Abstract
The ubiquitous overexpression of agouti-signaling protein (ASP), a paracrine-signaling molecule that regulates pigment-type switching in the hair follicle of the mouse, is responsible for the obesity and yellow pelage of the Yellow mouse (Ay). Mahogany (Attractin, Atrn/mg) and mahoganoid (Mahogunin Ring Finger-1, Mgrn1/md) are mutations epistatic to Ay. These mutations have been described as suppressors of ASP action, blocking its antagonizing effects on the melanocortin 1 and 4 receptors (MC1R and MC4R) in the skin and the brain, respectively, via unknown mechanisms. Here, we describe the molecular bases for the md- and mg-dependent rescue of the Ay phenotype at the MC4R. We show that overexpression of ASP inhibits the rise in cAMP levels in response to α-melanocyte-stimulating hormone, an MC4R agonist, by blocking ligand binding and by directing MC4R trafficking to the lysosome. Loss-of-function of either attractin or MGRN1 blocks ASP-dependent MC4R degradation and promotes increased trafficking of internalized MC4R to the cell surface, but it does not restore α-melanocyte-stimulating hormone-dependent cAMP signaling. We propose that MGRN1 and attractin are components of an evolutionarily conserved receptor trafficking pathway and that the md and mg mutations rescue the Ay phenotypes by a primarily cAMP-independent mechanism promoting trafficking of MC4R and likely MC1R away from the lysosome toward the cell surface.
Keywords: Cell Surface Receptor, E3 Ubiquitin Ligase, Obesity, Receptor Recycling, Trafficking, Agouti, Attractin, Mahogunin, Melanocortin
Introduction
The cardinal phenotypes of the yellow obese mouse (Ay) are due, at a molecular level, to antagonism of melanocortin 1 and 4 receptors by continuous overexpression of agouti-signaling protein (ASP)3 in the skin (MC1R) and brain (MC4R) (1, 2). Hypomorphic mutations in either mahogany (Atrn) or mahoganoid (Mgrn1) are essentially fully epistatic to Ay, resulting in a dark coat and normal body weight in Ay animals. The molecular bases for these epistatic effects are largely unknown but have important implications for the normal physiology of the regulation of these receptors. This study addresses the cellular mechanisms for these effects on MC4R, a key hypothalamic regulator of energy homeostasis in mice and humans.
The five G-protein-coupled receptors of the melanocortin receptor (MCR) family regulate diverse physiological processes, including pigmentation, corticosteroid production, food intake and energy metabolism, sexual behavior, inflammation, and exocrine gland function (3–10). Naturally occurring MCR agonists are derived from proopiomelanocortin, a precursor protein that is post-translationally processed in a tissue-specific manner by the proprotein convertases PC1 and PC2 (11). At least 10 different biologically active peptides are processed from proopiomelanocortin; among them are α-melanocyte-stimulating hormone (α-MSH), which binds to and activates MC1R, MC3R, MC4R, and MC5R, and adrenocorticotropic hormone (ACTH), which exclusively stimulates MC2R. MC1R, MC3R, and MC4R are unique in the G-protein-coupled receptor family by virtue of having naturally occurring protein antagonists, i.e. ASP and agouti-related protein (AGRP) (12–14). ASP binds with high affinity to MC1R and MC4R and less avidly to MC3R; by contrast, AGRP binds only MC3R and MC4R and not MC1R. In addition to competitive antagonism, ASP and AGRP also act as inverse agonists, decreasing basal cAMP levels in cells expressing MCRs (15).
ASP is a paracrine-signaling molecule normally produced intermittently by dermal cells at the base of a growing hair shaft (16). Binding of ASP to the melanocyte MC1R produces the common “agouti” coat color pattern, an otherwise black or dark colored hair with subapical yellow or light colored bands. The intermittent synthesis of ASP during the hair growth cycle switches pigment production alternatively from the black-brown pigment, eumelanin, to the yellow-red pigment, pheomelanin. Mutations at the agouti locus resulting in the ubiquitous overexpression of ASP (e.g. Ay and Avy) produce a pleiotropic syndrome that is characterized not only by the anticipated yellow (pheomelanin) coat color but also by obesity secondary to hyperphagia (17, 18). In Ay and Avy mice, ASP produced ectopically in the hypothalamus mimics the normal function of AGRP and inhibits signaling through the MC4R, causing increased food intake and decreased energy expenditure.
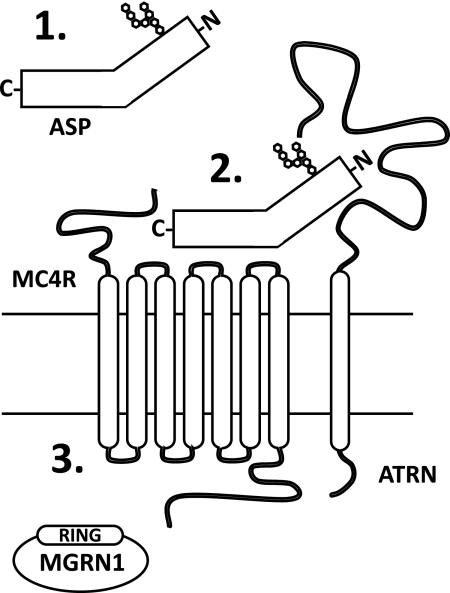
The mouse mutations mahogany (Atrnmg) and mahoganoid (Mgrn1md) were first described and named in the 1960s for their identical effects on coat-related pigment-type switching in mice (19, 20). Both mutations darken the Agouti wild-type coat color similar to MC1R gain-of-function and ASP loss-of-function mutations (19, 20). Null mutations of either mahogany (Atrnmg−3J) or mahoganoid (Mgrn1md−nc) completely suppress the effects of the Ay mutation on coat color and obesity; the double mutants are black and lean. However, neither the Atrnmg−3J nor the Mgrn1md alleles (Mgrn1md is a hypomorphic allele of Mgrn1) can rescue the yellow coat color caused by extension, an MC1R null mutation (Mc1re), or the obesity associated with MC4R loss-of-function (21, 22). Consequently, Atrn and Mgrn1 must function “downstream” of Agouti and “upstream” or at the level of MC1R/MC4R, possibly affecting ASP secretion and/or processing, the interaction of ASP with these receptors, or regulation of receptor number/function (Fig. 1).
FIGURE 1.
MGRN1 and ATRN are required for ASP function. Based on genetic crosses of Ay and Mc4r-null mice with mahoganoid (Mgrn1md) and mahogany (Atrnmg) mutant animals, MGRN1 and ATRN could affect the following: 1) ASP secretion and/or processing; 2) the interaction of ASP with the MC4R; or 3) MC4R regulation/function in response to ASP binding. RING, really interesting new gene.
Atrn (Attractin; formerly mahogany, mg) encodes attractin, a widely expressed type I single-pass transmembrane-spanning protein with a large putative extracellular domain and a short cytoplasmic tail (23, 24). The extracellular domain of ATRN binds with low affinity, 500-fold weaker than the interaction of the ASP C-terminal domain with MC1R, to a stretch of basic amino acids in the N-terminal domain of ASP, suggesting that ATRN may act as a co-receptor for ASP (25, 26). ATRN shows no affinity for AGRP, and Atrn mutations do not suppress the obese phenotype associated with a ubiquitously expressed Agrp transgene (26).
Mahogunin Ring Finger-1 (Mgrn1; formerly mahoganoid, md) is an E3 ubiquitin ligase (22, 27). MGRN1 (mahogunin) multiubiquitinates tumor suppressor gene 101 (Tsg101), an essential component of the endosomal sorting complex required for transport-I (ESCRT-I) that is necessary for the proper trafficking of monoubiquitinated cell surface receptors from the plasma membrane to the lysosome for degradation (28, 29). The precise cell physiological consequences of MGRN1-dependent ubiquitination of TSG101 are unknown. Like ATRN, MGRN1 is absolutely necessary for ASP function at MC1R/MC4R; a possible requirement for MGRN1 in AGRP signaling has not been explored.
MGRN1 and ATRN are part of an evolutionarily conserved molecular pathway. MGRN1 and ATRN orthologs have been identified in Caenorhabditis elegans and Drosophila melanogaster, predating the emergence of the melanocortin-signaling system and suggesting that both proteins have functions independent of ASP and the MCRs; there are no readily apparent orthologs in yeast (26, 27). Both genes show nearly identical patterns of mRNA distribution and share similar loss-of-function phenotypes.
Here, we show that transgenic overexpression of ASP, but not AGRP, promotes MC4R degradation in the lysosome and that siRNA-mediated knockdown of MGRN1 or ATRN prevents this process. Reduction of MGRN1 and ATRN function restores MC4R levels in ASP-overexpressing cells. Thus, these studies identify the cell molecular basis for the epistatic effects of hypomorphic alleles of Mgrn1 and Atrn on the effects of ASP at MC4R and provide insights into the mechanism by which MC4R signaling is regulated at the cellular level.
EXPERIMENTAL PROCEDURES
Cell Lines, Culture, and Transfection
Neuro-2a cells were obtained from the ATCC, cultured in DMEM (Invitrogen) supplemented with 10% fetal bovine serum (Gemimi Bio-Products), and transfected with Lipofectamine 2000 or RNAiMAX (Invitrogen) according to the manufacturer's suggestions. Stably transfected cells were selected with 1 mg/ml G418 (Invitrogen) for at least 2 weeks.
Plasmids
The mCherry-TSG101 plasmid was obtained through Addgene (Addgene plasmid 21505) from Dr. James Hurley (30). MGRN1(III)-GFP was generated by amplifying the Mgrn1 coding sequences from a C57BL/6J cDNA pool and cloning the fragments into the pcDNA3.1/CT-GFP-TOPO vector (Invitrogen). Specific primer pairs were used to isolate Mgrn1 isoform III by colony PCR. The HA-His-ubiquitin construct was kindly provided by Dr. Wei Gu (Columbia University) (31). The ASP expression constructs were created by amplifying the Asp coding sequence, with or without the endogenous stop codon, from a C57BL/6J cDNA pool and directionally cloning into the pcDNA3.1/V5-His-TOPO vector (Invitrogen). The MC4R-eGFP plasmid was obtained from Dr. Christian Vaisse (University of California, San Francisco) (32). HA-MC4R-eGFP was created through sequential rounds of site-directed mutagenesis, using the GeneTailor site-directed mutagenesis kit (Invitrogen), to insert the HA epitope sequence (YPYDVPDYA) at the N terminus of MC4R-eGFP. All plasmid sequences were verified by DNA sequencing to contain no coding sequence mutations compared with the UCSC reference genomes.
