogenic differentiation, we found a novel activity of MEK1 for MyoD. Here, we describe that activated MEK1 directly phosphorylates MyoD at Tyr-156 in helix 2 of the HLH domain, increasing its stability during skeletal myogenesis.
EXPERIMENTAL PROCEDURES
Reagents and Antibodies
U0126 and lactacystin were obtained from Biomol (Plymouth Meeting, PA). PD184352 was provided by R. Marquez (University of Dundee). Cycloheximide was purchased from Calbiochem. Protease inhibitor mixture and other chemical reagents were obtained from Sigma. Anti-MyoD (C-20), anti-ERK1/2 (K-23), and anti-pERK1/2 (E-4) antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA), anti-MEK1, anti-pMEK1 (Ser-217/Ser-221), anti-pTyr (P-Tyr-100), anti-pThr (42H4), anti-ubiquitin and anti-Myc (9B11) antibodies were from Cell Signaling Technology (Beverly, MA), anti-pSer (PSR-45), anti-FLAG (M2), and anti-α-tubulin (DM1A) antibodies were from Sigma, and monoclonal anti-MyoD (clone 5.8A) antibody was from BD Biosciences. Anti-pMEK1 (Ser-217/Ser-221) used for immunostaining was purchased from BioVision (Mountain View, CA). Anti-GST and anti-HA (12CA5) antibodies were obtained from Amersham Biosciences and Roche Diagnostics, respectively. The anti-MHC (MF20) supernatant was used at a 1:5 dilution, depending upon the preparation. Anti-MAFbx/AT-1 antibody was produced by immunizing rabbits with the synthetic peptide KKRKKDIQNSKTKTQY corresponding to amino acids 62–77 of MAFbx/AT-1. The antibody was affinity-purified against antigenic peptides (Peptron, Korea). Anti-pMyoD-Y156 antibody was generated by immunizing rabbits with the synthetic peptide AIR*YIEGL corresponding to amino acids 153–160 of MyoD with a phosphotyrosine at position 156 (*Y). The antibody was purified from serum by two rounds of affinity chromatography on a phospho-Tyr-156 peptide column, followed by a non-phosphopeptide column (Peptron).
Cell Culture
C2C12 myoblast cells (CRL-1772, ATCC, Manassas, VA) and C3H10T1/2 fibroblast cells (CCL-226, ATCC) were grown in growth medium (GM), DMEM supplemented with 10% fetal bovine serum, penicillin (50 units/ml), and streptomycin (100 μg/ml) (all from Invitrogen). Cells were maintained at 37 °C in humidified atmosphere with 95% air and 5% CO2. To induce muscle differentiation, C2C12 cells were allowed to grow ∼70–80% confluence in GM and then switched to DM (DMEM with 2% horse serum).
Transient Transfection and Luciferase Assays
Cells were grown to ∼80% confluence in 12-well plates (for reporter assay), on coverslips (for immunostaining), in six-well plates (for immunoblotting) or in 60-mm dishes (for immunoprecipitation) and transiently transfected with various combinations of plasmids as indicated in the figure legends using Lipofectamine (Invitrogen) according to the manufacturer's instructions. The total amount of DNA used for each well or plate was normalized with the relevant mock vectors. Luciferase activity was measured using the Luciferase Assay System (Promega, Madison, WI) and a luminometer (LB 9505, Berthold, Germany). Transfection efficiency was normalized with β-galactosidase activity.
Plasmids
EMSV-MyoD, HA-MEK1, and 4RE (4× E box) tk-CAT plasmids were kind gifts from M. A. Rudnicki (McMaster University; 24). Expression plasmids of the Gal4 DNA-binding domain fusion protein (pSG424) and Myc6-MAFbx/AT1 were generous gifts from B. R. Cullen (Duke University; 27) and A.L. Goldberg (Harvard Medical School; 28), respectively. Gal4-Luc (pFR-Luc) reporter plasmid was purchased from Stratagene (La Jolla, CA). To construct plasmids expressing FLAG-, Gal4-, and bacterial GST-MyoD, the MyoD gene was amplified by PCR using the EMSV-MyoD plasmid as a template and cloned into pCMV-tag2C (Stratagene), pSG424, and pGEX-3X (Amersham Biosciences), respectively. MyoD mutants (Y30V, Y156V, Y156F, Y192F, Y156E, S200G/Y156F, and L164Q/Y156F) were prepared using the QuikChange site-directed mutagenesis kit (Stratagene). The plasmid expressing His6-MEKEE (S218E/S222E) was constructed by amplifying the MEK1 gene by PCR using the MEKEE plasmid as a template and cloning into pQE-30 vector (Qiagen, Valencia, CA). Nuclear export signal (residues 32–51; 29, 30) deletion mutants of MEK1, HA-MEKEE (ΔN, S218E/S222E), and HA-MEKAA (ΔN, S218A/S222A), were constructed using the PCR-based gene fusion technique (31). To prepare plasmids expressing GFP-MEKEE (ΔN) and mCherry-MyoD, MEKEE (ΔN) and MyoD genes were amplified by PCR from HA-MEKEE (ΔN) and EMSV-MyoD plasmids, and the PCR products were cloned into pEGFP-N1 (Clontech, Mountain View, CA) and pmCherry (Clontech), respectively. The 4REtk-Luc reporter plasmid was constructed by subcloning the E box region four times from 4REtk-CAT into pGL2-Basic (Promega). Muscle creatine kinase-Luc reporter plasmid was previously described by Jo et al. (26). All constructs were confirmed by sequencing.
Immunoblotting
Cells were washed once with PBS and lysed with modified radioimmune precipitation assay buffer (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1 mm EGTA, 1% Nonidet P-40, 0.25% sodium deoxycholate, 0.1% (w/v) SDS, 1 mm NaF, 1 mm Na3VO4, 1× protease inhibitor mixture). Proteins were extracted on ice with periodic vortexing for 30–40 min, and lysates were cleared by centrifugation at 10,000 × g for 10 min at 4 °C. An aliquot (50 μg) of protein was separated on 10% SDS-PAGE and were electrotransferred onto a 0.2-μm nitrocellulose membrane in Towbin transfer buffer (192 mm glycine, 25 mm Tris, 20% (v/v) methanol, pH 8.3). The membrane was preincubated with PBS containing 5% (w/v) skim milk, and probed with a primary antibody in PBS containing 5% skim milk for 1 h at room temperature. The membrane was then washed with PBS containing 0.03% (v/v) Tween 20 and incubated with a corresponding HRP-conjugated secondary antibody (Amersham Biosciences). After several washes, the blot was developed using an ECL (Amersham Biosciences) according to the manufacturer's instructions. The protein concentration was determined by the BCA method (Sigma).
