Abstract
NDRG1 and KAI1 belong to metastasis suppressor genes, which impede the dissemination of tumor cells from primary tumors to distant organs. Previously, we identified the metastasis promoting transcription factor, ATF3, as a downstream target of NDRG1. Further analysis revealed that the KAI1 promoter contained a consensus binding motif of ATF3, suggesting a possibility that NDRG1 suppresses metastasis through inhibition of ATF3 expression followed by activation of the KAI1 gene. In this report, we found that ectopic expression of NDRG1 was able to augment endogenous KAI1 gene expression in prostate cancer cell lines, whereas silencing NDRG1 was accompanied with significant decrease in KAI1 expression in vitro and in vivo. In addition, our results of ChIP analysis indicate that ATF3 indeed bound to the promoter of the KAI1 gene. Importantly, our promoter-based analysis revealed that ATF3 modulated KAI1 transcription through cooperation with other endogenous transcription factor as co-activator (ATF3-JunB) or co-repressor (ATF3-NFκB). Moreover, loss of KAI1 expression significantly abrogated NDRG1-mediated metastatic suppression in vitro as well as in a spontaneous metastasis animal model, indicating that KA11 is a functional downstream target of the NDRG1 pathway. Our result of immunohistochemical analysis showed that loss of NDRG1 and KAI1 occurs in parallel as prostate cancer progresses. We also found that a combined expression status of these two genes serves as a strong independent prognostic marker to predict metastasis-free survival of prostate cancer patients. Taken together, our result revealed a novel regulatory network of two metastasis suppressor genes, NDRG1 and KAI1, which together concerted metastasis-suppressive activities through an intrinsic transcriptional cascade.
Keywords: Chromatin Immunoprecipitation (ChIP), NF-kB Transcription Factor, Transcription Regulation, Tumor Metastases, Tumor Suppressor, Molecular Mechanism Of Metastasis, Tumor Metastasis Diagnosis
Introduction
Although significant advances have been made in reducing mortality rates, the majority of cancer patients are still diagnosed at an advanced stage and ultimately die from sequelae of metastatic disease. Metastasis involves a complex process through which malignant cancer cells leave a primary organ site, journey to a distant site via circulation, and finally establish a clinically detectable mass in a distant organ, and therefore, the metastatic progression requires dysregulation of a series of genes and related signaling. Metastasis suppressor genes are negative regulators of metastasis, which inhibit metastasis but do not affect the ability of the transformed cells to generate a tumor at the primary site (1–4). More than 20 metastasis suppressors have been discovered so far, and they appear to be involved in several pivotal steps of metastasis, including invasion (NM23, DLC1, KAI1, and NDRG1), dissemination (KAI1, CD44), survival (BRMS1, caspase-8), and growth in distant sites (NM23, KAI1, RHOGD12, KISS1, Raf kinase inhibitor protein, and MKK4/6) (4–7). However, the detailed molecular mechanism of how these genes are regulated and their functions are less elucidated.
KAI1, also known as CD82, was discovered initially from T-cell activation study and was identified later as a prostate-specific tumor metastasis suppressor gene (8–11). It is ubiquitously expressed in normal tissues with high mRNA levels in spleen, placenta, kidney, and prostate, whereas decrease or loss of its expression is constantly found in the clinically advanced cancers (12). Consistently, inverse correlations between KAI1 expression and the invasive and metastatic potential as well as poor survival of patients have been observed frequently in a wide range of malignancies (12). KAI1 belongs to the tetraspanin transmembrane protein superfamily, and it is found to be associated with other tetraspanins (CD9 and CD81), integrins (β1 and β2), immunoreceptors (MHCI and II, EWI1/PGRL, CD4, CD8, CD19, and CD46), growth factors and receptors (EGF and EGFR), as well as intracellular signaling proteins (PKC) (12). KAI1 was found previously to inhibit cell motility by regulating the biological activities of its associated proteins and/or reorganizing plasma membrane microdomains (12). This process occasionally induces apoptosis by releasing intracellular glutathione and accumulating intracellular reactive oxygen intermediates (13, 14). Moreover, we have demonstrated recently that KAI1 exerts its metastasis suppressor function by directly binding to Duffy blood group, chemokine receptor on endothelial cells, thereby inducing senescence signaling in tumor cells in circulation (15). Because mutations and loss of heterozygosity of the KAI1 gene is rare, the down-regulation of this gene is not likely because of genetic alterations but is rather due to modulation of transcriptional and post-transcriptional regulation (16–20, 43). However, how KAI1 is down-regulated in metastatic cancer cells is largely unknown. Previously, we showed that p53 is able to bind to the KAI1 promoter and turn on its transcription (21, 22). Other molecules involved in KAI1 transcriptional regulation include NFκB, β-catenin/Reptin, Tip60/Fe65, N-CoR/TAB2/HDAC3 and AP-1 (23–28). Interestingly, these transcription factors were frequently found to coordinately regulate the expression of KAI1 as either co-activator or co-repressor. Therefore, studies on these transcription factors may aid in elucidating the mechanism leading to KAI1 suppression and following metastatic progression.
NDRG1 (N-Myc downstream regulated gene 1) was originally isolated as a novel gene that was induced strongly during differentiation of colon carcinoma cell lines (29). Recent studies demonstrated that the NDRG1 gene is controlled by a variety of factors and stimuli related to cancer progression, including oncogenes, tumor suppressors, hypoxic microenvironment, and hormone dysregulation (30, 31). Clinical studies also provided compelling evidence that reduced expression of the NDRG1 gene was significantly associated with poor overall survival rate in pancreatic ductal adenocarcinoma, glioma, prostate, breast, and colorectal cancers (32). The significant inverse correlation of NDRG1 expression with the extent of metastasis in a clinical setting raised an important question as to whether the down-regulation of NDRG1 is cause or result of metastases. To address this issue, we overexpressed the NDRG1 gene in a highly metastatic prostate cell line AT6.1 and implanted it into severe combined immunodeficiency mice (33). Our results indicate that NDRG1 has the ability to suppress the metastatic process of prostate cancer cells without affecting tumorigenicity in vivo. Similar metastasis suppressor effect of NDRG1 was also observed in other animal models including colon, bladder, and pancreatic carcinoma cells (34–36). Furthermore, NDRG1 was shown to suppress the invasive and angiogenic abilities of aggressive cancer cells (36–38). A recent study employing whole genome gene array analysis in examining the functions of NDRG1 in a number of different cancer cells strongly indicate the pleiotropic nature of NDRG1 in suppressing metastasis (39).