Reagents
The polyclonal anti-MGRN1 antibody was purchased from Proteintech Group. The anti-β-actin antibody was from Abcam. Polyclonal anti-GFP and monoclonal anti-V5 antibodies were from Invitrogen. Anti-mCherry was from Clontech. Rat anti-HA and rat anti-HA-POD antibodies were purchased from Roche Applied Science. α-MSH was from Phoenix Pharmaceuticals. Control, MGRN1, and ATRN siRNA probes were purchased from Invitrogen and Sigma (sequences available on request). All other chemicals and reagents were obtained from Sigma.
MGRN1 Detection in Neuro2a Cells
Total RNA was extracted from Neuro2a cells using an RNeasy kit (Qiagen) and converted to cDNA using the Transcriptor first strand cDNA synthesis kit (Roche Applied Science). Quantitative RT-PCR of Mgrn1 isoforms was performed in a LightCycler 2.0 (Roche Applied Science) according to the manufacturer's instructions. Samples were heated at 95 °C for 10 min, followed by 45 cycles of 95 °C for 10 s, 61 °C for 5 s, and 72 °C for 10 s. Primers for the Mgrn1 isoforms and control genes were as follows: Mgrn1 isoform I, F, gatgagcactctagttctgacagt, and R, cctcctctataccaacagagcacga; Mgrn1 isoform II, F, tgtccctttaaaaaatcaaa, and R, cctcctctataccaacagagcacga; Mgrn1 isoform III, F, gatgagcactctagttctgacagt, and R, cgaatggggtcagctcagcaactc; Mgrn1 isoform IV, F, tgtccctttaaaaaatcaaa, and R, cgaatggggtcagctcagcaactc; Actb, F, agccatgtacgtagccatcc, and R, ctctcagctgtggtggtgaa. Transcript quantification was performed using the absolute quantification mode of the LightCycler software. Standard curves were calculated for each set of primers using Neuro2a cDNA and had amplification efficiencies of 1.8 or greater. For detection of MGRN1 protein, Neuro2a cells were rinsed with cold PBS and lysed on ice in lysis buffer (150 mm NaCl, 20 mm Tris, 1% Igepal, and HALT protease/phosphatase inhibitor (Pierce)) for 30 min. Samples were centrifuged at 4 °C at 12,000 × g for 10 min, and the protein concentration of the supernatant was determined by the BCA method. Equal amounts of protein were subjected to SDS-PAGE and Western blotting using standard protocols. For confocal microscopy, 175,000 cells (Countess Automated Cell Counter, Invitrogen) were plated on poly-d-lysine-coated culture slides (BD Biosciences) and stained 24 h later. Cells were rinsed with PBS, fixed with freshly prepared 3.7% paraformaldehyde, and permeabilized with 0.1% Triton X-100 for 3 min. The samples were blocked with 5% goat serum containing 1% bovine serum albumin (BSA) for at least 30 min at room temperature and labeled with anti-MGRN1 primary antibody in 0.5% BSA overnight at 4 °C. The cells were incubated with the appropriate secondary antibody for 30 min at room temperature and mounted with SlowFade reagent containing DAPI (Invitrogen). The images were collected using an inverted Zeiss LSM 5 confocal microscope and acquired using a 63× oil immersion objective with 2× optical zoom with Zeiss Pascal software. Image processing was done with Adobe Photoshop/Illustrator CS3 Premium software (Adobe Systems).
TSG101 Immunoprecipitation, Ubiquitination, and Localization
For co-immunoprecipitation, Neuro2a cells were transiently transfected with empty vector, MGRN1(III)-GFP, and mCherry-TSG101 for 24 h. Cells were rinsed with cold PBS and lysed in lysis buffer on ice for 30 min. Samples were centrifuged at 4 °C at 12,000 × g for 10 min; equal amounts of total protein were incubated with anti-GFP or anti-mCherry overnight at 4 °C with gentle rocking. Immunocomplexes were recovered with recombinant protein G-agarose beads (Pierce) for 2 h at room temperature. After several washes in lysis buffer, the complexes were dissociated by boiling in lithium dodecyl sulfate sample buffer and analyzed by SDS-PAGE and immunoblotting. For ubiquitination, Neuro2a cells previously transfected with control, or Mgrn1 siRNA were transfected with mCherry-TSG101 and HA-His-ubiquitin for 18 h. Cells were lysed in lysis buffer supplemented with 0.1% Triton X-100 and 10 mm iodoacetamide, and equal amounts of total protein were subjected to immunoprecipitation with anti-mCherry antibody as described above. TSG101 ubiquitination was detected following SDS-PAGE and immunoblotting with anti-HA antibodies. Confocal microscopy was performed as described above. All results are representative of at least two independent experiments.
ASP-V5/His Secretion
Neuro2a.MC4R-eGFP cells (250,000 cells/well) were plated in 6-well plates and transfected 24 h later with control, Mgrn1, or Atrn siRNA using RNAiMAX. After 24 h, cells were transfected with empty vector or ASP-V5/His with Lipofectamine-2000; experiments were performed 24 h later. Immunoprecipitation buffer was added directly to equal volumes of cell culture media (820 μl) to a final concentration of 500 mm NaCl, 20 mm Tris (pH 8.0), 3 mm CaCl2, and 20 mm imidazole (binding buffer). A slurry of equilibrated Ni-NTA beads (Qiagen) equal to 5% of the total volume of the immunoprecipitation was added, and the sample was gently rocked overnight at 4 °C. Beads were washed five times with ice-cold binding buffer and eluted by heating in LDS loading buffer (Invitrogen). Transfected cells were rinsed with cold PBS and lysed in lysis buffer on ice for 30 min. Samples were centrifuged at 4 °C at 12,000 × g for 10 min to remove insoluble material. Protein concentrations were determined by the BCA method (Pierce). Results are representative of at least three independent experiments.
AgRP Secretion
Neuro2a.MC4R-eGFP cells (75,000 cells/well) were plated on poly-d-lysine-coated 24-well plates (BD Biosciences) and transfected 24 h later with control, Mgrn1, or Atrn siRNA using RNAiMAX. After 24 h, cells were transfected with empty vector or AgRP with Lipofectamine-2000. Equal volumes of cell culture media were collected and analyzed for AgRP using the Quantikine human AgRP ELISA kit (R&D Systems).
Measurement of cAMP Levels
Plasmid and siRNA transfections were performed as described previously. cAMP was measured with the direct cAMP EIA kit according to the manufacturer's instructions (Assay Designs). Cells were treated with 3-isobutyl-1-methylxanthine (Sigma) 10 min before adding 100 μm α-MSH for 15 min. Total cell protein was determined by the BCA method.
Determination of Cell Surface and Total HA-MC4R-eGFP Levels
HA-MC4R-eGFP levels were determined according to previously described methods with the following modifications (33, 34). Briefly, to quantify MC4R cell surface levels, 75,000 Neuro2a.HA-MC4R-eGFP cells were plated on poly-d-lysine-coated 24-well plates and sequentially transfected with RNAi and plasmids as described previously. Cells were rinsed twice with cold PBS and fixed with 3.7% freshly prepared paraformaldehyde for 15 min on ice, washed three times with PBS, and blocked with PBS containing 5% goat serum and 1% BSA for 30 min. The cells were then incubated with an anti-HA-POD antibody (diluted 1:1000) for 1 h at room temperature in PBS with 1% BSA. The cells were washed five times with PBS, and POD activity was measured by incubating the cells with 400 μl of o-phenylenediamine dihydrochloride reagent (Sigma) for 30 min at room temperature. The reaction was stopped with 100 μl of 3 m HCl, and the product was detected by reading its absorbance at 492 nm. Native Neuro2a cells were used to determine nonspecific binding of the antibody and background absorbance. Total HA-MC4R-eGFP levels were determined using an antibody feeding approach as described by Mohammad et al. (33). Neuro2a.HA-MC4R-eGFP cells were transfected as indicated above, and anti-HA-POD antibodies were added directly to the cell culture media for 1 h at 37 °C. Cells were rinsed, fixed, and washed as above and then permeabilized with 0.2% Triton X-100 for 30 min at room temperature. POD activity was determined as described previously.
MC4R Lysosomal Localization
For confocal microscopy, 175,000 Neuro2a.HA-MC4R-eGFP cells transfected with empty vector or ASP were plated on poly-d-lysine-coated culture slides (BD Biosciences). Cells were incubated with 100 μm cycloheximide (Sigma) and Lysotracker Red DND-99 according to the manufacturer's recommendations (Molecular Probes) for 1 h at 37 °C. Cells were rinsed with PBS, fixed with freshly prepared 3.7% paraformaldehyde, and mounted with SlowFade reagent (Invitrogen). The images were collected using an inverted Zeiss LSM 5 confocal microscope and acquired using a 63× oil immersion objective with 2× optical zoom with Zeiss Pascal software. Image processing was done with Adobe Photoshop/Illustrator CS3 Premium software (Adobe Systems).
Statistical Analysis
All experiments were performed at least twice; the numbers of independent samples comprising each study are indicated in the figure legends. For most experiments, the effects of empty vector, AGRP, or ASP expression between control and Mgrn1/Atrn knockdown cells were compared by using the paired Student's t test.