Immunofluorescence
Cells grown on coverslips were fixed with 4% (w/v) paraformaldehyde in PBS, followed by a 10-min permeabilization in 0.2% (v/v) Triton X-100 in PBS at 25 °C. pMEK1 and MyoD was detected using anti-pMEK1 (Ser-217/Ser-221, 1:200, BioVision) and anti-MyoD (clone 5.8A, 1:100, BD Biosciences) primary antibodies and Alexa Fluor 488- and 594-conjugated secondary antibodies (1:200, Invitrogen), respectively. Images were photographed using a confocal microscope (Carl Zeiss LSM710).
Preparation of Fusion Proteins and GST Pulldown Assay
Recombinant His6-MEKEE, GST, and GST-MyoD proteins expressed in Escherichia coli were purified using a NTA column (Qiagen) or glutathione-Sepharose (Amersham Biosciences) according to the manufacturers' instructions. For the GST pulldown assay, equal quantities (5 μg) of purified GST or GST-MyoD were bound to 20-μl glutathione beads (50% (w/v) slurry) by incubation for 1 h at 4 °C in GST binding buffer (50 mm Tris-HCl, pH 8.0, 10 mm MgCl2, 0.1% (v/v) Nonidet P-40, 0.5 mm EDTA, 1 mm DTT, and 1× protease inhibitor mixture). Then, equal quantities (3 μg) of purified, recombinant His6-MEKEE were added to GST- or GST-MyoD-loaded beads with or without 1 mm ATP. The reaction mixtures were additionally incubated for 2 h at 4 °C. GST-bound complexes were washed three times with GST binding buffer and subjected to immunoblotting.
IP and IP Kinase Assays
Cells were extracted in Nonidet P-40 lysis buffer (0.5% Nonidet P-40, 50 mm Tris-HCl, pH 8.0, 150 mm NaCl, 10 mm sodium pyrophosphate, 1 mm EDTA, 0.1 m NaF, 1 mm Na3VO4, 1× protease inhibitor mixture). Equal amounts of lysates were incubated with 2 μg of antibody and 20 μl of protein A-conjugated Sepharose beads (50% (w/v) slurry; Roche Diagnostics) at 4 °C overnight on a rotating platform. Beads were washed with three changes of NETN wash buffer (0.1% Nonidet P-40, 50 mm Tris-HCl, pH 8.0, 150 mm NaCl, 1 mm EDTA) and boiled in SDS sample buffer for 5 min. Proteins were separated by SDS-PAGE and immunoblotted as described above. For immunoprecipitation (IP) kinase assays, cells were extracted with TNN lysis buffer (50 mm Tris-HCl, pH 8.0, 0.5% Nonidet P-40, 120 mm NaCl, 1 mm Na3VO4, and 1× protease inhibitor mixture). Immunoprecipitates were washed twice with TNN lysis buffer and then washed twice with 1× kinase buffer (Cell Signaling Technology). The immunoprecipitates were resuspended in an equal volume of kinase buffer containing 5 μCi of [γ-32P]ATP and 2 μg of GST-MyoD and incubated at 37 °C for 30 min. Reactions were stopped by boiling in SDS sample buffer for 5 min. The reaction products were separated by SDS-PAGE, and phosphorylated proteins were detected by autoradiography.
Mass Spectrometry
GST-MyoD protein (10 μg) was phosphorylated with immunoprecipitated HA-MEKEE (ΔN) as described previously in the IP kinase assay. Phosphorylated GST-MyoD protein was digested by incubation with trypsin (Promega) in 10 mm NH4HCO3, pH 7.8, overnight. After diluting 1:1 with matrix solution (20 mg/ml 2,5-dihydroxylbenzoic acid in aqueous solution of 50% (v/v) acetonitrile containing 1% formic acid and 1% phosphoric acid), the samples were loaded onto a plate and allowed to dry. The digested peptides were analyzed on an Applied Biosystems (Foster City, CA) Voyager-DE STR MALDI-TOF-MS operated in the reflector mode. MALDI spectra were calibrated internally using trypsin autolysis products to get accurate monoisotopic masses of all the tryptic peptides.
RESULTS
Activated MEK1 Associates with MyoD in Nucleus
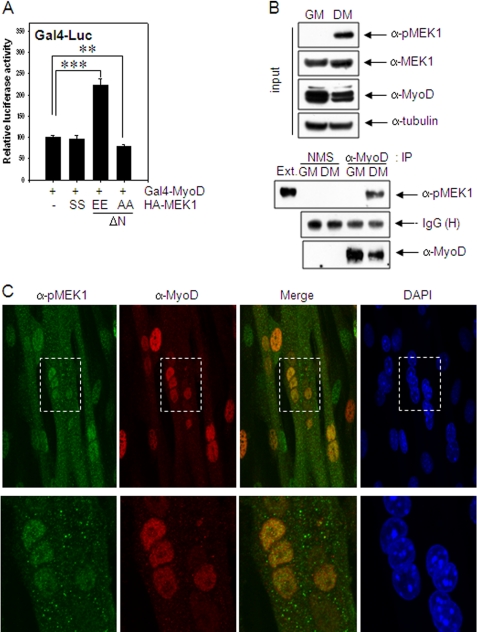
We observed previously that endogenous MEK1 is activated in the differentiating myoblasts and localized in the nucleus of the differentiating myoblasts (26). To examine whether activated MEK1 associates with MyoD in differentiating myoblasts, we used the modified mammalian two-hybrid, Gal4-Luc system in which the transcriptional activity of Gal4-MyoD depends on the association of a molecule binding to the fused bait protein Gal4-MyoD. Gal4-Luc activity of Gal4-MyoD was increased in differentiating C2C12 cells by the transfection of constitutively active MEK1 (HA-MEKEE (ΔN), S218E/S222E), whereas this response was reversed by dominant-negative MEK1 (HA-MEKAA (ΔN), S218A/S222A) (Fig. 1A).
FIGURE 1.
MEK1 associates and co-localizes with MyoD in the nucleus. A, C2C12 cells grown in GM were co-transfected with Gal4-Luc vector and Gal4-MyoD along with either HA-MEK1, HA-MEKEE (ΔN, S218E/S222E), or HA-MEKAA (ΔN, S218A/S222A), maintained in DM for 2 days and assayed for luciferase activity. B, C2C12 cells grown in GM or DM for 2 days were extracted, and cell extracts were immunoprecipitated with anti-MyoD (clone 5.8A) antibody or normal mouse serum (NMS). Co-precipitation of endogenous, activated MEK1 was analyzed by immunoblotting using anti-pMEK1 antibody. C, C2C12 cells grown to ∼70–80% confluence in GM were switched to DM to induce differentiation. Immunoreactivities of pMEK1 and MyoD in differentiating myotubes (3 days in DM) were examined by immunostaining using anti-pMEK1 and anti-MyoD antibodies, respectively, and visualized with a confocal microscope (Carl Zeiss LSM710) at ×400 (upper panel) or x630 (lower panel) magnifications. The area of dashed boxes in upper panels is enlarged and shown in lower panels. The results show the mean ± S.D. of triplicate experiments. **, p < 0.01; ***, p < 0.001.