We previously performed the Affymetrix gene array analysis and found that NDRG1 suppressed metastasis of prostate tumor cells by inhibiting the transcription factor ATF3 (15). Consistent with our observation, Ishiguro et al. (40) showed that ectopic expression of ATF3 converted the low metastatic potential melanoma cell line to become highly metastatic. Moreover, ATF3 expression appears to be required for the maintenance of a high metastatic state of melanoma and colon cancer cells (41). ATF3 is a member of cAMP-responsive element binding protein (ATF/CREB) family of basic leucine zipper transcription factors (42). Emerging evidence suggests that ATF3 plays a critical role in metastatic progression in a cell context-dependent manner. To gain further mechanistic insight into the functional role of NDRG1, we sought to identify and characterize the possible downstream targets of ATF3 that are involved in tumor metastasis. The result of our bioinformatic analysis for the promoters of metastasis-related genes revealed that there were a number of genes whose promoter contained the ATF3-responsive consensus sequence, TGACGTCA. Among these, we identified proapoptotic gene GADD153/CHOP10, cell adhesion molecular E-selectin, tumor suppressor p53, and metastasis suppressor KAI1 as potential targets of ATF3. These clues prompted us to examine a possible link between two metastasis suppressor genes, NDRG1 and KAI1, through ATF3 in the current study.
EXPERIMENTAL PROCEDURES
Cell Culture
Human prostate cancer cell line PC3mm and rat prostatic carcinoma cell line AT6.1 were kindly provided by Drs. I. J. Fidler (The University of Texas MD Anderson Cancer Center, Houston, TX) and C. Rinker-Schaeffer (University of Chicago), respectively. The PC3mm/Tet2 cell line was established previously as a derivative of PC3mm and contains a tetracycline-inducible suppressor. The human prostate cancer cell line DU145, prostate epithelial cell line RWPE1, and colon cancer cell line HT38 were obtained from the American Type Culture Collection (Manassas, VA). NDRG1- and KAI1-overexpressing stable clones of AT6.1 cells were established as described previously (33). For AT6.1/NDRG1/shKAI1, shRNA for KAI1 (Open Biosystems) or the vector alone was transfected into AT6.1/NDRG1 cells, and the puromycin-resistant clones were selected. The expression of KAI1 in the cloned cells was tested by RT-PCR. All cells were maintained in RPMI 1640 medium supplemented with 10% fetal calf serum, streptomycin (100 μg/ml), penicillin (100 units/ml), and 250 nm dexamethasone (Sigma) and grown at 37 °C in a 5% CO2 atmosphere. For all transfection experiments, Lipofectamine 2000 (Invitrogen) was used according to the manufacturer's protocol.
Reporter Assay
The luciferase activities were measured by using the Dual-Luciferase Reporter Assay System (Promega, Madison, MI) and a luminometer (Berthold Detection Systems, Huntsville, Alabama). For each transfection experiment, the Renilla expression plasmid phRG-TK (Promega) was co-transfected as an internal control, and the promoter activities were normalized accordingly.
Immunohistochemistry
Immunohistochemical analysis on paraffin-embedded, surgically resected specimens of prostate and breast was carried out using anti-NDRG1 rabbit polyclonal antibody (from Dr. Commes) and anti-KAI1 antibody (kindly provided by Dr. Yoshie). Briefly, sections were deparaffinized, rehydrated, and heated at 80 °C for 20 min in 25 mm sodium citrate buffer (pH 9) for antigen exposure. They were then treated with 3% H2O2 to block endogenous peroxidase activity and further incubated with primary antibody for 1 h at 24 °C. After washing with Tris-buffered saline/0.1% Tween 20, the sections were incubated with horseradish peroxidase-conjugated rabbit-specific IgG (Dako). The sections were washed extensively, and 3,3-diaminobenzidine substrate chromogen solution was applied followed by counterstaining with hematoxylin.
Bioinformatics and Statistical Analysis
The GEO database (accession no. GSE21034, n = 218) was used to evaluate the clinical relevance of NDRG1 and KAI1 in prostate cancer. The data were log2-transformed, with the median set as zero and with S.D. set as one. Each patient was assigned to have positive or negative expression of each gene and was matched with metastasis-free survival. The gene expressions in normal, primary tumor, or metastatic sites in patients who have either metastatic or nonmetastatic disease were compared using Box-and-whisker plot analysis and evaluated by the Mann-Whitney test. The association between genes and clinical outcomes was calculated by the Pearson χ2 test. The Kaplan-Meier method was used to calculate the overall survival rate, and prognostic significance was evaluated by the log-rank test. Multivariate analysis for the prognostic value of gene signatures was performed by the Cox proportional hazard regression model. For all statistical analysis, Prism and SPSS software were used. For in vitro experiments and animal studies, results are reported as mean ± S.D. (or mean ± S.E.) as indicated in the figure legends. Statistical significance was determined by a two-sided Student's test.
Animal Studies
4–6-Week-old severe combined immunodeficiency mice (Harlan Sprague-Dawley, Indianapolis, IN) were used for the spontaneous metastasis studies. 0.5 × 106 cells in 0.1 ml of PBS were injected subcutaneously into the dorsal flank of mice. Mice were monitored daily, and the tumor volume was measured as an index of the growth rate and calculated as (width + length)/2 × width × length × 0.5236. Mice were sacrificed 30 days after the inoculation of cells, and metastatic lesions were counted macroscopically.