RESULTS
mRNA and Protein Isoforms of MGRN1 in Neuro2a Cells
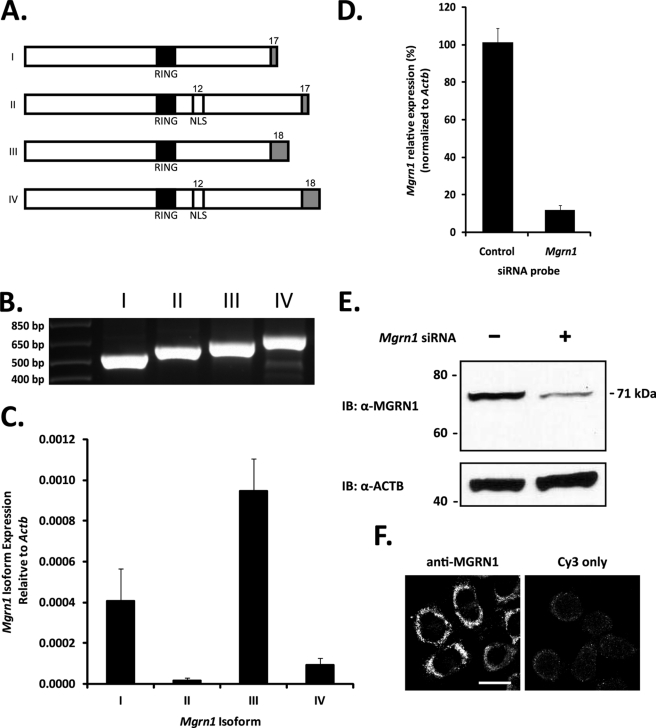
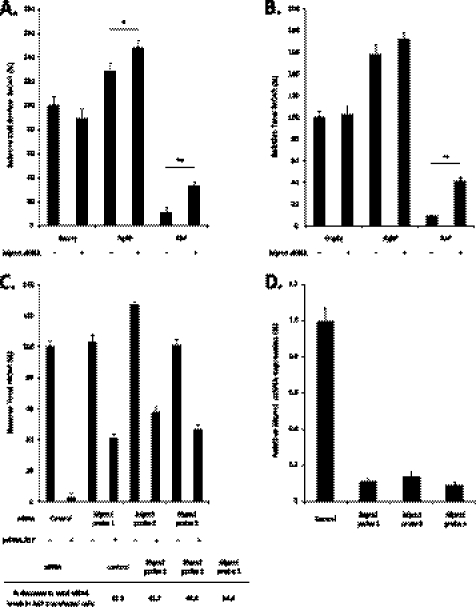
Alternative splicing of Mgrn1 produces four major MGRN1 isoforms (Fig. 2A) (35). The inclusion of exon 12 in isoforms II and IV results in the insertion of a bipartite nuclear localization sequence; alternative splicing of exon 17 results in the final stop codon containing exon of isoforms I and II to differ from isoforms III and IV. In a previous report, no endogenous MGRN1 protein was found in Neuro2a cells (mRNA levels were not reported) (36). However, we recovered all four Mgrn1 isoforms by RT-PCR (Fig. 2B) and detected MGRN1 protein by Western blot (Fig. 2, B and E). Isoform III was the most highly expressed Mgrn1 transcript, followed by isoform I; isoform II was expressed at low levels, and isoform IV was nearly undetectable (Fig. 2C). Mgrn1 transcripts were reproducibly depleted by greater than 85% by transient siRNA transfection (Fig. 2D) and confirmed by Western blotting with a commercially available antibody (Fig. 2E). Endogenous MGRN1 in Neuro2a cells has an apparent molecular mass of ∼71 kDa, larger than the predicted molecular mass of the 555 amino acids of isoform III (61 kDa); occasionally, a second band ∼2 kDa lower in molecular mass, possibly representing the less abundant MGRN1 isoform I (predicted molecular mass of 58.5 kDa), was detected. The subcellular localization of endogenous MGRN1 to discrete punctate structures (probably early and late endosomes, see below) throughout the cytoplasm was consistent with previously published results (Fig. 2F) (29, 37).
FIGURE 2.
MGRN1 expression in Neuro2a cells. A, Mgrn1 encodes an E3 ubiquitin ligase containing a centrally located RING domain (indicated by black box). Alternative splicing produces four Mgrn1 transcripts identified as isoforms I, II, III, and IV. Inclusion of exon 12 in isoforms II and IV introduces a bipartite nuclear localization sequence (NLS). Alternative splicing of the stop codon containing exons, exons 17 and 18 (indicated by gray boxes), causes the final 13 amino acids of isoforms I and II to differ from the last 37 amino acids of isoforms III and IV. B, exon splice site-spanning primers were designed to specifically amplify each Mgrn1 transcript by PCR. All four Mgrn1 transcripts are expressed in Neuro2a cells. C, quantitative real time-PCR analysis of Mgrn1 isoform expression in Neuro2a cells. Expression levels are relative to Actb levels. Expression levels of isoform I are 60% lower than isoform III; isoforms II and IV, the nuclear localization sequence-containing transcripts, are expressed at relatively low levels (98 and 90% lower, respectively, than isoform III). Data are expressed as mean ± S.D. of samples from four different cell passages run in triplicate. D, Neuro2a cells were transfected with either a nontargeting control or Mgrn1 siRNA for 48 h. Mgrn1 knockdown was confirmed by quantitative real time-PCR using primers common to all four Mgrn1 isoforms. Data are expressed as mean ± S.D. of three biological replicates from three independent experiments. E, cells transfected with siRNAs as outlined above were lysed in 1% Igepal buffer, and whole cell lysates were subjected to SDS-PAGE followed by Western blot analysis for MGRN1. The membrane was stripped and re-probed for ACTB as a loading control. F, MGRN1 was visualized in Neuro2a cells by confocal microscopy. IB, immunoblot. Bar, 10 μm.
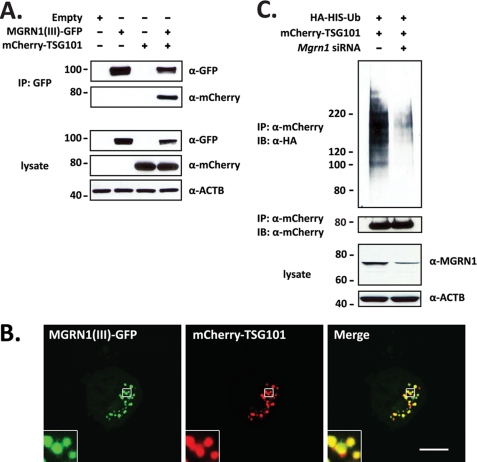
MGRN1 Interacts with and Ubiquitinates TSG101
TSG101 is an MGRN1 substrate (28, 29). MGRN1 interacts with the ubiquitin E2 variant domain of TSG101 via a P(S/T)AP motif (a “late viral domain”) found in the C-terminal half of all vertebrate MGRN1 orthologs; invertebrate MGRN1 proteins lack this motif. Co-immunoprecipitation of MGRN1-GFP followed by immunoblotting for mCherry-TSG101 shows that the proteins interact in Neuro2a cells (Fig. 3A). Transient co-transfection of MGRN1-GFP isoform III and mCherry-TSG101 shows that the proteins co-localize on endosomal structures in the cytoplasm (Fig. 3B).
FIGURE 3.
TSG101 is an MGRN1 ubiquitination substrate in Neuro2a cells. A, MGRN1 interacts with TSG101. Neuro2a cells expressing MGRN1-GFP (isoform III) and mCherry-tagged TSG101 were immunoprecipitated with anti-GFP antibody followed by immunoblotting with anti-GFP and anti-mCherry. Cell lysates were probed with anti-GFP, anti-mCherry, and anti-ACTB. *, p < 0.05. B, MGRN1 co-localizes with TSG101. MGRN1-GFP and mCherry-TSG101 localizations were directly visualized by confocal microscopy in Neuro2a cells by GFP and mCherry fluorescence. Both proteins localized to enlarged endosomal structures in the cytoplasm. Insets show 2-fold magnification of the outlined area. Bar, 10 μm. C, MGRN1 ubiquitinates TSG101. Lysates from Neuro2a cells transfected with mCherry-tagged TSG101, HA-His-tagged ubiquitin, and control or Mgrn1 siRNA were immunoprecipitated with anti-mCherry antibody. Ubiquitinated TSG101 was detected by immunoblotting with anti-HA antibody. Cells transfected with Mgrn1 siRNA showed only low levels of ubiquitinated mCherry-TSG101. IP, immunoprecipitation; IB, immunoblot.
To confirm that endogenous MGRN1 ubiquitinates TSG101, Neuro2a cells were transfected with HA-His-ubiquitin, mCherry-TSG101, and control or Mgrn1 siRNAs. TSG101 ubiquitination was decreased (∼85%) in cells transfected with Mgrn1 siRNA (Fig. 3C). The blot was stripped and re-probed for the mCherry tag to confirm that equal amounts of TSG101 were transfected into and immunoprecipitated from each sample. This result confirms that MGRN1-dependent ubiquitination of TSG101 does not affect the absolute amount of TSG101 present in the cell, which would be anticipated if MGRN1 was targeting TSG101 for degradation in the proteasome. MGRN1 may regulate the solubility and/or endosomal membrane targeting of TSG101 (28), but the exact role of MGRN1 in TSG101 function is unknown.
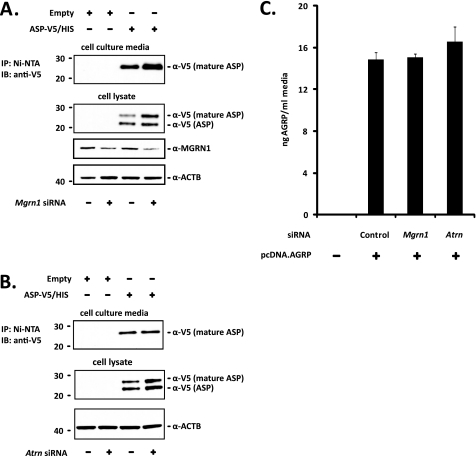
ASP Processing and Secretion Are Unaffected by MGRN1 or ATRN Depletion
The correspondence between MGRN1 levels and TSG101 ubiquitination suggests that the mahoganoid mutation rescues MC1R and MC4R function in Ay mice through a direct effect on endosomal activity. Phenotypes of the mahoganoid and mahogany mutations segregating in Ay mice place the functions of MGRN1 and ATRN downstream of agouti (ASP) transcription and upstream or at the level of MC4R function. A simple explanation for the rescue of MC1R and MC4R function in Mgrn1md and Atrnmg mice ubiquitously overexpressing ASP would be reduced ASP protein production. To assess the processing and secretion of ASP in MGRN1- and ATRN-depleted cells, Neuro2a cells were transfected with Mgrn1 and Atrn siRNAs and V5/His-tagged ASP. The V5/His tag was placed at the C terminus of ASP to avoid interference with signal peptide processing. Two V5-positive bands were detected in the lysate of Neuro2a cells transfected with ASP-V5/His (Fig. 4, A and B). The predicted molecular mass of ASP after signal peptide cleavage is 11.8 kDa; however, Western blotting of mouse skin extracts for endogenous ASP detects a protein of ∼21.5 kDa (38). The upper V5-positive band migrates at ∼25 kDa, the anticipated molecular mass of the ASP-V5/His construct; the lower band is probably ASP that has not been fully processed (i.e. glycosylated). ASP secretion into the cell culture media was assessed by Western blot for the V5 epitope following immunoprecipitation with Ni-NTA beads. Only the upper band, corresponding to the molecular weight of fully processed ASP detected in mouse skin extracts, was recovered from the cell culture media (Fig. 4, A and B). No bands were detected in any of the controls transfected with empty vector. Neither MGRN1 nor ATRN depletion appeared to significantly affect the post-translational processing or secretion of ASP (Fig. 4, A and B), indicating that mahoganoid and mahogany block ASP function by preventing the ASP-MC4R interaction and/or by altering the response of MC4R to ASP binding.