The association of MEK1 with MyoD was also demonstrated by co-precipitation of endogenous phosphorylated MEK1 (pMEK1) with anti-MyoD antibody in differentiating muscle cells (Fig. 1B). Moreover, the immunoreactivity of pMEK1 overlapped that of MyoD in the nucleus (Fig. 1C). When EGFP-MEKEE (ΔN) and mCherry-MyoD were expressed in 10T1/2 cells by transient transfection, tagged MEK1 and MyoD were also co-localized in differentiating myotube, especially in the nuclei (supplemental Fig. S1). Because ChIP analysis demonstrated that muscle creatine kinase promoter was immunoprecipitated with anti-MyoD antibody but not with anti-pMEK1 antibody, activated MEK1 was likely to interact with free MyoD in the nucleus rather than to be recruited to a MyoD-responsive promoter (supplemental Fig. S2).
Activated MEK1 Phosphorylates MyoD Directly
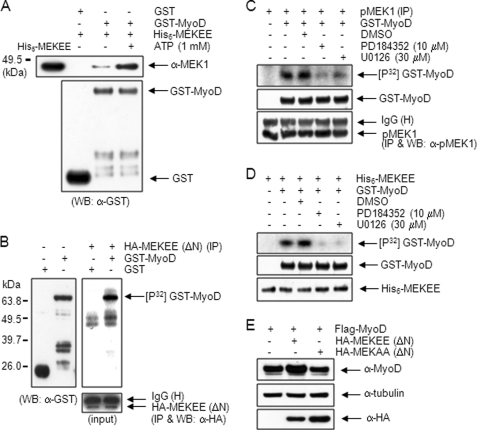
We performed the GST pulldown assay using recombinant His6-tagged constitutively active MEK1 (His6-MEKEE, S218E/S222E) and GST-MyoD proteins to investigate direct interaction of activated MEK1 with MyoD. Notably, His6-MEKEE protein was co-precipitated with GST-MyoD but not with GST. The amount of His6-MEKEE protein co-precipitated was further increased in the presence of 1 mm ATP (Fig. 2A). Thus, these results suggest that the activated MEK1 interacts directly with MyoD.
FIGURE 2.
Activated MEK1 interacts directly with and phosphorylates MyoD. A, equal quantities (5 μg) of purified GST or GST-MyoD protein was prebound to glutathione beads and incubated with an equivalent amount (3 μg) of purified, recombinant His6-MEKEE (S218E/S222E) protein with or without 1 mm ATP. Precipitated protein was immunoblotted with anti-MEK1 antibody; His6-MEKEE protein was included as a control (lane 1). B, 10T1/2 cells were transfected with HA-MEKEE (ΔN). After 36 h, cells were extracted, and the immunoprecipitates were prepared using anti-HA antibody were incubated with 2 μg of either GST or GST-MyoD protein in a kinase buffer, including 5 μCi of [γ-32P]ATP at 37 °C for 30 min. The mixtures were separated by 10% SDS-PAGE, and the gel was dried and exposed to x-ray film (right panel). The positions of GST and GST-MyoD (∼65 kDa) were identified by immunoblotting using anti-GST antibody (left panel). The same amount of HA-MEKEE (ΔN) was used in the kinase assay (lower panel). C, endogenous, activated MEK1 in C2C12 cells kept in DM for 2 days were immunoprecipitated using anti-pMEK1 antibody, and the immune complex was assayed in the absence or presence of specific MEK inhibitors, PD18435 (10 μm) and U0126 (30 μm). D, the kinase activity of recombinant His6-MEKEE protein (50 ng) toward GST-MyoD was examined in the absence or presence of PD18435 (10 μm) and U0126 (30 μm). E, 10T1/2 cells were co-transfected with FLAG-MyoD together with either empty vector, HA-MEKEE (ΔN) or HA-MEKAA (ΔN). After 36 h, cell extracts were immunoblotted with anti-MyoD (C-20) antibody. WB, Western blot.
The association between MEK1 and MyoD depends on the activation of MEK1 (Fig. 1, A and B), and their interaction increases in the presence of ATP (Fig. 2A). Thus, we performed an in vitro kinase assay to examine whether active MEK1 phosphorylates MyoD. 10T1/2 cells transfected with HA-MEKEE (ΔN) were immunoprecipitated with anti-HA antibody, and the immune complex was then incubated with GST or GST-MyoD protein as a substrate in the presence of [γ-32P]ATP. As shown in Fig. 2B, a 32P-labeled protein band appeared at ∼65 kDa, which corresponded to the GST-MyoD protein, as determined by immunoblotting using anti-GST antibody (left panel). The amount of immunoprecipitated HA-MEKEE (ΔN) used in the kinase assay was equal (low panel). In contrast, the corresponding 32P-labeled protein band was not observed in the cells transfected with HA-MEKAA (ΔN) (supplemental Fig. S3).
We also examined the possibility of MyoD phosphorylation by endogenous MEK1 activated during myogenic differentiation. Activated MEK1 immunoprecipitated using anti-pMEK1 antibody from the C2C12 cells kept in DM for 2 days was incubated with GST-MyoD protein and further assayed. A 32P-labeled GST-MyoD protein band appeared on the gel, and its phosphorylation was inhibited in the presence of PD184352 and U0126, specific MEK inhibitors, suggesting that endogenous activated MEK1 phosphorylates MyoD directly (Fig. 2C). In addition, a kinase assay using the recombinant His6-MEKEE protein, instead of the immune complexes, showed that the GST-MyoD protein was phosphorylated by His6-MEKEE, which was inhibited in the presence of PD184352 and U0126 (Fig. 2D). This result thus eliminated the possibility that any MEK1 up- or downstream kinase such as Mos (11, 12) or ERK is involved in MyoD phosphorylation. In addition, the intensity of the MyoD upper band in the 10T1/2 cells transfected with MyoD and HA-MEKEE (ΔN) increased compared with that of without HA-MEKEE (ΔN) or with HA-MEKAA (ΔN) (Fig. 2E). Together, these results demonstrate that MyoD is phosphorylated by activated MEK1 directly.