RESULTS
NDRG1 Up-regulates Expression of KAI1 Gene in Prostate Cancer Cells
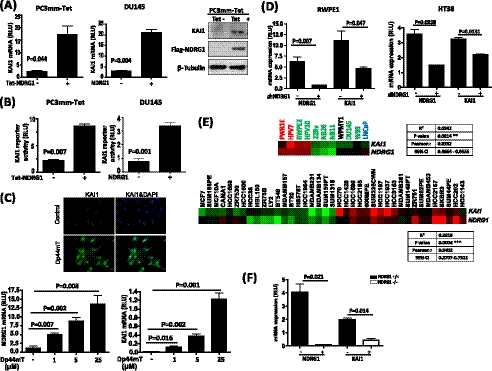
We first examined the effect of NDRG1 on the expression of the KAI1 gene in two human prostate cancer cell lines, DU145 and PC3mm/Tet-FLAG-NDRG1 (referred to as PC3mm/Tet), by quantitative RT-PCR and Western blot. NDRG1 expression was induced by tetracycline treatment in PC3mm-Tet cells. Alternatively, the NDRG1 expression plasmid was transfected into DU145 cells. We found that the expression of KAI1 gene was significantly up-regulated by ectopic expression of NDRG1 (Fig. 1A). To further examine whether NDRG1 has an effect on the transcription of the KAI1 gene, we transfected the KAI1 reporter plasmid into PC3mm/Tet and DU145 cells, which were then forced to overexpress the NDRG1 gene. As shown in Fig. 1B, our results showed that NDRG1 expression indeed significantly augmented KAI1 gene transcription. Moreover, when PC3mm cells were treated with Dp44mT, an NDRG1-specific agonist, the expression of the KAI1 gene was also significantly elevated in a dose-dependent manner (Fig. 1C, lower panel). The increased KAI1 expression in the Dp44mT-treated cells was also confirmed by immunofluorescence staining (Fig. 1C, upper panel). Furthermore, we introduced NDRG1 shRNA into nonmalignant prostate epithelial cell RWPE1 or NDRG1 siRNA into colon carcinoma cell HT38 because both cell lines are known to express the KAI1 gene. We found that the specific knockdown of NDRG1 indeed significantly down-regulated the expression of the KAI1 gene in these cell lines (Fig. 1D). We also examined the expression of KAI1 and NDRG1 in a panel of prostate and breast cancer cell lines (n = 11 and 39 for prostate and breast cancer cell lines, respectively). We found that KAI1 mRNA expression was significantly correlated with NDRG1 expression in multiple cancer cell lines (p = 0.0014 and 0.0004 for prostate (upper panel) and breast (lower panel) cancer cell lines, respectively, Fig. 1E), indicating that the expression of NDRG1 and KAI1 generally has a positive correlation in human prostate and breast cancer cells. To further examine the relevance of NDRG1 and KAI1 in vivo, we compared the expression of Ndrg1 and Kai1 in NDRG1 knock-out (Ndrg1−/−) and syngenic wild type mice (Ndrg1+/+). Consistent with the results of our in vitro studies, we found that Kai1 mRNA expressed at a significantly lower level in Ndrg1 knock-out mice compared with that in wild type mice (Fig. 1F).
FIGURE 1.
NDRG1 up-regulates KAI1 in prostate cancer cells. A and B, PC3mm/Tet-FLAG-NDRG1 cells were treated with or without tetracycline. DU145 cells were transfected with the NDRG1 expression plasmid or an empty vector. The expression of KAI1 mRNA was then examined by quantitative RT-PCR using primers for KAI1 (A, left and middle panels). Western blot analysis was also performed to confirm KAI1 expression (A, right panel). B, KAI1 promoter-driven luciferase reporter plasmid was transfected into PC3mm/Tet-FLAG-NDRG1 and DU145 cells. Luciferase activity was assayed and normalized by internal Renilla luciferase activity. Values are means ± S.D. of triplicate measurements. p values are based on a two-sided Student's test. C, PC3mm/Tet-FLAG-NDRG1 cells were treated with different doses of Dp44mT. Expressions of NDRG1 and KAI1 mRNA were examined by quantitative RT-PCR. Values are means ± S.D. of triplicate measurements. p values are based on a two-sided Student's test. Immunocytochemical staining with antibody for KAI1 was also performed. D, RWPE1 cells were transfected with shRNA for NDRG1(left panel). HT38 cells were transfected with siRNA for NDRG1 (right panel). The expressions of NDRG1 and KAI1 mRNA were then examined by quantitative RT-PCR. E, KAI1 expression correlates with NDRG1 in a panel of prostate (upper panel, GSE9633) and breast (lower panel, Ref. 52) cancer cell lines. The heat map depicts the relative expression of normalized values of KAI1 and NDRG1 for each cell line. Green indicates low expression, and red indicates high expression. The correlation of two genes was analyzed by linear regression. F, the expression of Ndrg1 and Kai1 mRNA in NDRG1 knock-out and wild type mice were examined by quantitative RT-PCR. Values are means ± S.D. of triplicate measurements. p values are based on a two-sided Student's test. RLU, relative unit.
NDRG1 Suppresses ATF3 Signaling-mediated KAI1 Inhibition in Prostate Cancer Cells
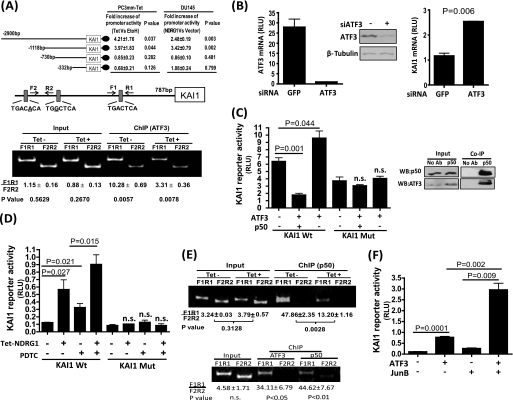
To identify the NDRG1-responsive sequence on the KAI1 promoter, we generated a series of luciferase reporter plasmids containing up to −2900, −1118, −730, and −332 bases of the KAI1 promoter, and luciferase reporter activities were measured in PC3mm/Tet cells with or without induction of NDRG1 expression. As shown in Fig. 2A, the deletion between −1118 and −730 bases of the KAI1 promoter region almost completely abolished the ability of this promoter to respond to NDRG1. We also examined the KAI1 promoter activity in DU145 cells, which was transfected with the NDRG1 expression plasmid and observed a similar result. These results suggest that the region between −1118 and −730 contains an NDRG1-responsive sequence. When we examined the sequence of this region, we found three potential binding sites for ATF3, and one of these was a complete match to the consensus sequence. To further determine whether ATF3 binds to the KAI1 promoter, ChIP assay was performed, and the results clearly showed that ATF3 indeed bound to the predicted region of the KAI1 promoter, whereas induction of NDRG1 expression significantly blocked this binding (Fig. 2A, lower panel). Consistently, we found that silencing the expression of ATF3 by siRNA in PC3mm cells resulted in significant up-regulation of KAI1 mRNA expression (Fig. 2B). Therefore, our results strongly suggest that NDRG1 regulates KAI1 expression by modulating ATF3 signaling pathway. Surprisingly, however, we found that ectopic expression of ATF3 in PC3mm cells significantly up-regulated KAI1 promoter activity and that the mutation of the ATF3-binding consensus sequence on the KAI1 promoter abolished this responsiveness to ATF3 (Fig. 2C, left panel).