FIGURE 4.
ASP processing and secretion are normal in MGRN1- and ATRN-depleted Neuro2a cells. A, MGRN1 depletion does not affect ASP post-translational processing or secretion into the cell culture media. Neuro2a cells stably expressing MC4R-eGFP (Neuro2a.MC4R-eGFP) previously transfected with control or Mgrn1 siRNA were transfected with empty vector or V5/His-tagged ASP (ASP-V5/His) for 24 h. ASP-V5/His was immunoprecipitated from equal volumes of cell culture media with Ni-NTA beads followed by immunoblotting with anti-V5 antibody. Cells were lysed in 1% Igepal lysis buffer and whole cell lysates probed for ASP-V5/His with anti-V5 antibody. The membrane was stripped and probed for MGRN1 to verify gene knockdown and β-actin (ACTB) as a loading control. B, Atrn knockdown has no affect on ASP-V5/His processing or secretion. The above experiment was repeated in cells transfected with control or Atrn siRNA. Atrn knockdown was confirmed by quantitative RT-PCR to be greater than 80% (data not shown). C, AGRP secretion was not disrupted in Mgrn1 or Atrn-depleted Neuro2a cells. Full-length AGRP was overexpressed in Neuro2a.MC4R-eGFP cells previously transfected with control, Mgrn1, or Atrn siRNA for 24 h and amount secreted into the cell culture media determined by ELISA. Data are expressed as mean ± S.D. (n = 6 biological replicates) of samples run in duplicate. There were no significant differences among the groups. IP, immunoprecipitation; IB, immunoblot.
As a positive control, full-length AGRP was overexpressed in Mgrn1 and Atrn siRNA-transfected Neuro2a cells, and protein levels were measured by ELISA in equal volumes of the culture media. There were no significant differences in the amount of AGRP in the media versus the control following knockdown of either Mgrn1 or Atrn (Fig. 4C). AGRP levels in the media were ∼15–17 ng/ml or 2.5–3.0 nm (based on the assumption that full-length AGRP is processed in the cell to AGRP(82–131)), which is similar to the amount of recombinant AGRP(82–131) generally used in in vitro experiments of AGRP(82–131) action at MC4R (1–10 nm), and are considered adequate to block α-MSH function (33, 34).
α-MSH-mediated Activation of cAMP and Effects of ASP
The observation that ASP processing and secretion were independent of either MGRN1 or ATRN function suggested that md and mg effects on ASP action could be at the level of the ASP-MC4R interaction. ATRN has been predicted to act as a co-receptor for ASP, specifically binding to a region rich in arginine and lysine residues in the N-terminal domain of ASP. Atrn-null animals are completely resistant to ASP effects in vivo (26). Thus, ATRN may be necessary for ASP binding to the MC4R. MGRN1 is an intracellular protein and not likely to be directly involved in the ASP-MC4R interaction in the extracellular space; however, based on the nearly identical phenotypic effects of the mahogany and mahoganoid mutations in Ay mice, ATRN and MGRN1 are likely components of a conserved biochemical pathway involving ASP and MC4R.
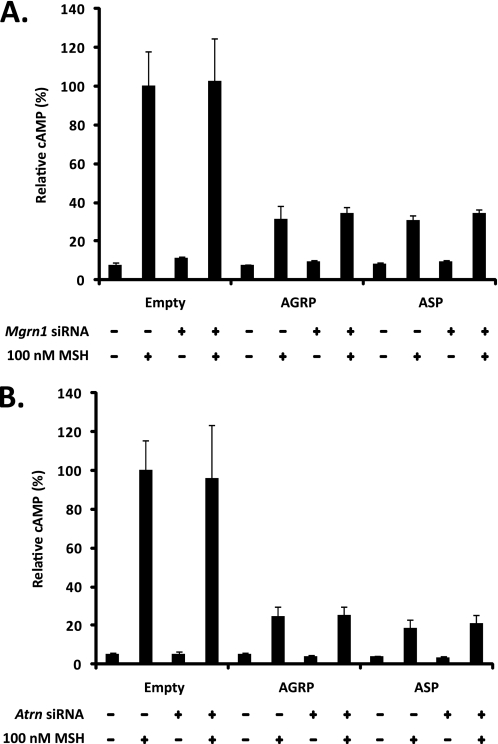
ASP binding to MC4R was indirectly assessed by measuring cAMP levels following α-MSH treatment of Neuro2a cells stably expressing HA-MC4R-eGFP or MC4R-eGFP and transfected with control, Mgrn1, or Atrn siRNAs 24 h before a second round of transfections with an empty vector or untagged ASP. Asp expression was confirmed by RT-PCR (data not shown). AGRP overexpression was included as a positive control as neither Mgrn1 nor Atrn loss-of-function affects its activity at MC4R. Attachment of a GFP tag to the C terminus of MC4R does not alter the ability of the receptor to respond to α-MSH (33, 34, 39).
ASP and AGRP overexpression impaired α-MSH-induced activation of cAMP levels; the α-MSH-dependent cAMP response was consistently 3–5-fold lower in AGRP- and ASP-transfected cells than in cells transfected with the empty vector (Fig. 5, A and B). This result confirms that the transfected ASP is functional and suggests that full-length AGRP is processed to AGRP(82–131) in Neuro2a cells; full-length AGRP is 10-fold less effective than AGRP(82–131) at blocking the α-MSH-dependent generation of cAMP (40). Interestingly, neither Mgrn1 nor Atrn depletion restored α-MSH-dependent cAMP generation in the ASP-overexpressing cells (Fig. 5, A and B). As expected, Mgrn1/Atrn depletion had no effect on AGRP action. These findings show that ASP binding to the MC4R and antagonism of the α-MSH-dependent cAMP pathway are essentially independent of MGRN1 and ATRN function, implying that there must be a cAMP-independent component of ASP function at the MC4R that is dependent on both MGRN1 and ATRN. Similar results have been observed at MC1R in immortalized melanocytes from Atrnmg−3J/mg−3J and Mgrn1md−nc/md−nc mice (41).
FIGURE 5.
Mgrn1 and Atrn depletion do not restore α-MSH-dependent generation of cAMP in ASP-overexpressing cells. A, ASP antagonism of α-MSH-dependent cAMP pathway is independent of MGRN1 function. Empty vector, AGRP, or ASP was overexpressed in Neuro2a.HA-MC4R-eGFP cells previously transfected with control or Mgrn1 siRNA for 24 h. Cells were treated with 0.5 mm 3-isobutyl-1-methylxanthine for 10 min and then stimulated with 100 nm α-MSH for 15 min at 37 °C. The amount of cAMP generated was normalized to total protein levels by the BCA method. Data are expressed as mean ± S.D. of six independent samples relative to empty vector transfected controls. B, ASP antagonism of the α-MSH-dependent cAMP pathway is also independent of ATRN function. The above experiment was repeated in cells transfected with control or Atrn siRNA.
Depletion of MGRN1 or ATRN Blocks ASP-dependent MC4R Trafficking to the Lysosome
Because neither Mgrn1 nor Atrn depletion interrupts ASP processing, secretion, or binding to MC4R, the mahoganoid and mahogany mutations must rescue MC4R function in Ay mice at the level of ASP-dependent MC4R regulation (see Fig. 1). Pharmacological effects of ASP antagonism on the melanocortin receptor present in isolated Xenopus melanophores indicate that ASP action involves both time- and temperature-dependent inhibition of melanocortin signaling (38). Similar conclusions were reached in B16-F1 murine melanoma cells that express MC1R (42). These findings suggest that, like a traditional receptor antagonist, ASP blocks agonist binding but also, unconventionally, down-regulates receptor levels.
In Neuro2a cells, α-MSH down-regulates cell surface levels of MC4R by sequestering the ligand-bound receptor in a cytoplasmic compartment; when the agonist is removed from the media, the receptor is efficiently recycled back to the cell surface (33). The effects of ASP and AGRP on MC4R trafficking in Neuro2a cells have not been thoroughly investigated. Neuro2a cells stably expressing HA-MC4R-eGFP were generated to characterize the distribution and trafficking of MC4R in Mgrn1- and Atrn-depleted cells in response to ASP and AGRP overexpression. Attachment of an HA tag to the N terminus of MC4R does not appear to alter α-MSH binding or change the ability of the receptor to generate cAMP (34). Again, AGRP-overexpressing cells were included as positive controls because neither MGRN1 nor ATRN are expected to affect AGRP function (26, 43, 44). HA-MC4R-eGFP at the cell surface was detected by an enzyme-linked immunoassay using anti-HA-POD as described previously (33, 34). siRNA-mediated reduction of MGRN1 did not have a significant effect on cell surface levels of MC4R in cells transfected with empty vector (Fig. 6A). ASP overexpression for 24 h decreased the abundance of MC4R on the cell surface by greater than 90% compared with the empty vector in control siRNA cells; in MGRN1-knockdown cells, surface MC4R was ∼60% lower in ASP-transfected cells versus empty vector-transfected cells, resulting in ∼4-fold more MC4R on the cell surface in ASP overexpressing Mgrn1-depleted cells versus siRNA control (Fig. 6A). No significant differences in cell number or total cell protein between any of the conditions tested (siRNA or plasmid) were observed (supplemental Figs. S1 and S2). This finding confirms that ASP down-regulates MC4R surface levels in Neuro2a cells by a mechanism that is at least partially dependent on intact MGRN1 function.
FIGURE 6.