Tyr-156 of MyoD Is Phosphorylated by Activated MEK1
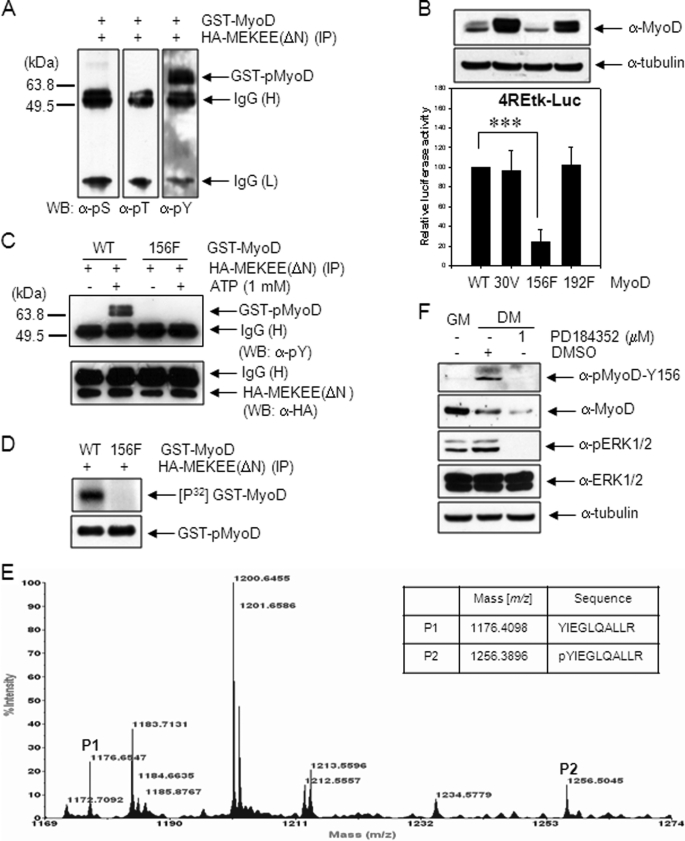
To determine the amino acid residue of MyoD phosphorylated by activated MEK1, GST-MyoD protein was phosphorylated with HA-MEKEE (ΔN) immunoprecipitated as described in Fig. 2B, and the phosphorylated residue was screened by immunoblotting using phospho-specific antibodies (anti-phosphoserine, anti-phosphothereonine, and anti-phosphotyrosine antibodies). As shown in Fig. 3A, GST-MyoD was phosphorylated by active MEK1 only at a tyrosine residue(s).
FIGURE 3.
MyoD Tyr-156 is phosphorylated by active MEK1. A, 10T1/2 cells were transfected with HA-MEKEE (ΔN, S218E/S222E). After 36 h, cells were extracted, and the immunoprecipitates prepared with anti-HA antibody were incubated with 2 μg of GST-MyoD protein in a kinase buffer, including 1 mm ATP, at 37 °C for 30 min. The mixtures were immunoblotted using anti-pSer (pS), anti-pThr (pT), or anti-pTyr (pY) antibody. B, 10T1/2 cells were co-transfected with 4REtk-Luc reporter vector and EMSV-MyoD (wild type, Y30V (30V), Y156F (156F), or Y192F (192F)), maintained in DM for 2 days, and assayed for luciferase activity. The expression levels of MyoD proteins were examined by immunoblotting using anti-MyoD (C-20) antibody (upper panel). C, to examine whether recombinant, mutant GST-MyoD-Y156F protein could be phosphorylated by active MEK1, the immune complex prepared in A was incubated with wild type GST-MyoD or mutant GST-MyoD-Y156F protein, and the reaction mixtures were analyzed by immunoblotting using anti-pTyr antibody as described in A. In D, the immune complex was incubated with wild type GST-MyoD or mutant GST-MyoD-Y156F in the presence of 5 μCi of [γ-32P]ATP instead of cold ATP. The reaction mixtures were separated by 10% SDS-PAGE, and the gel was dried and exposed to x-ray film. E, GST-MyoD protein (10 μg) was phosphorylated with immunoprecipitated HA-MEKEE (ΔN) as described previously in A. The tryptic digest of phosphorylated GST-MyoD was analyzed by MALDI-TOF as described under “Experimental Procedures.” The table shows the expected masses of two tryptic peptides, YIEGLQALLR (P1) and pYIEGLQALLR (P2) (ProteinProspector). F, phosphorylation at Tyr-156 of endogenous MyoD in proliferating or differentiating C2C12 cells kept in DM for 3 days was examined by immunoblotting using anti-pMyoD-Y156 antibody. C2C12 cells in lane 3 were treated with PD184352, a MEK inhibitor, for 24 h before cell extraction. The results show the mean ± S.D. of triplicate experiments. ***, p < 0.001. WB, Western blot. H, heavy chain; L, light chain.
Because the MEK1-induced phosphorylation of MyoD enhanced heterodimerization with E proteins,3 we reasoned that the tyrosine residue (s) activated by MEK1 could be located in the HLH domain, which participates in heterodimerization with E proteins. Among three tyrosine residues (Tyr-156, Tyr-178, and Tyr-192) in the HLH domain, Tyr-178 was excluded because it is not conserved among several species including human, mouse, and rat. Thus, Tyr-156, Tyr-192, and Tyr-30, which are known to be phosphorylated by activated c-Abl1 tyrosine kinase (8), were tested. These three tyrosine residues were replaced with Phe or Val by the site-directed mutagenesis. Then, the transcriptional activity of MyoD was measured in 10T1/2 cells after transfection of 4REtk-Luc together with either wild type or mutant MyoD plasmids. Compared with wild type MyoD, no significant change in luciferase activity was observed with mutant MyoD-Y30V and MyoD-Y192F, whereas dramatic reduction was seen with mutant MyoD-Y156F. In contrast, the protein level of mutant MyoD-Y30V and MyoD-Y192F increased, whereas that of mutant MyoD-Y156F (Fig. 3B) reduced. These results suggest that phosphorylation at Tyr-156, but not Tyr-30 and Tyr-192, is involved in MyoD transcriptional activity.
Thus, Tyr-156 residue was presumed to be the residue phosphorylated by MEK1. This was evidenced by an observation that mutant GST-MyoD-Y156F protein was resistant to phosphorylation by immunoprecipitated HA-MEKEE (ΔN) (Fig. 3, C and D).
The phosphorylation of MyoD at the Tyr-156 residue was further confirmed by MALDI-TOF analysis. The phosphorylated GST-MyoD protein was digested with trypsin, and the molecular masses of the fragments were analyzed by mass spectrometry. We observed P1 (156YIEGLQALLR, m/z 1176.6547) fragment and P2 (m/z 1256.5045) fragment, which corresponded to the calculated mass of phosphorylated P1 (156pYIEGLQALLR), thus demonstrating that the Tyr-156 residue of MyoD was indeed phosphorylated (Fig. 3E). Moreover, the tyrosine phosphorylation of endogenous MyoD increased with differentiation, as measured by immunoblotting with a specific anti-pMyoD-Y156 antibody; however, this response was not observed in the presence of PD184352, a specific MEK inhibitor (Fig. 3F). Together, these results clearly indicated that the Tyr-156 residue of MyoD is phosphorylated directly by activated MEK1.