FIGURE 2.
KAI1 expression is regulated through ATF3. A, luciferase reporter constructs with various lengths of the KAI1 promoter were transfected to PC3mm/Tet-FLAG-NDRG1 and DU145 cells. PC3mm/Tet-FLAG-NDRG1were cultured with or without tetracycline. DU145 cells were transfected with the NDRG1 expression plasmid or an empty vector. Cells were then assayed for luciferase activities (upper panel). For ChIP assay (lower panel), PC3mm/Tet-FLAG-NDRG1 cells were cultured with or without tetracycline. Precipitated DNA was subjected to quantitative PCR using primers specific for the ATF3 binding consensus sequence on the KAI1 promoter (F1R1) as well as a control primer set (F2R2). The ratio of the DNA was calculated based on cyclic threshold value for each reaction. B, siRNA for ATF3 or GFP was transfected to PC3mm cells, and the expressions of ATF3 (left) and KAI1 (right) mRNA were examined by quantitative RT-PCR. The expression of ATF3 was also confirmed by Western blot analysis. C, left panel, wild type or a mutant of KAI1 luciferase reporter plasmids were transfected into PC3mm cells that were then transfected with expression plasmids of ATF3 and/or p50 and vector control. Cells were then assayed for luciferase activates. Right panel, co-immunoprecipitation (Co-IP) analysis for the interaction of ATF3 and p50 in PC3mm cells. D, wild type or a mutant of KAI1 luciferase reporter plasmids were transfected into PC3mm/Tet-FLAG-NDRG1 cells that were cultured with or without tetracycline followed by treatment with or without PDTC. Cells were then assayed for luciferase activates. E, PC3mm/Tet-FLAG-NDRG1 cells were cultured with or without tetracycline (upper panel). RWPE1 cells was infected with Lenti-p50 virus (lower panel). Cells were lysed and precipitated with ATFe or p50 antibody. Precipitated DNA was then subjected to quantitative PCR using primers as described above. F, KAI1 luciferase reporter plasmid was transfected into PC3mm cells that were then transfected with expression plasmids of ATF3 and JunB or vector control. Cells were then assayed for luciferase activates. For all experiments, values are means ± S.D. of triplicate measurements. p values are based on a two-sided Student's test. Mut, mutant. RLU, relative unit; PDTC, pyrrolidine dithiocarbamate; n.s., no significance.
ATF3 is known to be a bidirectional transcription factor, which either activates or represses transcription dependent on its dimerized partner in a cell context-dependent manner (42). Interestingly, the NFκB subunit p50 was reported previously to be involved in a protein complex that was capable of inhibiting KAI1 transcription (25), and p50 is also known to be able to form a dimeric complex with ATF family members (53). Therefore, we examined a possibility that ATF3 coordinates with p50 to repress KAI1 transcription. We found that ATF3 was indeed pulled down with p50 (Fig. 2C, right panel) and works together to suppress KAI1 promoter activity after co-transfection of ATF3 and p50 expression plasmids in PC3mm cells (Fig. 2C, left panel). However, ATF3 and p50 failed to down-regulate KAI1 promoter activity when ATF3 binding sites were mutated (Fig. 2C, left panel), suggesting that p50 cooperates with ATF3 to suppress KAI1 expression. To further verify this result, we examined an effect of the p50 inhibitor, pyrrolidine dithiocarbamate, on the expression of the KAI1 gene in the presence or absence of NDRG1. As shown in Fig. 2D, either induction of NDRG1 or inhibition of p50 significantly up-regulated KAI1 transcriptional activity in the PC3mm cell, and a combination of NDRG1 and pyrrolidine dithiocarbamate further up-regulated KAI1 promoter activity. However, neither of the treatments induced suppression of KAI1 promoter activity when the ATF3 consensus binding site was mutated. These results strongly support our notion that NDRG1 up-regulates KAI1 by blocking the suppressor activity of the ATF3-p50 complex on the KAI1 promoter. Notably, it was reported previously that p50 dimerized and bound to a region between −6631 and −6996 bp upstream of the KAI1 gene transcription start site (23, 24). To further corroborate that p50 is involved in ATF3-mediated KAI1 transcriptional regulation, we performed a ChIP assay by precipitating the p50-chromatin complex followed by quantitative PCR analysis with the pairs of primers specific to the region of ATF3 binding site. We found that p50 indeed bound to the proximal region of ATF3 binding site on the KAI1 promoter by forming a complex with ATF3, which was negatively regulated by NDRG1 (Fig. 2E).
ATF3 can form a complex with various other members in the ATF family such as ATF2 and ATF4, as well as AP-1 proteins (27, 28). Interestingly, we found that ATF3, in combination with the AP-1 family protein JunB, significantly enhanced KAI1 transcriptional activity compared with each alone (Fig. 2F). Taken together, our result indicates that ATF3 modulates KAI1 expression through an intrinsic mechanism that is dependent on the dynamic cellular context of tumor progression.