MGRN1 depletion blocks ASP-dependent trafficking of MC4R to the lysosome. A, ASP-dependent decrease in HA-MC4R-eGFP cell surface levels is partially blocked by reduction of MGRN1. Neuro2a cells stably expressing HA-MC4R-eGFP (Neuro2a.HA-MC4R-eGFP) previously transfected with control or Mgrn1 siRNA were transfected with empty vector, AGRP-, or ASP-expressing plasmids for ∼24 h. Cells were fixed under nonpermeabilizing conditions, incubated with POD-conjugated anti-HA antibodies (anti-HA-POD), and washed extensively. HA-MC4R-eGFP cell surface levels were determined colorimetrically after incubation with the POD substrate o-phenylenediamine dihydrochloride. Unless otherwise indicated, data are presented relative to control siRNA/empty vector transfected cells (100%) and represent the mean ± S.D. of at least five independent samples. Both nonspecific binding and background were measured and subtracted from HA-MC4R-eGFP samples in all experiments. *, p < 0.05; **, p < 0.001. B, ASP overexpression leads to MGRN1-dependent MC4R degradation. Transfections were performed as above. Cells were incubated with anti-HA-POD antibodies for 1 h at 37 °C, fixed, and permeabilized to measure total HA-MC4R-eGFP with the o-phenylenediamine dihydrochloride reagent. C, MGRN1 function is necessary for ASP-dependent MC4R degradation. Upper panel, the total HA-MC4R-eGFP experiment in B was repeated with three different Mgrn1 siRNA probes against three different segments of the Mgrn1 sequence common to all four Mgrn1 isoforms. Lower panel, % decrease in total HA-MC4R-eGFP levels in Mgrn1 siRNA-transfected ASP-overexpressing cells versus controls. D, Mgrn1 knockdown in C was confirmed by quantitative real time-PCR using primers common to all four Mgrn1 isoforms. Data are expressed as mean ± S.D. of three biological replicates from cells co-transfected with those in experiment C.
Surprisingly, AGRP overexpression for 24 h increased the amount of MC4R on the cell surface by 30–40 and 50–60% in control and Mgrn1 siRNA-transfected cells, respectively (Fig. 6A). This result suggests that AGRP diverts MC4R trafficking away from normal lysosomal degradation and/or increases the number of receptors at the plasma membrane by depleting the intracellular MC4R pool. Increased HA-MC4R-eGFP cell surface levels in AGRP-overexpressing cells are unlikely to be related to transcriptional effects as MC4R expression is under the control of the cytomegalovirus promoter. Similar increases in MC4R cell surface levels in cells treated with AGRP have been observed in HEK293 cells overexpressing HA-MC4R (34); however, conflicting results in the same cell line have been reported (45).
Because a fraction of MC4R is located intracellularly at steady state in Neuro2a cells, the total amount of HA-MC4R-eGFP was measured by a previously established antibody-feeding approach (33). MGRN1 depletion had no effect on total MC4R levels in cells transfected with empty vector (Fig. 6B). ASP overexpression decreased the total amount of HA-MC4R-eGFP by more than 90% (Fig. 6B), implying that ASP binding to the MC4R traffics the receptor toward the lysosomal degradation pathway. ASP-dependent reduction in MC4R levels in Neuro2a cells was further confirmed by Western blot for the receptor in empty vector and ASP-overexpressing cells (supplemental Fig. S3). ASP-dependent MC4R trafficking to the lysosome appears to be an MGRN1-dependent process; MGRN1 depletion increased the total amount of HA-MC4R-eGFP in ASP-transfected cells ∼5-fold compared with control siRNA (Fig. 6B). MGRN1-dependent trafficking of MC4R to the lysosome in ASP-overexpressing cells was confirmed using three different Mgrn1 siRNA sequences; each probe decreased Mgrn1 mRNA levels by greater than 85% (Fig. 6D) and rescued ∼40–50% of HA-MC4R-eGFP from ASP-dependent lysosomal degradation (Fig. 6C).
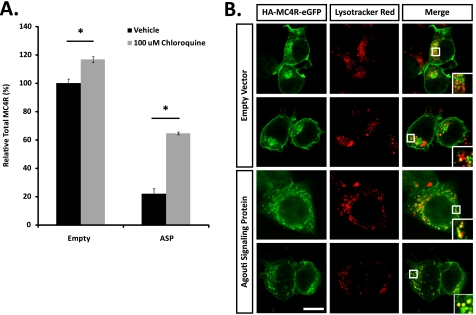
An 8-h incubation with chloroquine, a weak base that increases the pH of intracellular acidic compartments and thereby reduces lysosomal degradation, blocked MC4R degradation in ASP-transfected cells (Fig. 7A). Additionally, direct confocal microscopy of Neuro2a.HA-MC4R-eGFP cells transfected with empty vector shows MC4R predominantly localized to both the plasma membrane and an endosomal compartment with very little or no overlap with lysosomes, although in cells overexpressing ASP, the receptor is largely intracellular and displays extensive co-localization with the lysosomal compartment (Fig. 7B). These results strongly suggest that ASP overexpression targets MC4R for lysosomal degradation.
FIGURE 7.
MC4R is degraded in the lysosome in ASP-overexpressing Neuro2a cells. A, lysosomal inhibition blocks ASP-dependent MC4R degradation. Neuro2a.HA-MC4R-eGFP cells transfected with empty vector or ASP for 16 h were incubated with 100 μm chloroquine for 8 h to block protein degradation in the lysosome. Total HA-MC4R-eGFP levels were measured as indicated above. Data represent the mean ± S.D. of eight independent samples. *, p < 0.05. B, MC4R co-localizes with lysosomes in ASP-overexpressing cells. Neuro2a.HA-MC4R-eGFP cells transfected with empty vector or ASP were incubated with 100 μm cycloheximide and Lysotracker Red for 1 h at 37 °C. MC4R and lysosome co-localization was identified by confocal microscopy via direct fluorescence of GFP and Lysotracker Red. HA-MC4R-eGFP in cells transfected with empty vector (upper two lines) localizes to the plasma membrane and an unidentified endosomal compartment, while the receptor shows strong co-localization with lysosomes in cells overexpressing ASP (bottom two lines). Bar, 10 μm.
The total amount of HA-MC4R-eGFP in both control and Mgrn1 siRNA-transfected AGRP-overexpressing cells was 60–70% higher than in empty vector controls, suggesting that AGRP enhances MC4R cell surface levels (Fig. 6A) primarily by reducing receptor degradation and not by redistributing the receptor from the intracellular pool to the cell surface. This result is confirmed by Western blot for HA-MC4R-eGFP in empty vector and AGRP-overexpressing cells (supplemental Fig. 3).
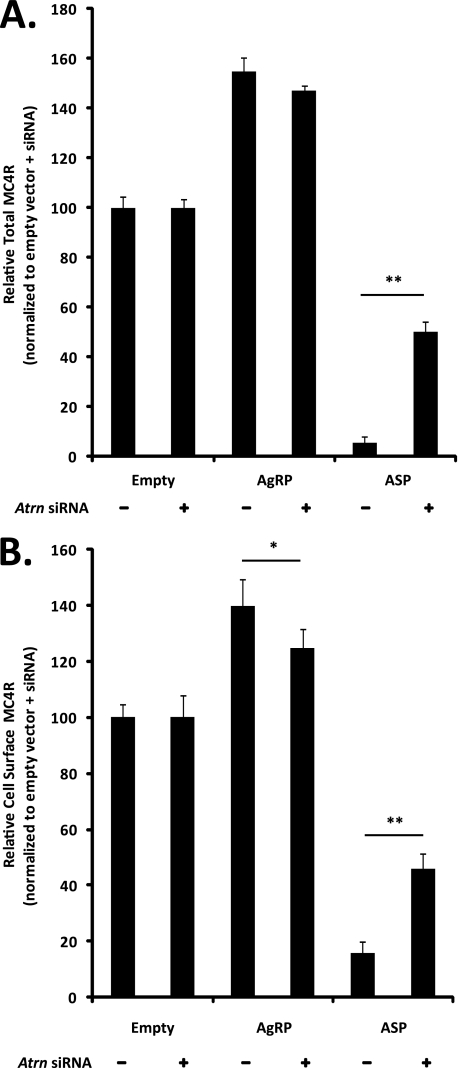
Because genetic analysis indicates that MGRN1 and ATRN are components of a conserved biochemical pathway, the effect of Atrn depletion on ASP-dependent MC4R trafficking to the lysosome was also examined. Atrn knockdown for 48 h increased the absolute amount of HA-MC4R-eGFP present in empty vector, AGRP-, and ASP-transfected cells by at least 2-fold without affecting cell number (data not shown), suggesting that reduction of ATRN function disrupts normal receptor turnover in the lysosome but does not affect cell survival or replication. For this reason, changes in HA-MC4R-eGFP levels in Atrn siRNA-transfected cells overexpressing AGRP or ASP are expressed relative to Atrn siRNA cells transfected with empty vector rather than to control (scrambled) siRNA. After 24 h of ASP overexpression, both cell surface and total HA-MC4R-eGFP levels were decreased by 85–95%. Atrn knockdown partially blocked ASP-dependent MC4R trafficking to the lysosome; both cell surface and total HA-MC4R-eGFP levels were ∼50% of the empty vector-transfected controls (Fig. 8, A and B). These results suggest that lysosomal targeting of MC4R in ASP-overexpressing cells is dependent on the presence of both MGRN1 and ATRN.
FIGURE 8.
Atrn knockdown prevents ASP-dependent MC4R degradation in the lysosome. A, ASP overexpression stimulates ATRN-dependent MC4R degradation. Atrn depletion increases total HA-MC4R-eGFP levels in ASP-transfected cells relative to control siRNA. Total HA-MC4R-eGFP levels were determined as described previously in cells transfected with control or Atrn siRNA. Atrn deficiency increases the absolute amount of both total and cell surface HA-MC4R-eGFP by ∼2-fold (see text); therefore, data are presented relative to control or Atrn siRNA empty vector transfected cells (each set to 100%). B, Atrn depletion increases HA-MC4R-eGFP cell surface levels in ASP-transfected cells relative to control siRNA. Cell surface HA-MC4R-eGFP was measured as described in Fig. 6, and results are presented as described above. *, p < 0.05; **, p < 0.001.
DISCUSSION
Mahogany (Atrnmg) and mahoganoid (Mgrn1md), loss-of-function mutations in ATRN and MGRN1, rescue the functional consequences at MC1R and MC4R of the constitutive overexpression of ASP that results from gain-of-function mutations at the agouti locus (22, 23). However, neither mg nor md can rescue the obesity associated with MC4R-null mutations (21), and neither mutant is protected from obesity caused by ubiquitous overexpression of AGRP,4 the physiological MC4R antagonist. Thus, ATRN and MGRN1 are specifically required for ASP-mediated effects on MC4R (and MC1R) signaling. Based on genetic models, MGRN1, an E3 ubiquitin ligase, and ATRN, a transmembrane protein, must act downstream of ASP and upstream or at the level of the MC4R to enable ASP-dependent obesity. Consistent with this premise, we show here that MGRN1 and ATRN are required for ASP-dependent MC4R degradation in the lysosome.