MyoD Phosphorylation at Tyr-156 Increases Stability
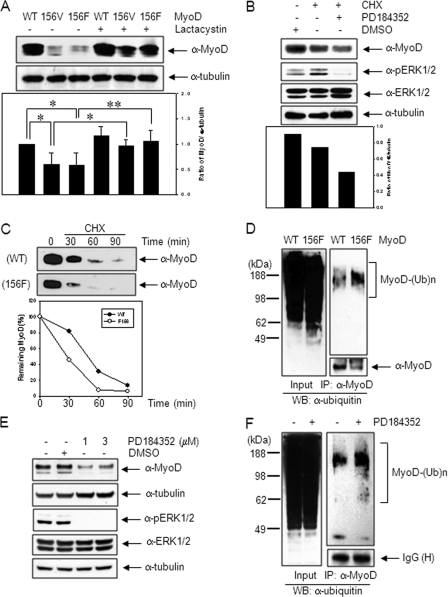
The level of MyoD protein increased in the presence of active MEK1, whereas that decreased in the presence of dominant negative MEK1 (Fig. 2E). In addition, the protein level of mutant MyoD-Y156F decreased compared with wild type (Fig. 3B). Thus, the phosphorylation at Tyr-156 of MyoD is likely to affect its protein stability. To clarify this issue, the protein levels of wild type and mutant MyoD were measured in 10T1/2 cells following transfection with either wild type or mutant MyoD plasmids. As shown in Fig. 4A, the band intensities of MyoD-Y156V and -Y156F proteins were significantly lower than that of the wild type. However, the reduced protein levels of the mutant MyoD recovered after treatment for 8 h with lactacystin (10 μm), a proteasome inhibitor, suggesting the involvement of Tyr-156 residue in protein stability. This was confirmed by an experiment measuring the residual protein level of MyoD after blocking protein synthesis by cycloheximide (10 μg/ml) treatment; the MyoD protein level was 83% of control cells, but in the cells pretreated with PD184352, a MEK1-specific inhibitor, it was 49% of control cells (Fig. 4B). In addition, the half-life of mutant MyoD-Y156F (27.5 min) was reduced compared with the wild type (48.5 min), when measured after blocking protein synthesis by cycloheximide (Fig. 4C). The amount of polyubiquitinated MyoD protein in the cells transfected with MyoD-Y156F plasmid was significantly higher than that of the wild type (Fig. 4D).
FIGURE 4.
MyoD-Y156 phosphorylation is required for protein stabilization. A, 10T1/2 cells transfected with wild type MyoD or mutant MyoD (-Y156V (156V) or -Y156F (156F)) were kept in the absence or presence of lactacystin (10 μm), a proteasome inhibitor for 8 h, extracted, and analyzed by immunoblotting, using anti-MyoD (C-20) antibody. The ratio of MyoD protein to α-tubulin is shown in the lower panel. B, 10T1/2 cells were transiently transfected with MyoD expression plasmid. On the next day, the cells were pretreated with either PD184352 (1 μm), a specific MEK inhibitor for 30 min or not, and the protein synthesis was blocked by treatment of cycloheximide (CHX; 10 μg/ml), a protein synthesis inhibitor for an additional 30 min. The MyoD protein level was analyzed by immunoblotting using anti-MyoD (C-20) antibody. Reduction of pERK1/2 intensity showed that the MEK inhibitor were effective. C, 10T1/2 cells transfected with either wild type or mutant MyoD were treated with cycloheximide (10 μg/ml) for the indicated time. MyoD protein levels were analyzed by immunoblotting, using anti-MyoD (C-20) antibody. Quantitation of MyoD turnover following cycloheximide treatment is shown in the bottom panel. D, 10T1/2 cells transfected with either wild type or mutant MyoD-Y156F were treated with lactacystin (5 μm) for 12 h. Cell extracts were immunoprecipitated with anti-MyoD (C-20) antibody, and the polyubiquitination levels of endogenous MyoD were analyzed by immunoblotting, using anti-ubiquitin (Ub) antibody. E, C2C12 cells incubated in DM for 1 day were treated with PD184352 (1 or 3 μm) for 2 days, and the MyoD protein level was analyzed by immunoblotting using anti-MyoD (C-20) antibody. F, differentiating C2C12 myoblasts kept in DM for 1 day were treated with lactacystin (5 μm) for 12 h in the presence or absence of PD184352 (1 μm). Cell extracts were immunopreciptated with anti-MyoD (C-20) antibody, and the polyubiquitination levels of endogenous MyoD were analyzed by immunoblotting using anti-ubiquitin antibody. The results show the mean ± S.D. of triplicate experiments. *, p < 0.05; **, p < 0.01. WB, Western blot.
Next, we examined whether activated MEK1 enhances endogenous MyoD protein stability. As seen in Fig. 4E, the level of MyoD protein in differentiating C2C12 myoblasts decreased in the presence of PD184352, a MEK-specific inhibitor. In addition, we tested whether MEK1 gene knockdown decreases MyoD protein stability. As shown in supplemental Fig. S4, phosphorylated MyoD at Tyr-156 disappeared in the cells transfected with siRNA targeting MEK1, and total MyoD was reduced significantly. No change was seen in cells transfected with scrambled siRNA used as a control. The polyubiquitination of MyoD in differentiating C2C12 cells increased in the presence of PD184352 (Fig. 4F). Together, these results suggest that the MEK1-mediated phosphorylation of MyoD at Tyr-156 increases its stability possibly by interfering with ubiquitin/proteasome-mediated degradation.
MyoD Phosphorylation at Tyr-156 Makes MyoD Escape from MAFbx/AT-1, an E3-ubiquitin Ligase Binding
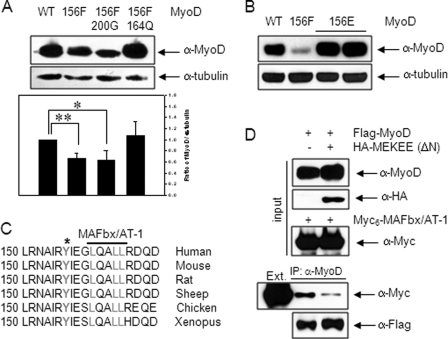
To understand the molecular mechanism of MyoD stabilization by Tyr-156 phosphorylation, we examined the protein levels of two mutants, MyoD-S200G/Y156F and MyoD-L164Q/Y156F, following transfection. As shown in Fig. 5A, the additional L164Q mutation, which causes a lack of interaction with MAFbx/AT-1 (19), restored the protein level to that of the wild type; however, the additional mutation at Ser-200 (S200G), which is the site phosphorylated by Cdk1/2/cyclin E (5, 6), showed no change in protein level compared with that of the Y156F mutation. Also, the protein level of Y156E mutant that contains the negative charge and thus mimics the constitutive phosphorylation at the 156 residue was highly stable (Fig. 5B). Thus, phosphorylation at Tyr-156 appeared to affect the MAFbx/AT-1-dependent proteasome-mediated degradation of MyoD.
FIGURE 5.
MyoD-Y156 phosphorylation increases its stability by interfering with MAFbx/AT-1 binding. A, 10T1/2 cells transfected with either wild type or mutant MyoD (Y156F, S200G/Y156F, L164Q/Y156F) were extracted and analyzed by immunoblotting using anti-MyoD (C-20) antibody. The ratio of MyoD protein to α-tubulin is shown in the lower panel. B, 10T1/2 cells transfected with either wild type or mutant MyoD (Y156F, Y156E) were extracted and analyzed by immunoblotting using anti-MyoD (C-20) antibody. C, the Tyr-156 residue (*) and the interacting region (boldface line) of MAFbx/AT-1 in MyoD are conserved among several species. D, 10T1/2 cells were transfected with FLAG-MyoD and HA-MEKEE (ΔN) and separately with Myc6-MAFbx/AT-1. After 24 h, each cell extract (Ext.) was prepared separately, mixed, and immunoprecipitated with anti-MyoD (C-20) antibody. The immunoprecipitates were analyzed by immunoblotting with anti-Myc antibody. The results show the mean ± S.D. of triplicate experiments. *, p < 0.05; **, p < 0.01.