KAI1 Is Involved in Downstream Signal of NDRG1-mediated Metastasis Suppression
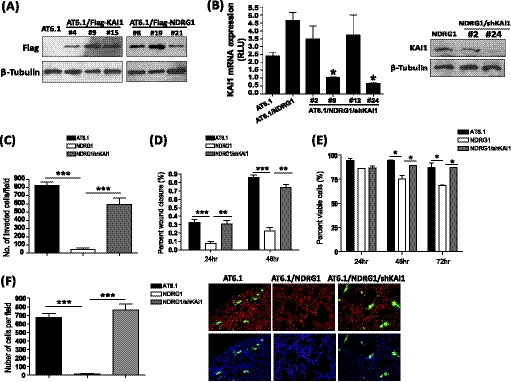
The positive regulatory role of NDRG1 in KAI1 expression prompted us to examine the functional relevance of these two metastasis suppressors in tumor progression. We established permanent cell lines expressing NDRG1 with or without knocking down the KAI1 gene using the highly metastatic prostate cancer cell line AT6.1 (Fig. 3, A and B). These cell lines were then examined for their metastatic behavior through a series of functional assays. As we have reported previously, the expression of NDRG1 significantly reduced the invasive ability of prostate cancer cells through Matrigel; however, silencing of the KAI1 gene significantly abolished NDRG1-induced suppression of invasion (Fig. 3C). Moreover, the expression of NDRG1 significantly reduced the migration ability of AT6.1 cells but not in the cells which KAI1expression was knocked down (Fig. 3D). Because we have shown that NDRG1 did not affect the growth rate of prostate cancer cells in vitro and in primary tumors (33), we examined the effect of NDRG1 on tumor cells that were grown in suspension. As shown in Fig. 3E, we found that the majority of AT6.1 cells (88.9% ± 3.05) maintained their viability during the 72 h of culture in suspension, and NDRG1-overexpressing cells showed significant deficiency in survival under such conditions (66.64% ± 2.07). However, the knockdown of the KAI1 gene was able to restore anoikis resistance to the same level as the control (87.72% ± 0.78). Anoikis can be stimulated by detachment from the extracellular matrix in the primary site, which serves as an important barrier to prevent metastatic tumor cell dissemination. Therefore, metastatic tumor cells need to acquire both invasive and anoikis resistance abilities, which aid in thriving during the early stage of metastasis. To directly assess whether KAI1, in conjunction with NDRG1, affects early seeding of tumor cells in vivo, we intravenously injected AT6.1 or AT6.1 carrying the expression vector of NDRG1 with or without shRNA to the KAI1 gene, into nude mice. After 48 h, mice were sacrificed, and the lung sections were examined. We found that the number of NDRG1 expressing cells (17.09 ± 3.69) was significantly lower compared with parental AT6.1 cells (672.2 ± 47.38), whereas KAI1 knockdown (764.1 ± 69.78) substantially abrogated the inhibitory effect of NDRG1 (Fig. 3F). Taken together, our results revealed that NDRG1 exerts its metastatic function by up-regulating KAI1, which in turn suppresses various metastatic traits in cancer cells.
FIGURE 3.
KAI1 is involved in downstream signal of NDRG1-mediated metastasis suppression. A and B, establishing Rat prostate cancer cells AT6.1 sublines: AT6.1/NDRG1, AT6.1/KAI1, and AT6.1/NDRG1/shKAI1. Western blot (A) confirming overexpression of FLAG-NDRG1 and FLAG-KAI1 in AT6.1/NDRG1 and AT6.1/KAI1 cells. B, knockdown of expression of KAI1 was confirmed in AT6.1/NDRG1/shKAI1 cells by RT-PCR (left) and (right). C–F, rat prostate cancer cells, including the parental cell line (AT6.1), NDRG1-transfected sub-lines with (AT6.1/NDRG1/shKAI1) or without (AT6.1/NDRG1) shRNA of KAI1, were subjected to assays for invasion (C), migration (D), and anoikis resistance (E). F, AT6.1, AT6.1/NDRG1, and AT6.1/NDRG1/shKAI1 cells were labeled with CellTracker Green and injected into the lateral vein of nude mice (n = 3). Lungs were removed after 48 h and sectioned, stained by DAPI, followed by counting the number of tumor cells under the confocal microscope. Lung vasculature was visualized by staining with rhodamine-conjugated lectin. For all experiments, values are means ± S.D. of triplicate measurements. p values are based on a two-sided Student's test. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
NDRG1 and KAI1 Correlate in Clinical Setting and Predict Clinical Outcome of Prostate Cancer
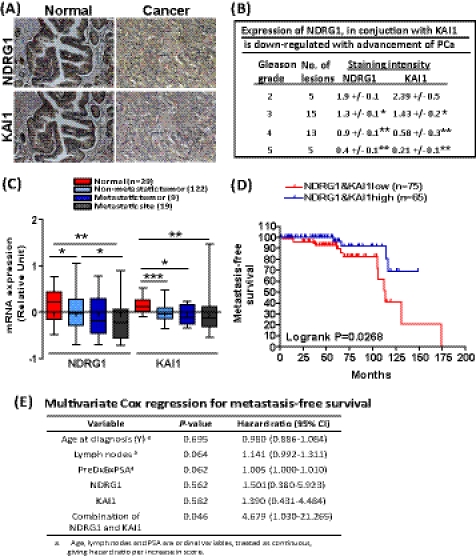
To further examine the relevance of NDRG1 and KAI1 in clinical setting, we performed immunohistochemical analysis with tumor specimens from prostate cancer patients (n = 19). The results showed that NDRG1 was strongly expressed in the epithelial cells of normal ducts and glands in prostate tissue, whereas it was significantly down-regulated as tumor grades increased (p = 0.0126, 0.0003, and 0.0001, respectively, Fig. 4, A and B). Similarly, KAI1 was also found to be highly expressed in normal prostate epithelium and mainly localized at membrane and cytoplasm, but it was significantly decreased in poorly differentiated tumors (p = 0.0441, 0.0051, and 0.0051, respectively, Fig. 4, A and B). Linear regression analysis revealed a significant positive correlation (p = 0.0045) between NDRG1 and KAI1 expression status in prostate cancer patients (Fig. 4B). Next, we examined the expression of NDRG1 and KAI1 in a GEO database that contains the largest number of prostate patient samples to date (accession no. GSE21034). The expression of NDRG1 and KAI1 were all significantly down-regulated in tumors and metastatic sites compared with that in normal tissues (Fig. 4C). Notably, there was no statistically significant difference between the level of both genes in tissues from metastatic primary tumors and metastatic sites, indicating that the expression of NDRG1 and KAI1 are already down-regulated in the primary tumor, which may trigger an activation of metastasis cascade at an early stage. Previously, we have shown that the status of NDRG1 and KAI1, individually, serves as a diagnostic and prognostic marker for disease-free survival of prostate cancer (21, 33). Considering the significant cross-talk between NDRG1 and KAI1 expression in our study, we next evaluated the prognostic value of a combination of these markers. Prostate cancer patients were stratified into two groups based on NDRG1/KAI1 expression and analyzed for their metastasis-free survival by the Kaplan-Meier analysis with a 15-year follow-up. As shown in Fig. 4D, patients with a low (n = 75) expression level of NDRG1 and KAI1 had significantly worse metastasis-free survival than those with a high (n = 65) expression (overall log-rank, p = 0.0268) of both genes. Importantly, the result of a multivariate Cox regression analysis indicates that the combination of NDRG1 and KAI1 was an independent prognostic marker in prostate cancer (p = 0.046) with a hazard ratio of 4.679, compared with 1.501 and 1.390 for NDRG1 and KAI1 alone, respectively (Fig. 4E). These data underscore the utility of the combination of NDRG1 and KAI1 as an independent prognostic marker for prostate cancer.