Several mechanisms have been proposed to account for the loss-of-function of ASP at the MC4R in md and mg mice. MGRN1 and ATRN may be required for the following: 1) ASP processing and/or secretion; 2) ASP binding to MC4R; or 3) ASP-dependent regulation of MC4R number/function (Fig. 1) (22, 27). We used Neuro2a cells stably expressing MC4R constructs as a model system to examine each of these possibilities. Reportedly, no MGRN1 is present in Neuro2a cells (36). However, we recovered mRNA for all four Mgrn1 isoforms, showed the expected decrease in protein levels following siRNA transfections, and, as reported by others, observed MGRN1 localization to the endosomal compartment (29). We and others have yet to find a cell line in which Mgrn1 is not present,4 strongly suggesting that the protein performs an essential housekeeping function(s).
Mechanism 1
Perhaps the simplest explanation for the apparent loss-of-function of ASP in md and mg mice would be an MGRN1-/ATRN-dependent effect on the secretion and/or processing of ASP. ASP has been detected in the brains of ATRN-null Ay mice (23), arguing against mechanism 1. Although ASP is present in equal amounts in hypothalamic extracts of control and Atrn-null Ay animals (23), it could have been retained within the cells and never secreted, i.e. have been functionally inactive. We were able to recover sufficient ASP from the media of ASP-transfected cells, following siRNA-mediated MGRN1/ATRN depletion, to conclude that neither MGRN1 nor ATRN is primarily involved in ASP secretion. The apparent molecular weight of ASP immunoprecipitated from the media of Mgrn1 and Atrn siRNA-transfected cells was indistinguishable from that from control cells and consistent with ASP from mouse skin extracts (38), suggesting that MGRN1 and ATRN are not involved in the post-translational processing of ASP.
Mechanism 2
ATRN binds ASP with low affinity, possibly acting as a co-receptor to enhance ASP binding to MC4R and/or to increase the local concentration of the ligand in the vicinity of the receptor (26). Based on this observation, MGRN1 and ATRN loss-of-function mutations might prevent ASP from binding to the MC4R, allowing increased α-MSH access to the receptor. Surprisingly, neither MGRN1 nor ATRN depletion had any effect on the ability of ASP to reduce α-MSH-dependent cAMP levels, suggesting that ASP-MC4R signaling must include an MGRN1-/ATRN-dependent, cAMP-independent, signaling pathway. However, based on this negative result, we cannot exclude the possibility that small amounts of cAMP are generated in specific subcellular locations. Similar conclusions have been reached in experiments examining ASP-MC1R signaling in melanocytes isolated from MGRN1- and ATRN-null mice (41). Pérez-Oliva et al. (46) showed that MGRN1 overexpression in HEK293 cells transfected with MC1R or MC4R blocks α-MSH-dependent cAMP signaling by interfering with Gαs coupling to the receptors. Although interesting, this model predicts that MGRN1 loss-of-function mutations would up-regulate MC4R cAMP signaling, a result that we did not observe. Additionally, MGRN1 overexpression in mice fails to cause coat color darkening in wild-type (agouti) mice (37). Such darkening, which is indicative of hyperactivity of the MC1R pathway, would be predicted by the in vitro observations of Pérez-Oliva et al. (46). The discrepancies that exist between the work of Pérez-Oliva et al. (46) and those shown here and of others (37, 41) could be a consequence of studying MC1R/MC4R function in a cell type (HEK293) in which neither MC1R nor MC4R is endogenously expressed.
Mechanism 3
Based on the functional interaction between MGRN1 and TSG101 (28, 29), an essential component of the endosomal machinery required for receptor trafficking to the lysosome, we hypothesized that MGRN1 and likely ATRN are required for MC4R degradation. ASP binding to the melanocortin receptor in Xenopus melanophores (38) and to MC1R in B16 melanoma cells (42) blocks receptor signaling through both competitive antagonism and receptor down-regulation. The C-terminal end of ASP binds to MC1R/MC4R and blocks agonist binding; the structurally homologous domain of AGRP blocks agonist binding at MC4R but shows no affinity for MC1R (14). ASP-dependent MC1R down-regulation occurs via an unknown mechanism that requires both the C-terminal and N-terminal domains of ASP (AGRP lacks a homologous N-terminal domain (47)); MC4R down-regulation following ASP binding has not been investigated. We found that ASP overexpression in Neuro2a cells stably transfected with MC4R decreases both cell surface and total MC4R levels by stimulating receptor trafficking to the lysosome. This same response was not observed in cells overexpressing AGRP, suggesting that this effect is dependent on the N terminus of ASP. In agreement with the results of others (34), AGRP increased the amount of MC4R present at the cell surface independently of MC4R transcription. Furthermore, we showed that AGRP overexpression increased total MC4R levels, suggesting that AGRP blocks α-MSH signaling at the cell surface and, surprisingly, stabilizes overall receptor levels by diverting the MC4R away from degradation, a process independent of MGRN1 and ATRN function. As an inverse agonist, AGRP may bind to and stabilize the inactive conformation of MC4R (48) and that MC4R is trafficked away from the lysosome when locked in its inactive state. Whether C-terminal fragments of ASP (i.e. lacking the N-terminal domain necessary for lysosomal targeting) have effects on MC4R trafficking similar to those of AGRP is unknown. ASP-dependent MC4R lysosomal trafficking requires MGRN1 and ATRN and is partially blocked by suppressing the expression levels of either protein. Diversion of MC4R trafficking away from lysosomal degradation is likely to be directly related to the rescue of the Ay metabolic phenotypes in mahogany and mahoganoid mice.
Unexpectedly, suppression of ATRN greatly increased both cell surface and total MC4R levels, even in the absence of ASP overexpression. However, although the presence of MC4R on the plasma membrane was increased, cAMP levels in response to α-MSH were unchanged. Interestingly, mutations at the Atrn locus reportedly disrupt the structural integrity of specialized plasma membrane microdomains (e.g. lipid rafts) but have no effect on signal transduction pathways (49). The increased number of MC4Rs observed on the plasma membrane of Atrn siRNA-transfected cells may represent a compensatory mechanism for the loss of a specific signaling microenvironment in mahogany mutant cells, accounting to some extent for the absence of increased body weight in Ay/Atrnmg mice (26). Receptor clustering in lipid rafts following ligand binding is an important factor in the initiation of a variety of signal transduction events, including the generation of cAMP (50). Pretreatment of Neuro2a.MC4R-eGFP cells with methyl-β-cyclodextrin, a reagent that removes cholesterol from the plasma membrane and disrupts the formation of lipid rafts (51), reduced α-MSH-dependent cAMP generation by ∼70% (data not shown), suggesting a connection between lipid raft formation and normal MC4R signaling function.
We were unable to show a direct physical interaction between MGRN1 or ATRN and MC4R by co-immunoprecipitation in Neuro2a cells under basal conditions or transiently transfected with ASP (data not shown). This result suggests that ASP forms a bridge between ATRN and MC4R with the two proteins interacting exclusively through the N and C termini of ASP; ASP binding to ATRN may be too weak or too transient to detect using conventional immunoprecipitation approaches. It is also possible that additional molecules may mediate some parts of this interaction. For example, ATRNL1 (attractin-like 1), is an ATRN homolog that has been shown to interact with the C terminus of MC4R (52) and, when overexpressed, can compensate for loss of ATRN (53). However, because loss-of-function mutations in Atrnl1 show no pigmentary or body weight phenotypes (53) its direct role, if any, in melanocortin biology remains unclear.
Our inability to demonstrate a direct physical interaction between MGRN1 and MC4R disagrees with previous work (46); it is possible that our system was not sufficiently sensitive to detect this interaction and/or that the interaction is dependent on differences in the cellular models used. The MC4R is constitutively trafficked through the endosomal recycling pathway in Neuro2a cells (33), but it does not appear to be recycled in HEK293 cells (39). The physical interactions, if any, between MGRN1 and MC4R in Neuro2a cells may be more transient or less stringent than those observed in HEK293 cells (46), and they eluded our detection. Lack of a specific physical interaction between MGRN1/ATRN and the MC4R and the pleiotropic nature of the phenotypes associated with the mahogany and mahoganoid mutations that are independent of melanocortin signaling strongly suggest that MGRN1 and ATRN are part of a conserved protein trafficking pathway that is required for multiple signaling proteins. Further elucidation of this pathway will likely explain the association of the md and mg mutations with neurodegeneration (27, 54), mitochondrial dysfunction (55, 56), and abnormalities such as left-right embryonic patterning defects (particularly in the heart) in MGRN1-null mice (57) and compromised immune cell function in mahogany mutant animals (58).
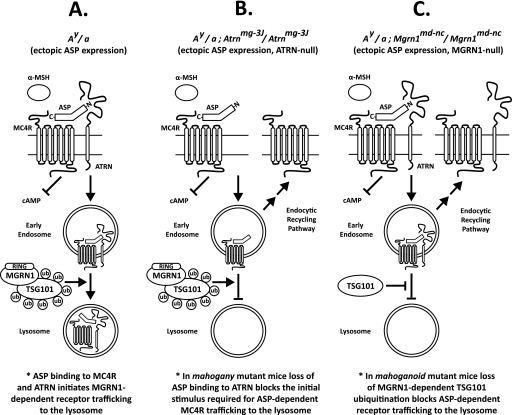
We propose the following model for ASP action at the MC4R in Ay mice (and possibly the physiological ASP-MC1R interaction in the skin) (Fig. 9A). At the cell surface, ASP antagonizes α-MSH signaling through direct binding of its C terminus to the MC4R, and it initiates receptor trafficking to the lysosome via the interaction of its N terminus with the extracellular domain of ATRN. This step results in the degradation of MC4R and recapitulates the obese phenotype observed in MC4R-null mice (7). The ASP-ATRN interaction could stimulate MC4R trafficking to the lysosome by inducing a conformational change in the receptor or by recruiting accessory proteins, such as MGRN1, that are necessary for lysosomal trafficking. Following receptor internalization, MC4R proceeds to the lysosome via the MGRN1-dependent multiubiquitination of TSG101, effectively blocking any further possibility of α-MSH signaling or recycling to the plasma membrane. It is likely that the MC1R is physiologically regulated (i.e. in the presence of ASP produced normally in skin) by similar mechanisms as MC4R in the brain. Increased degradation of the MC1/4R in the presence of ASP fully explains why ubiquitous overexpression of ASP in the Ay mouse produces phenotypes resembling those of null alleles for both MC1R and MC4R. Furthermore, this receptor degradation hypothesis is consistent with the epistatic relationships among the participating genes and explains why mg and md cannot rescue MC1R- and MC4R-null mutations; both ATRN and MGRN1 act at the level of these receptors. In mg mutant Ay mice, the interaction between the N terminus of ASP and the extracellular domain of ATRN is lost, eliminating the initial stimulus required for ASP-dependent MC4R trafficking to the lysosome (Fig. 9B). Loss of MGRN1 function in Ay md mice decreases TSG101 ubiquitination, which impairs the function of TSG101, and prevents MC4R trafficking from early endosomes to lysosomes (Fig. 9C).