Because the MAFbx/AT-1 binding domain (160-LXXLL-164) is located near MyoD-Y156 (Fig. 5C), phosphorylation at Tyr-156 could interfere with the binding of MAFbx/AT-1. To test this possibility, we examined the amount of MAFbx/AT-1 co-precipitated with MyoD in the presence or absence of HA-MEKEE (ΔN). As expected, we observed a band indicative of an interaction between MAFbx/AT-1 and MyoD. Notably, the amount of MAFbx/AT-1 co-precipitated with MyoD decreased significantly in cells co-transfected with HA-MEKEE (ΔN) (Fig. 5D). Together, these results suggested that MEK1-mediated phosphorylation at MyoD-Y156 hinders the binding of MAFbx/AT-1, thereby allowing MyoD to escape ubiquitin/proteasome-mediated degradation.
MyoD Phosphorylation at Tyr-156 Is Required for Skeletal Myogenic Differentiation
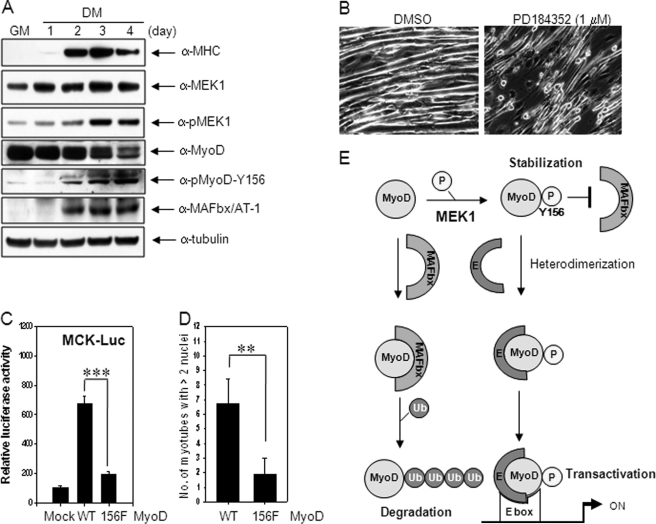
We measured the protein levels of MEK1, pMEK1, MyoD, pMyoD-Y156, and MAFbx/AT-1 in differentiating C2C12 cells. As shown in Fig. 6A, the MEK1 level showed almost no change with differentiation, whereas pMEK1 increased dramatically up to days 3 and 4. Phosphorylated MyoD-Y156 increased along with MEK1 activation during differentiation. In contrast, total MyoD decreased from day 2 when an increase of MAFbx/AT-1 appeared. Thus, the stable pMyoD-Y156 level increased in coordination with MEK1 activation while the unstable form (not phosphorylated at Tyr-156) reduced probably due to the presence of MAFbx/AT-1.
FIGURE 6.
MyoD phosphorylation at Tyr-156 is required for skeletal myogenic differentiation. A, C2C12 cells grown in GM were switched to DM to induce differentiation, and the protein levels of MHC, MEK1, pMEK1, MyoD, phosphorylated MyoD at Tyr-156, MAFbx/AT-1 in cell extracts were examined by immunoblotting using anti-MHC, anti-MEK1, anti-pMEK1, anti-MyoD (C-20), anti-pMyoD-Y156, and anti-MAFbx/AT-1 antibodies, respectively. B, C2C12 cells incubated in DM for 1 day were treated with PD184352 (1 μm), a MEK-specific inhibitor, for 2 days, and phase-contrast images of muscle cells are photographed. C, 10T1/2 cells were co-transfected with either muscle creatine kinase-Luc reporter vector together with MyoD constructs (mock, wild type MyoD, or MyoD-Y156F), maintained in DM for 2 days, and assayed for luciferase activity. D, 10T1/2 cells were transfected with wild type MyoD or mutant MyoD-Y156F along with pEGFP-N1 plasmid (supplemental Fig. S5). The number of myotubes with above two nuclei per field was counted, and >10 fields randomly chosen were averaged under a fluorescent microscope (Carl Zeiss AX10, ×200 magnifications). The results show the mean ± S.D. of triplicate experiments. **, p < 0.01; ***, p < 0.001. E, schematic diagram showing the mechanism of MEK1-mediated stimulation of MyoD transactivation. E, E proteins; Ub, ubiquitin.
To measure the effect of phosphorylated MyoD-Y156 on myogenic differentiation, differentiating myoblasts were treated with PD184352, a specific MEK inhibitor, on day 1 in DM for 2 days and observed for differentiation. Fully differentiated, long myotubes were rarely observed in the presence of PD184352 (1 μm; Fig. 6B). In addition, we performed transient transfection of wild type MyoD or mutant MyoD-Y156F to 10T1/2 cells, which lack endogenous MyoD, and examined the muscle creatine kinase transcriptional activity and the number of myotubes. The luciferase activity by wild MyoD was enhanced dramatically but not much by the mutant (29% of wild type) (Fig. 6C). Similarly, the number of myotubes with more than two nuclei in MyoD-Y156F was only 28% of wild type (Fig. 6D). Taken together, these results suggest that MEK1-mediated MyoD phosphorylation at Tyr-156 is essential for skeletal muscle differentiation.
DISCUSSION
Many extracellular growth factors and intracellular signaling pathways are involved in controlling the growth and differentiation of skeletal muscle cells (2, 32). The ERK signaling pathway is one of such signaling pathways, but there have been conflicting reports about its role in skeletal myogenesis (21–25). With this regard, we recently presented the data that activated MEK1 plays contrary stage-specific roles in myogenic differentiation; activation of MEK1 in the early stage inhibits skeletal muscle differentiation, whereas that in the mid-stage stimulates it (26), explaining at least partially the discrepancy reported from several laboratories. In an effort to elucidate the precise role of MEK1 activation in mid- and late stage of myogenesis, we investigated the interaction of MEK1 to MyoD, a master regulator in skeletal myogenesis. We found a novel activity of MEK1 for MyoD; active MEK1 phosphorylates MyoD at Tyr-156 in helix 2 of the HLH domain. This phosphorylation increases the stability of MyoD protein via interfering the binding of MAFbx/AT-1, a key E3-ubiquitin ligase necessary for MyoD degradation (19), thus enhances the MyoD/E protein heterodimerization (data not shown) and thereby participates in the positive regulation of skeletal muscle differentiation (Fig. 6E).