FIGURE 4.
KAI1 and NDRG1 are down-regulated in prostate cancer and a combination of two genes expression significantly correlates with patient survival. A, immunohistochemical analysis was performed for NDRG1 and KAI1 with prostate tumor specimens from 19 patients. Representative images for immunohistochemical analysis using anti-NDRG1 (upper panels) and anti-KAI1 (lower panels) antibodies were shown. B, staining intensities of both NDRG1 and KAI1 for the specimens were calculated (on a scale of 0–3, with 3 being the highest) for each group of Gleason grade. C, expression of NDRG1 and KAI1 in tissues from normal, tumor, and distant metastatic sites were examined using the cohort dataset of GSE12034. p values are based on a Mann-Whitney Test. *, p < 0.05; **, p < 0.01; ***, p < 0.001. D, A Kaplan-Meier analysis was done to determine the diagnostic value of the combined expression of NDRG1 and KAI1. The p value was determined by log-rank test. E, a multivariate Cox regression analysis was conducted to assess the contribution of the indicated variables to disease prognosis.
KAI1 Knockdown Abrogated Suppressive Effect of NDRG1 on Metastasis in Animals
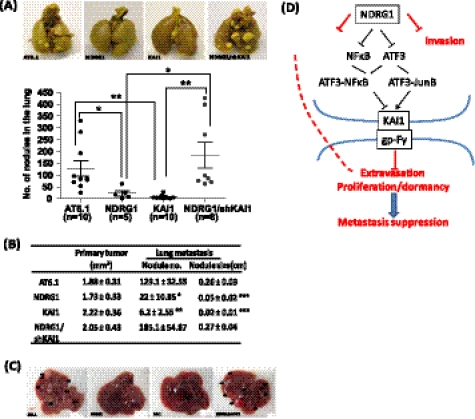
To further verify our notion that NDRG1 suppresses metastasis through activation of KAI1 in vivo, the metastatic cell line AT6.1 and its derivatives, AT6.1/NDRG1, AT6.1/KAI1, and AT6.1/NDRG1/shKAI1, were individually inoculated subcutaneously into the dorsal flanks of severe combined immunodeficiency mice. The mice were monitored for the formation of primary tumors and were sacrificed after 30 days. The lungs were then removed, and the number of metastatic lesions was grossly counted. We found that all cell lines formed a primary tumor with similar size (Fig. 5B), confirming the notion that NDRG1 and KAI1 genes do not have an apparent effect on tumorigenesis. However, cells with overexpressing the NDRG1 or KAI1 gene showed a significantly lower lung metastases compared with the parental cell line AT6.1. As shown in Fig. 5, A and B, AT6.1/NDRG1 and AT6.1/KAI1 cells generated not only a significantly fewer number of lesions (p = 0.042, and 0.0014, respectively) but also a smaller size (p < 0.0001 and 0.0001, respectively) of nodules in the lungs, suggesting that NDRG1 or KAI1 alone can functionally attenuate the seeding and colonization abilities of tumor cells. Importantly, knockdown of KAI1 expression in NDRG1-overexpressing cells almost completely abrogated the metastasis suppressive ability of the NDRG1 gene, which is consistent with our observation in in vitro studies. Interestingly, we found that mice (two of ten) inoculated with AT6.1 parental cells developed extensive liver metastasis in addition to lung metastasis, whereas none of the mice that received AT6.1 carrying the NDRG1- or KAI1-overexpressing vector developed liver metastasis. On the other hand, knockdown of KAI1 expression restored the ability of AT6.1 to develop liver metastasis. Therefore, KAI1 serves as an essential downstream factor of NDRG1-mediated metastasis suppression in our animal model.
FIGURE 5.
KAI1 expression is indispensible for NDRG1-mediated metastasis suppression in animal. Rat prostate cancer cells including the parental cell line (AT6.1), NDRG1-transfected sub-lines with (AT6.1/NDRG1/shKAI1) or without (AT6.1/NDRG1) shRNA of KAI1, and KAI1-transfected sub-line were injected subcutaneously into SCID mice. After 30 days, the mice were sacrificed, and lungs were removed. Tumor nodules on the lungs were counted macroscopically. A, upper panels show representative images of lung metastases. The lower panel shows the number of tumor-bearing mice and nodules on the lungs. Sizes of primary tumor and nodules on the lungs are shown in B. Data are presented as the mean nodule number or size. Error bars represent the S.E. p values are based on a two-sided Student's test. *, p < 0.05; **, p < 0.01. C, representative images of liver metastases. D, concerted activities of NDRG1 and KAI1-mediated metastasis suppression.