FIGURE 9.
Proposed model. ASP-dependent MC4R trafficking to the lysosome in Ay mice requires both ATRN and MGRN1 function. A, C-terminal domain of ASP binds to MC4R at the cell surface, preventing α-MSH access to the receptor and blocking the generation of cAMP. A basic amino acid-rich domain in the N terminus of ASP interacts with the extracellular domain of ATRN and initiates MC4R trafficking to the lysosome. Following internalization, MGRN1-dependent TSG101 multiubiquitination is required for MC4R trafficking from early endosomes to the lysosome for degradation (see under “Discussion” for further details). Similar mechanisms are likely to regulate ASP-dependent MC1R degradation in the skin. B, in mg mutant mice, the interaction between the ASP N terminus and the extracellular domain of ATRN is lost eliminating the initial stimulus required for MC4R trafficking to the lysosome. Internalized MC4R is then preferentially directed toward the default endosomal recycling pathway and returned to the cell surface. C, in md mutant mice, TSG101 is not multiubiquitinated, and ASP-bound MC4R is diverted away from the lysosomal pathway and returned to the plasma membrane. Thus, both of these loss-of-function mutations produce gain-of-function MC4R phenotypes in Ay mice overexpressing ASP. Similar mechanisms are probably relevant at MC1R. In both mg and md mice, the C terminus of ASP binds MC4R and prevents α-MSH-dependent cAMP generation. Ub, ubiquitin.
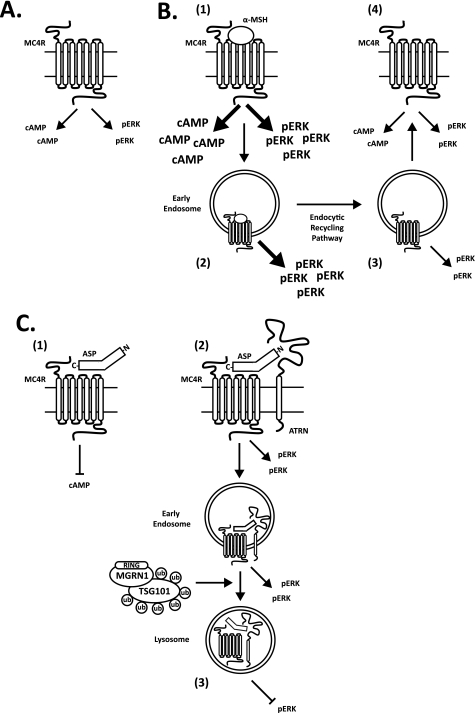
Additional work is necessary to determine how the restoration of MC4R levels in mahogany and mahoganoid mutant Ay mice allows MC4R to resume its biological function(s). We propose dual signaling mechanisms for MC4R (see Fig. 10 for details) as follows: 1) the classical cAMP-dependent pathway that is inhibited by the C terminus of ASP, and 2) a second signaling pathway that is blocked, independently, by the N terminus of ASP. A similar mechanism has been recently proposed by Bennett and co-workers (41) for the MC1R. Considering our results, this second signaling pathway would be attenuated following ASP binding to MC4R via receptor degradation in the lysosome. Multiple signaling intermediates are reportedly activated by MC4R signaling (59), the MAPK pathway being a strong candidate as a second messenger in our proposed cAMP-independent system. Following EGF binding at the EGF receptor, ERK1/2 signaling is initiated at the cell surface and continues from within the cell until the EGF receptor is fully encapsulated within a multivesicular body en route to the lysosome (60); a similar signaling pathway could exist for MC4R. A “second candidate signaling pathway” (represented by pERK in Fig. 10) could, hypothetically, be activated by the β-defensins, additional melanocortin receptor ligands encoded by the K locus (61). Transgenic mice expressing β-defensins have a black coat color and reduced body weight, mimicking the biological function of α-MSH at the MC1R and MC4R; these effects are conveyed by cAMP-independent mechanisms (61).
FIGURE 10.
Dual MC4R signaling model to account for “rescue” of Ay phenotype by alterations in MC4R trafficking. A, MC4R is shown with constitutive cAMP-dependent and hypothetical cAMP-independent activity (possibly the signaling pathway activated by β-defensins and represented in the figure by pERK). B, α-MSH up-regulates both the cAMP-dependent and -independent signaling pathways (step 1) (arrow/text size indicates direction of effect). cAMP is generated only while the MC4R is on the plasma membrane; pERK remains up-regulated following receptor internalization (step 2) until ligand and receptor dissociate along the endocytic recycling pathway (step 3). Upon α-MSH withdrawal, cAMP and pERK return to basal levels (step 4). C, ASP antagonizes cAMP signaling through its C terminus (step 1) and pERK signaling through its N terminus (step 2) (requiring MGRN1 and ATRN). pERK is only completely attenuated following ASP-dependent MC4R degradation in the lysosome (step 3). Loss-of-function mutations in ATRN or MGRN1 impair the ASP-dependent lysosomal trafficking of MC4R, preventing the full reduction of pERK signaling and allowing basal cAMP-independent activity to be maintained. The hypothetical pERK signaling pathway is analogous to that of the activated EGF receptor; ERK signaling initiated at the cell surface continues from within the cell until the EGF receptor enters the lysosome.
Further understanding of the complex trafficking pathways of the MC4R, including those involving MGRN1 and ATRN, could provide help in the treatment of human obesity. Mutations in the MC4R coding sequence account for up to 5–6% of severe obesity in humans (62). Functional characterization of the many mutations that have been detected in MC4R in obese humans indicates that reduced cell surface expression secondary to intracellular retention is among the most common molecular defects (59). These results should be interpreted with caution, however, as most MC4R mutations have been studied in HEK293 cells, a cell line in which MC4R function may not be fully represented either qualitatively or quantitatively. Our results and the findings of others (33) show that MC4R endosomal trafficking is dynamic; MC4R shows three unique signaling/trafficking patterns in response to three separate ligands as follows: 1) α-MSH, the physiological agonist of the receptor, induces signaling and causes the receptor to be retained intracellularly; 2) AGRP, the naturally occurring antagonist, blocks agonist binding and stabilizes receptor levels by diverting the MC4R away from the lysosome; and 3) ASP, an antagonist only present in agouti mutant animals, blocks agonist binding and, as opposed to AGRP, promotes receptor degradation in the lysosome.
Supplementary Material
Acknowledgment
We are grateful to Dr. Timothy McGraw (Cornell University) for helpful advice with regard to some of the experiments reported here and for comments on an earlier draft of this manuscript.
This work was supported in part by National Institutes of Health Grants R01-DK52431 and P30-DK26687.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
Teresa M. Gunn, personal communication.
- ASP
- agouti-signaling protein
- AGRP
- agouti related protein
- α-MSH
- α-melanocyte-stimulating hormone
- ATRN
- attractin
- MCR
- melanocortin receptor
- POD
- peroxidase
- F
- forward
- R
- reverse
- Ni-NTA
- nickel-nitrilotriacetic acid.