An interesting finding from our study is the identification of a new substrate, MyoD, for MEK1. Until now, ERK is the only substrate known to be phosphorylated by MEK1 although GSK-3β could be another (38). Because MEK1 is a well known dual-specificity kinase, it could not be impossible that MEK1 phosphorylates a tyrosine residue of MyoD. In addition, it is reported that dual-specificity kinases phosphorylate various target proteins, including transcription factors in addition to downstream kinases (39). A recent study reported that MEK1 associates with STAT5 and induces its tyrosine phosphorylation and DNA binding activity (40). The fact that activated nuclear MEK1 reduces the transcriptional activity of PPARγ by associating with it, followed by a rapid nuclear export of the complex also supports the unexpected function of MEK1 for MyoD phophorylation in the nucleus (41).
Because activated MEK1 phosphorylates directly MyoD (Fig. 2), it is interesting to know how MEK1 interacts with MyoD. MEK1 has a conserved motif (human, 3-KKK-5) at the N terminus that mediates the high affinity interaction with ERK. This motif is characterized by a cluster of positively charged amino acids (33). Thus, the docking site of MEK1 on ERK is assumed to be a cluster of negatively charged amino acids as suggested that two aspartic acids of ERK are essential for docking MEK1 (34). Because the N terminus of MyoD contains many aspartic acid residues and exhibits a sequence similarity to the docking domain of ERK, the interaction of activated MEK1 with MyoD may occur through the N-terminal transactivation domain of MyoD, as suggested by Perry et al. (24).
It is noteworthy that the biological significance of MyoD-Y156 phosphorylation is protein stabilization. The protein level of MyoD increased in the cells transfected with active MEK1 (Fig. 2E), and the amount of MAFbx/AT-1 co-precipitated with MyoD decreased significantly in cells co-transfected with HA-MEKEE (ΔN) (Fig. 5D). Moreover, the amount of polyubiquitinated MyoD protein in the cells transfected with MyoD-Y156F plasmid was significantly higher than that of the wild type (Fig. 4D). Thus, MEK1-mediated phosphorylation at MyoD-Y156 somehow allows MyoD to escape ubiquitin/proteasome-mediated degradation.
Here, we propose that MAFbx/AT-1, an E3-ubiquitin ligase, plays a role in MyoD-Y156 phosphorylation-mediated stabilization. A recent study suggested that MyoD is a substrate of MAFbx/AT-1 and the LXXLL conserved motif sequence (residues 160–164) in the helix 2 region of the MyoD HLH domain is essential for MAFbx/AT-1 binding (19). Because MyoD-Y156 is located near the MAFbx/AT-1 binding motif (LXXLL, 160–164) (Fig. 5C), it is expected that the phosphorylation at Tyr-156 may interfere the binding of MAFbx/AT-1 with MyoD. As expected our results showed that the protein level of MyoD-Y156E mutant to mimic constitutive phosphorylation at Tyr-156 was highly stable. Moreover, introduction of an additional mutation at Leu-164, a crucial residue for MAFbx/AT-1 binding to MyoD-Y156F mutant (double mutant MyoD-Y156F/L164Q) recovered the protein level to that of wild type (Fig. 5, A and B). Together, these results clearly demonstrate that MEK1-mediated MyoD-Y156 phosphorylation inhibits its ubiquitination through interfering the binding of MAFbx/AT-1, thus resulting in its stabilization. Our observation also explains how active MEK1 increases the protein level and transcriptional activity of MyoD in Xenopus (22). A similar mechanism is found in case of p53 accumulation in the response of DNA damage. The p53 phosphorylation at Ser-20 plays a critical role in its stabilization through interrupting the binding of Mdm2, an E3-ubiquitin ligase for p53 (35, 36). Thus, this is the case that a protein is stabilized by phosphorylation, which blocks the binding of its E3-ubiquitin ligase and subsequently inhibits its ubiquitination and degradation through the ubiquitin-proteasome pathway. As both the protein level and the promoter activity of MAFbx/AT-1 increase during myogenic differentiation (Fig. 6A) (19, 37), stabilization of MyoD through this mechanism may play a crucial role in fully differentiated myotube formation.
It is reported that MyoD is essential for the function of myogenic stem cell in adult skeletal muscle (42). MyoD also is required to maintain aging muscle homeostasis and plays a role in skeletal muscle plasticity in response to hypertrophic or denervation stimuli (43–45). Also, inhibition of MAFbx/AT-1-dependent proteolysis of MyoD prevents skeletal muscle atrophy in vivo (46). Although we are not sure at the moment whether MEK1 activation is required for preventing skeletal muscle atrophy in vivo, it is expected that the reduction of polyubiquitination of MyoD by MEK1 activation plays an important role in muscle differentiation and proper muscle physiology.
Supplementary Material
Acknowledgments
We thank Drs. M. A. Rudnicki (McMaster University), B. R. Cullen (Duke University), and A. L. Goldberg (Harvard Medical School) for providing the plasmids and Dr. R. Marquez (University of Dundee) for providing PD184352.
This work was supported by an intramural research fund (4800-4845-300-210) from the National Institute of Health (Korea) and conducted by the Research Fund of Dankook University in 2010.

The on-line version of this article (available at http://www.jbc.org) contains supplemental “Experimental Procedures” and Figs. S1–S5.
C. Jo, S.-J. Cho, and S. A. Jo, unpublished data.
- MRF
- myogenic regulatory factor
- MEK1
- mitogen-activated protein kinase kinase 1
- MEKAA
- S218A/S222A
- MEKEE
- S218E/S222E
- GM
- growth medium
- Cdk
- cyclin-dependent kinase
- MAFbx/AT-1
- muscle atrophy F-box/Astrogin-1
- MuRF1
- muscle ring finger 1
- Luc
- luciferase
- EGFP
- enhanced GFP
- HLH
- helix-loop-helix
- DM
- differentiation medium.