DISCUSSION
In this report, we examined the regulatory network of two metastasis suppressor genes, NDRG1 and KAI1, and demonstrated that NDRG1 up-regulated KAI1 expression through modulation of ATF3 signaling in prostate cancer cells. Our results also indicate that KAI1 is a critical downstream target in the NDRG1 pathway and that loss of KAI1 expression significantly abrogates NDRG1-mediated metastatic suppression in vitro and in vivo. The concerted metastasis suppressor activity of NDRG1 and KAI1 was also evidenced by the result of clinical analysis that a combined expression status of these two genes serves as an independent diagnostic marker to predict metastasis-free survival of prostate cancer patients.
Down-regulated KAI1 expression has been observed in a variety of cancers; however, genetic alterations such as loss of heterozygosity and a mutation within the coding region or epigenetic modification of this gene is a rare event (16–20). Rather, it is likely that the replacement of transcriptional activators for the KAI1 promoter by the dominant co-repressor complex serves as a major mechanism to repress KAI1 expression (43). The KAI1 promoter region contains putative binding motifs for various transcription factors. The NFκB p50 subunit was found to bind the region between −6631 and −6996 bp upstream to the KAI1 gene transcription start site (23, 24). However, several lines of evidence indicate that NFκB alone does not affect KAI1 transcription. Kim et al. (25) demonstrated recently that β-catenin in combination with the Reptin chromatin remodeling complex replaced the Tip60-Pontin co-activator complex on the KAI1 promoter, where the p50 homodimer serves as a dock for both the β-catenin-Reptin and Tip60-Pontin complex during this binding complex switching process. In another study in determining a role of IκB kinase α (IKKα, an NFκB signaling activator) in prostate cancer metastasis, Luo et al. (44) generated a variant of TRAMP mice with IKKα deficiency (TRAMP IKKαAA/AA) and prevented activation of NFκB signaling. They found that WT/TRAMP mice developed prostate cancer and metastasis significantly earlier compared with TRAMP IKKαAA/AA; however, KAI1 expression was not different in both mice, thus strongly indicating that NFκB alone does not regulate KAI1 transcription.
Marreiros et al. (27, 28) previously identified a region of the KAI1 promoter that contained binding motifs for AP-2 (α, β, or γ), p53, and AP-1 (Jun and Fos) that is essential for a high level promoter activity in bladder and prostate cancer cells. Detailed examination of KAI1 promoter activity in a series of prostate cancer cells with low to high malignancy showed the requirement of three different transcription factors for the maximum promoter activity, whereas decrease or loss of functional p53 in conjunction with differential levels of AP-1 and AP-2 proteins in highly malignant cells frequently led to formation of a co-repressor complex to suppress KAI1 promoter activity. In our study, we showed that deletion of a similar region in the KAI1 promoter containing AP-2, p53, and AP-1 sites significantly abolished the NDRG-induced activation of the KAI1 promoter in PC3mm and DU145 prostate cells. Because both cell lines are absent of p53 function, our observation is likely to be independent of p53 activity, and it is rather considered to be a result of an interplay between other transcription factors. Of note, we found that knockdown of ATF3 significantly enhanced the KAI1 mRNA level; however, in contrast to our expectations, ectopic expression of ATF3 increased KAI1 promoter activity. We attributed this apparent discrepancy to the effect of differential experimental settings of silencing the endogenous expression and forced expression of ATF3, which may substantially alter cellular endogenous environment. We propose that silencing ATF3 expression directly affects the ATF3 co-repressor complex such as ATF3-p50 on the KAI1 promoter, thus releasing KAI1 from such suppressive signals. On the other hand, forced expression of ATF3 forms a co-activator complex with endogenous AP-1 proteins JunB (existing in PC3mm cells, see Ref. 27) and promotes KAI1 transcription activity. We have indeed shown that co-transfection of p50 and ATF3 tilted the balance of endogenous transcription factors and led to formation of ATF3-p50 co-repressor complex, which significantly suppressed KAI1 promoter activity.
It is noteworthy that, in our analysis for prostate cancer cohort data (GEO accession no. GSE21034), expression of the NFKB1 gene was found to be increasingly elevated with clinical grade becoming higher. Interestingly, we also found that the JunB expression was maintained at a relatively high level in low grade tumors, and it substantially decreased in high grade lesions (data not shown). Consistently, Konishi et al. (45) examined the expression status of JunB by immunohistochemical analysis and found that JunB expression was decreased significantly in high grade lesions with Gleason score ≥6 and in metastatic lesions. A more recent study revealed that JunB was a Smad1-responsive gene and that it was involved in suppression of prostate cancer metastasis (46). On the other hand, the role of NFκB transcription factors has been well established in the development and progression of prostate cancer through its effect on apoptosis, invasion, and inflammation and particularly on androgen-independent and recurrent prostate cancer (47–50). Therefore, ATF3 acts as a modulator of KAI1 transcription through coordinating other endogenous transcription factors as either co-activator or co-repressor, and this mechanism is dependent on the tumor stage and genetic background of tumors.