REFERENCES
- 1. Gantz I., Miwa H., Konda Y., Shimoto Y., Tashiro T., Watson S. J., DelValle J., Yamada T. (1993) J. Biol. Chem. 268, 15174–15179 [PubMed] [Google Scholar]
- 2. Lu D., Willard D., Patel I. R., Kadwell S., Overton L., Kost T., Luther M., Chen W., Woychik R. P., Wilkison W. O., et al. (1994) Nature 371, 799–802 [DOI] [PubMed] [Google Scholar]
- 3. Abdel-Malek Z. A. (2001) Cell. Mol. Life Sci. 58, 434–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Artigas R. A., Gonzalez A., Riquelme E., Carvajal C. A., Cattani A., Martínez-Aguayo A., Kalergis A. M., Pérez-Acle T., Fardella C. E. (2008) J. Clin. Endocrinol. Metab. 93, 3097–3105 [DOI] [PubMed] [Google Scholar]
- 5. Balthasar N., Dalgaard L. T., Lee C. E., Yu J., Funahashi H., Williams T., Ferreira M., Tang V., McGovern R. A., Kenny C. D., Christiansen L. M., Edelstein E., Choi B., Boss O., Aschkenasi C., Zhang C. Y., Mountjoy K., Kishi T., Elmquist J. K., Lowell B. B. (2005) Cell 123, 493–505 [DOI] [PubMed] [Google Scholar]
- 6. Chen W., Kelly M. A., Opitz-Araya X., Thomas R. E., Low M. J., Cone R. D. (1997) Cell 91, 789–798 [DOI] [PubMed] [Google Scholar]
- 7. Huszar D., Lynch C. A., Fairchild-Huntress V., Dunmore J. H., Fang Q., Berkemeier L. R., Gu W., Kesterson R. A., Boston B. A., Cone R. D., Smith F. J., Campfield L. A., Burn P., Lee F. (1997) Cell 88, 131–141 [DOI] [PubMed] [Google Scholar]
- 8. Robbins L. S., Nadeau J. H., Johnson K. R., Kelly M. A., Roselli-Rehfuss L., Baack E., Mountjoy K. G., Cone R. D. (1993) Cell 72, 827–834 [DOI] [PubMed] [Google Scholar]
- 9. Star R. A., Rajora N., Huang J., Stock R. C., Catania A., Lipton J. M. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 8016–8020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Van der Ploeg L. H., Martin W. J., Howard A. D., Nargund R. P., Austin C. P., Guan X., Drisko J., Cashen D., Sebhat I., Patchett A. A., Figueroa D. J., DiLella A. G., Connolly B. M., Weinberg D. H., Tan C. P., Palyha O. C., Pong S. S., MacNeil T., Rosenblum C., Vongs A., Tang R., Yu H., Sailer A. W., Fong T. M., Huang C., Tota M. R., Chang R. S., Stearns R., Tamvakopoulos C., Christ G., Drazen D. L., Spar B. D., Nelson R. J., MacIntyre D. E. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 11381–11386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhou A., Bloomquist B. T., Mains R. E. (1993) J. Biol. Chem. 268, 1763–1769 [PubMed] [Google Scholar]
- 12. Barsh G. S., Ollmann M. M., Wilson B. D., Miller K. A., Gunn T. M. (1999) Ann. N.Y. Acad. Sci. 885, 143–152 [DOI] [PubMed] [Google Scholar]
- 13. Ollmann M. M., Lamoreux M. L., Wilson B. D., Barsh G. S. (1998) Genes Dev. 12, 316–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ollmann M. M., Wilson B. D., Yang Y. K., Kerns J. A., Chen Y., Gantz I., Barsh G. S. (1997) Science 278, 135–138 [DOI] [PubMed] [Google Scholar]
- 15. Chai B. X., Neubig R. R., Millhauser G. L., Thompson D. A., Jackson P. J., Barsh G. S., Dickinson C. J., Li J. Y., Lai Y. M., Gantz I. (2003) Peptides 24, 603–609 [DOI] [PubMed] [Google Scholar]
- 16. Millar S. E., Miller M. W., Stevens M. E., Barsh G. S. (1995) Development 121, 3223–3232 [DOI] [PubMed] [Google Scholar]
- 17. Michaud E. J., Bultman S. J., Stubbs L. J., Woychik R. P. (1993) Genes Dev. 7, 1203–1213 [DOI] [PubMed] [Google Scholar]
- 18. Miller M. W., Duhl D. M., Vrieling H., Cordes S. P., Ollmann M. M., Winkes B. M., Barsh G. S. (1993) Genes Dev. 7, 454–467 [DOI] [PubMed] [Google Scholar]
- 19. Lane P. M. (1960) Mouse News Lett. 22, 35 [Google Scholar]
- 20. Lane P. W., Green M. C. (1960) J. Hered. 51, 228–230 [Google Scholar]
- 21. Miller K. A., Gunn T. M., Carrasquillo M. M., Lamoreux M. L., Galbraith D. B., Barsh G. S. (1997) Genetics 146, 1407–1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Phan L. K., Lin F., LeDuc C. A., Chung W. K., Leibel R. L. (2002) J. Clin. Invest. 110, 1449–1459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gunn T. M., Miller K. A., He L., Hyman R. W., Davis R. W., Azarani A., Schlossman S. F., Duke-Cohan J. S., Barsh G. S. (1999) Nature 398, 152–156 [DOI] [PubMed] [Google Scholar]
- 24. Nagle D. L., McGrail S. H., Vitale J., Woolf E. A., Dussault B. J., Jr., DiRocco L., Holmgren L., Montagno J., Bork P., Huszar D., Fairchild-Huntress V., Ge P., Keilty J., Ebeling C., Baldini L., Gilchrist J., Burn P., Carlson G. A., Moore K. J. (1999) Nature 398, 148–152 [DOI] [PubMed] [Google Scholar]
- 25. Duke-Cohan J. S., Kim J. H., Azouz A. (2004) J. Environ. Pathol. Toxicol. Oncol. 23, 1–11 [DOI] [PubMed] [Google Scholar]
- 26. He L., Gunn T. M., Bouley D. M., Lu X. Y., Watson S. J., Schlossman S. F., Duke-Cohan J. S., Barsh G. S. (2001) Nat. Genet. 27, 40–47 [DOI] [PubMed] [Google Scholar]
- 27. He L., Lu X. Y., Jolly A. F., Eldridge A. G., Watson S. J., Jackson P. K., Barsh G. S., Gunn T. M. (2003) Science 299, 710–712 [DOI] [PubMed] [Google Scholar]
- 28. Jiao J., Sun K., Walker W. P., Bagher P., Cota C. D., Gunn T. M. (2009) Biochim. Biophys. Acta 1792, 1027–1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kim B. Y., Olzmann J. A., Barsh G. S., Chin L. S., Li L. (2007) Mol. Biol. Cell 18, 1129–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lee H. H., Elia N., Ghirlando R., Lippincott-Schwartz J., Hurley J. H. (2008) Science 322, 576–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li M., Brooks C. L., Wu-Baer F., Chen D., Baer R., Gu W. (2003) Science 302, 1972–1975 [DOI] [PubMed] [Google Scholar]
- 32. Lubrano-Berthelier C., Durand E., Dubern B., Shapiro A., Dazin P., Weill J., Ferron C., Froguel P., Vaisse C. (2003) Hum. Mol. Genet. 12, 145–153 [DOI] [PubMed] [Google Scholar]
- 33. Mohammad S., Baldini G., Granell S., Narducci P., Martelli A. M., Baldini G. (2007) J. Biol. Chem. 282, 4963–4974 [DOI] [PubMed] [Google Scholar]
- 34. Shinyama H., Masuzaki H., Fang H., Flier J. S. (2003) Endocrinology 144, 1301–1314 [DOI] [PubMed] [Google Scholar]
- 35. Bagher P., Jiao J., Owen Smith C., Cota C. D., Gunn T. M. (2006) Pigment Cell Res. 19, 635–643 [DOI] [PubMed] [Google Scholar]
- 36. Chakrabarti O., Hegde R. S. (2009) Cell 137, 1136–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jiao J., Kim H. Y., Liu R. R., Hogan C. A., Sun K., Tam L. M., Gunn T. M. (2009) Genesis 47, 524–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ollmann M. M., Barsh G. S. (1999) J. Biol. Chem. 274, 15837–15846 [DOI] [PubMed] [Google Scholar]
- 39. Gao Z., Lei D., Welch J., Le K., Lin J., Leng S., Duhl D. (2003) J. Pharmacol. Exp. Ther. 307, 870–877 [DOI] [PubMed] [Google Scholar]
- 40. Jackson P. J., Douglas N. R., Chai B., Binkley J., Sidow A., Barsh G. S., Millhauser G. L. (2006) Chem. Biol. 13, 1297–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hida T., Wakamatsu K., Sviderskaya E. V., Donkin A. J., Montoliu L., Lynn Lamoreux M., Yu B., Millhauser G. L., Ito S., Barsh G. S., Jimbow K., Bennett D. C. (2009) Pigment Cell Melanoma Res. 22, 623–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Siegrist W., Willard D. H., Wilkison W. O., Eberle A. N. (1996) Biochem. Biophys. Res. Commun. 218, 171–175 [DOI] [PubMed] [Google Scholar]
- 43. Barsh G. S., He L., Gunn T. M. (2002) J. Recept. Signal. Transduct. Res. 22, 63–77 [DOI] [PubMed] [Google Scholar]
- 44. Phan L. K., Chung W. K., Leibel R. L. (2006) Am. J. Physiol. Endocrinol. Metab. 291, E611–E620 [DOI] [PubMed] [Google Scholar]
- 45. Breit A., Wolff K., Kalwa H., Jarry H., Büch T., Gudermann T. (2006) J. Biol. Chem. 281, 37447–37456 [DOI] [PubMed] [Google Scholar]
- 46. Pérez-Oliva A. B., Olivares C., Jiménez-Cervantes C., García-Borrón J. C. (2009) J. Biol. Chem. 284, 31714–31725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Creemers J. W., Pritchard L. E., Gyte A., Le Rouzic P., Meulemans S., Wardlaw S. L., Zhu X., Steiner D. F., Davies N., Armstrong D., Lawrence C. B., Luckman S. M., Schmitz C. A., Davies R. A., Brennand J. C., White A. (2006) Endocrinology 147, 1621–1631 [DOI] [PubMed] [Google Scholar]
- 48. Milligan G., Bond R. A., Lee M. (1995) Trends Pharmacol. Sci. 16, 10–13 [DOI] [PubMed] [Google Scholar]
- 49. Azouz A., Gunn T. M., Duke-Cohan J. S. (2007) Exp. Cell Res. 313, 761–771 [DOI] [PubMed] [Google Scholar]
- 50. Oshikawa J., Toya Y., Fujita T., Egawa M., Kawabe J., Umemura S., Ishikawa Y. (2003) Am. J. Physiol. Cell Physiol. 285, C567–C574 [DOI] [PubMed] [Google Scholar]
- 51. Rodal S. K., Skretting G., Garred O., Vilhardt F., van Deurs B., Sandvig K. (1999) Mol. Biol. Cell 10, 961–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Haqq A. M., René P., Kishi T., Khong K., Lee C. E., Liu H., Friedman J. M., Elmquist J. K., Cone R. D. (2003) Biochem. J. 376, 595–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Walker W. P., Aradhya S., Hu C. L., Shen S., Zhang W., Azarani A., Lu X., Barsh G. S., Gunn T. M. (2007) Genesis 45, 744–756 [DOI] [PubMed] [Google Scholar]
- 54. Gunn T. M., Inui T., Kitada K., Ito S., Wakamatsu K., He L., Bouley D. M., Serikawa T., Barsh G. S. (2001) Genetics 158, 1683–1695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Paz J., Yao H., Lim H. S., Lu X. Y., Zhang W. (2007) Neurobiol. Aging 28, 1446–1456 [DOI] [PubMed] [Google Scholar]
- 56. Sun K., Johnson B. S., Gunn T. M. (2007) Neurobiol. Aging 28, 1840–1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Cota C. D., Bagher P., Pelc P., Smith C. O., Bodner C. R., Gunn T. M. (2006) Dev. Dyn. 235, 3438–3447 [DOI] [PubMed] [Google Scholar]
- 58. Duke-Cohan J. S., Gu J., McLaughlin D. F., Xu Y., Freeman G. J., Schlossman S. F. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 11336–11341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Tao Y. X. (2010) Endocr. Rev. 31, 506–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Raiborg C., Rusten T. E., Stenmark H. (2003) Curr. Opin. Cell Biol. 15, 446–455 [DOI] [PubMed] [Google Scholar]
- 61. Candille S. I., Kaelin C. B., Cattanach B. M., Yu B., Thompson D. A., Nix M. A., Kerns J. A., Schmutz S. M., Millhauser G. L., Barsh G. S. (2007) Science 318, 1418–1423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Farooqi I. S., Keogh J. M., Yeo G. S., Lank E. J., Cheetham T., O'Rahilly S. (2003) N. Engl. J. Med. 348, 1085–1095 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.