REFERENCES
- 1. Buckingham M. (2001) Curr. Opin. Genet. Dev. 11, 440–448 [DOI] [PubMed] [Google Scholar]
- 2. Puri P. L., Sartorelli V. (2000) J. Cell. Physiol. 185, 155–173 [DOI] [PubMed] [Google Scholar]
- 3. Black B. L., Olson E. N. (1998) Annu. Rev. Cell Dev. Biol. 14, 167–196 [DOI] [PubMed] [Google Scholar]
- 4. Rudnicki M. A., Schnegelsberg P. N., Stead R. H., Braun T., Arnold H. H., Jaenisch R. (1993) Cell 75, 1351–1359 [DOI] [PubMed] [Google Scholar]
- 5. Song A., Wang Q., Goebl M. G., Harrington M. A. (1998) Mol. Cell. Biol. 18, 4994–4999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kitzmann M., Vandromme M., Schaeffer V., Carnac G., Labbé J. C., Lamb N., Fernandez A. (1999) Mol. Cell. Biol. 19, 3167–3176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Reynaud E. G., Pelpel K., Guillier M., Leibovitch M. P., Leibovitch S. A. (1999) Mol. Cell. Biol. 19, 7621–7629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Puri P. L., Bhakta K., Wood L. D., Costanzo A., Zhu J., Wang J. Y. (2002) Nat. Genet. 32, 585–593 [DOI] [PubMed] [Google Scholar]
- 9. Liu L. N., Dias P., Houghton P. J. (1998) Cell Growth Differ. 9, 699–711 [PubMed] [Google Scholar]
- 10. Leibovitch S. A., Guillier M., Lenormand J. L., Leibovitch M. P. (1991) Oncogene 6, 1617–1622 [PubMed] [Google Scholar]
- 11. Lenormand J. L., Benayoun B., Guillier M., Vandromme M., Leibovitch M. P., Leibovitch S. A. (1997) Mol. Cell. Biol. 17, 584–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pelpel K., Leibovitch M., Fernandez A., Leibovitch S. A. (2000) FEBS Lett. 474, 233–237 [DOI] [PubMed] [Google Scholar]
- 13. Abu Hatoum O., Gross-Mesilaty S., Breitschopf K., Hoffman A., Gonen H., Ciechanover A., Bengal E. (1998) Mol. Cell. Biol. 18, 5670–5677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lingbeck J. M., Trausch-Azar J. S., Ciechanover A., Schwartz A. L. (2003) J. Biol. Chem. 278, 1817–1823 [DOI] [PubMed] [Google Scholar]
- 15. Bodine S. C., Latres E., Baumhueter S., Lai V. K., Nunez L., Clarke B. A., Poueymirou W. T., Panaro F. J., Na E., Dharmarajan K., Pan Z. Q., Valenzuela D. M., DeChiara T. M., Stitt T. N., Yancopoulos G. D., Glass D. J. (2001) Science 294, 1704–1708 [DOI] [PubMed] [Google Scholar]
- 16. Gomes M. D., Lecker S. H., Jagoe R. T., Navon A., Goldberg A. L. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 14440–14445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Glass D. J. (2003) Nat. Cell Biol. 5, 87–90 [DOI] [PubMed] [Google Scholar]
- 18. Murton A. J., Constantin D., Greenhaff P. L. (2008) Biochim. Biophys. Acta 1782, 730–743 [DOI] [PubMed] [Google Scholar]
- 19. Tintignac L. A., Lagirand J., Batonnet S., Sirri V., Leibovitch M. P., Leibovitch S. A. (2005) J. Biol. Chem. 280, 2847–2856 [DOI] [PubMed] [Google Scholar]
- 20. Shaul Y. D., Seger R. (2007) Biochim. Biophys. Acta 1773, 1213–1226 [DOI] [PubMed] [Google Scholar]
- 21. Gredinger E., Gerber A. N., Tamir Y., Tapscott S. J., Bengal E. (1998) J. Biol. Chem. 273, 10436–10444 [DOI] [PubMed] [Google Scholar]
- 22. Zetser A., Frank D., Bengal E. (2001) Dev. Biol. 240, 168–181 [DOI] [PubMed] [Google Scholar]
- 23. Dorman C. M., Johnson S. E. (1999) Oncogene 18, 5167–5176 [DOI] [PubMed] [Google Scholar]
- 24. Perry R. L., Parker M. H., Rudnicki M. A. (2001) Mol. Cell 8, 291–301 [DOI] [PubMed] [Google Scholar]
- 25. Jo C., Kim H., Jo I., Choi I., Jung S. C., Kim J., Kim S. S., Jo S. A. (2005) Biochim. Biophys. Acta 1743, 187–197 [DOI] [PubMed] [Google Scholar]
- 26. Jo C., Jang B. G., Jo S. A. (2009) Cell Signal 21, 1910–1917 [DOI] [PubMed] [Google Scholar]
- 27. Blair W. S., Bogerd H. P., Madore S. J., Cullen B. R. (1994) Mol. Cell. Biol. 14, 7226–7234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sandri M., Sandri C., Gilbert A., Skurk C., Calabria E., Picard A., Walsh K., Schiaffino S., Lecker S. H., Goldberg A. L. (2004) Cell 117, 399–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fukuda M., Gotoh I., Adachi M., Gotoh Y., Nishida E. (1997) J. Biol. Chem. 272, 32642–32648 [DOI] [PubMed] [Google Scholar]
- 30. Jaaro H., Rubinfeld H., Hanoch T., Seger R. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 3742–3747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jo C., Kim J., Jo S. A. (2005) Protein Pept. Lett. 12, 631–633 [DOI] [PubMed] [Google Scholar]
- 32. Chargé S. B., Rudnicki M. A. (2004) Physiol. Rev. 84, 209–238 [DOI] [PubMed] [Google Scholar]
- 33. Bardwell L., Thorner J. (1996) Trends Biochem. Sci 21, 373–374 [PubMed] [Google Scholar]
- 34. Tanoue T., Adachi M., Moriguchi T., Nishida E. (2000) Nat. Cell Biol. 2, 110–116 [DOI] [PubMed] [Google Scholar]
- 35. Unger T., Juven-Gershon T., Moallem E., Berger M., Vogt Sionov R., Lozano G., Oren M., Haupt Y. (1999) EMBO J. 18, 1805–1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chehab N. H., Malikzay A., Stavridi E. S., Halazonetis T. D. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 13777–13782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhao W., Wu Y., Zhao J., Guo S., Bauman W. A., Cardozo C. P. (2005) J. Cell. Biochem. 96, 209–219 [DOI] [PubMed] [Google Scholar]
- 38. Takahashi-Yanaga F., Shiraishi F., Hirata M., Miwa Y., Morimoto S., Sasaguri T. (2004) Biochem. Biophys. Res. Commun. 316, 411–415 [DOI] [PubMed] [Google Scholar]
- 39. Galceran J., de Graaf K., Tejedor F. J., Becker W. (2003) J. Neural. Transm. Suppl. 67, 139–148 [DOI] [PubMed] [Google Scholar]
- 40. Maki K., Ikuta K. (2008) J. Immunol. 181, 494–502 [DOI] [PubMed] [Google Scholar]
- 41. Burgermeister E., Chuderland D., Hanoch T., Meyer M., Liscovitch M., Seger R. (2007) Mol. Cell. Biol. 27, 803–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Megeney L. A., Kablar B., Garrett K., Anderson J. E., Rudnicki M. A. (1996) Genes Dev. 10, 1173–1183 [DOI] [PubMed] [Google Scholar]
- 43. Hughes S. M., Koishi K., Rudnicki M., Maggs A. M. (1997) Mech. Dev. 61, 151–163 [DOI] [PubMed] [Google Scholar]
- 44. Ishido M., Kami K., Masuhara M. (2004) Acta Physiol. Scand. 180, 281–289 [DOI] [PubMed] [Google Scholar]
- 45. Ishido M., Kami K., Masuhara M. (2004) Am. J. Physiol. Cell Physiol. 287, C484–C493 [DOI] [PubMed] [Google Scholar]
- 46. Lagirand-Cantaloube J., Cornille K., Csibi A., Batonnet-Pichon S., Leibovitch M. P., Leibovitch S. A. (2009) PLoS One 4, e4973. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.