It is well documented that the expressions of metastasis suppressors were generally lost during tumor progression; however, a detailed mechanism of their loss is yet to be elucidated, and many critical questions remain unanswered. When does the expression of these metastasis suppressors start to diminish? Is it a sequential or dynamic process? Does loss of expression of one factor affect others and is there any functional complementary mechanism existing? In our in vitro experiment, we clearly showed that ectopic expression of NDRG1 induced up-regulation of KAI1 by indirectly inhibiting the transcription factor ATF3. Interestingly, previous studies reported that NDRG1attenuated the nuclear translocation of NFκB and its binding to the NFκB binding motif by suppressing the expression of the IκB kinase inhibitor IKKβ and also by blocking IκBα phosphorylation (38). It should be noted that the 5′-UTR region of the ATF3 gene contains the NFκB motif, which is involved in the signaling of stress stimuli (54), suggesting that NDRG1 inhibits ATF3 expression through modulation of NFκB. Therefore, a high level of NDRG1 expression seems to relieve KAI1 from the inhibitory signal of both ATF3 and NFκB. We also examined the relevance of NDRG1 and KAI1 in a broad range of prostate and breast cancer cell lines and in clinical samples, and our results strongly indicate that NDRG1 and KAI1 expression are correlated significantly to each other. Interestingly, we also found that down-regulation of both NDRG1 and KAI1 already took place in the primary tumor in patients without metastasis, although it will be more obvious in metastatic patients. Consistently, El Touny et al. (51) previously discovered an age-dependent down-regulation of KAI1 in the transgenic adenocarcinoma of the mouse prostate (TRAMP) mice model, which started from 24 weeks when the tumor developed but before metastasis occurred. We also found that Ndrg1 was down-regulated significantly in the hyperplasia stage of MMTV-Wnt1 and MMTV-neu transgenic mice (supplemental figure). Taken together, these results indicate that the loss of expression of both NDRG1 and KAI1 occurs in parallel, and it is considered to take place at a relatively early stage. Notably, we have shown that KAI1 is an essential downstream factor of NDRG1 signaling in our in vitro and animal experiments. Knocking down the expression of KAI1 not only abrogated NDRG1-mediated motility/invasion suppression of tumor cells but also restricted early seeding and later colonization of metastatic tumor cells in distant sites. Importantly, our results also suggest that the combination of NDRG1 and KAI1 expression status has a strong independent value for predicting patient outcome compared with either marker alone. Our results demonstrate that KAI1 is a downstream target and an effector of NDRG1, suggesting the concerted regulatory mechanism and activity of these metastasis suppressor genes. We also have shown that a small compound, Dp44mT, which activates the NDRG1 gene, significantly enhanced KAI1 expression. Considering that functionally reconstituting metastasis suppressor genes could be beneficial to clinical treatment of metastatic disease, our results provide a novel insight to develop strategies for metastasis suppressor-based therapies.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants R01CA124650 and R01CA129000 (to K. W.). This work was also supported by U. S. Department of Defense Grants BC044370 and PC061256 (to K. W.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental “Methods” and a figure.
- Tet
- tetracycline
- TRAMP
- transgenic adenocarcinoma of the mouse prostate.
REFERENCES
- 1. Berger J. C., Vander Griend D. J., Robinson V. L., Hickson J. A., Rinker-Schaeffer C. W. (2005) Cancer Biol. Ther. 4, 805–812 [DOI] [PubMed] [Google Scholar]
- 2. Nash K. T., Welch D. R. (2006) Front. Biosci. 11, 647–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Shevde L. A., Welch D. R. (2003) Cancer Lett. 198, 1–20 [DOI] [PubMed] [Google Scholar]
- 4. Steeg P. S., Ouatas T., Halverson D., Palmieri D., Salerno M. (2003) Clin. Breast Cancer 4, 51–62 [DOI] [PubMed] [Google Scholar]
- 5. Steeg P. S. (2003) Nat. Rev. Cancer 3, 55–63 [DOI] [PubMed] [Google Scholar]
- 6. Steeg P. S. (2006) Nature Med. 12, 895–904 [DOI] [PubMed] [Google Scholar]
- 7. Eccles S. A., Welch D. R. (2007) Lancet 369, 1742–1757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gaugitsch H. W., Hofer E., Huber N. E., Schnabl E., Baumruker T. (1991) Eur. J. Immunol. 21, 377–383 [DOI] [PubMed] [Google Scholar]
- 9. Ichikawa T., Ichikawa Y., Isaacs J. T. (1991) Cancer Res. 51, 3788–3792 [PubMed] [Google Scholar]
- 10. Ichikawa T., Ichikawa Y., Dong J., Hawkins A. L., Griffin C. A., Isaacs W. B., Oshimura M., Barrett J. C., Isaacs J. T. (1992) Cancer Res. 52, 3486–3490 [PubMed] [Google Scholar]
- 11. Dong J. T., Lamb P. W., Rinker-Schaeffer C. W., Vukanovic J., Ichikawa T., Isaacs J. T., Barrett J. C. (1995) Science 268, 884–886 [DOI] [PubMed] [Google Scholar]
- 12. Liu W. M., Zhang X.A. (2006) Cancer Lett. 240, 183–194 [DOI] [PubMed] [Google Scholar]
- 13. Ono M., Handa K., Withers D. A., Hakomori S. I. (1999) Cancer Res. 59, 2335–2339 [PubMed] [Google Scholar]
- 14. Schoenfeld N., Bauer M. K., Grimm S. (2004) FASEB J. 18, 158–160 [DOI] [PubMed] [Google Scholar]
- 15. Bandyopadhyay S., Wang Y., Zhan R., Pai S. K., Watabe M., Iiizumi M., Furuta E., Mohinta S., Liu W., Hirota S., Hosobe S., Tsukada T., Miura K., Takano Y., Saito K., Commes T., Piquemal D., Hai T., Watabe K. (2006) Cancer Res. 66, 11983–11990 [DOI] [PubMed] [Google Scholar]
- 16. Dong J. T., Suzuki H., Pin S. S., Bova G. S., Schalken J. A., Isaacs W. B., Barrett J. C., Isaacs J. T. (1996) Cancer Res. 56, 4387–4390 [PubMed] [Google Scholar]
- 17. Kawana Y., Komiya A., Ueda T., Nihei N., Kuramochi H., Suzuki H., Yatani R., Imai T., Dong J. T., Imai T., Yoshie O., Barrett J. C., Isaacs J. T., Shimazaki J., Ito H., Ichikawa T. (1997) Prostate 32, 205–213 [DOI] [PubMed] [Google Scholar]
- 18. Tagawa K., Arihiro K., Takeshima Y., Hiyama E., Yamasaki M., Inai K. (1999) Jpn. J. Cancer Res. 90, 970–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jackson P, Millar D, Kingsley E, Yardley G., Ow K., Clark S., Russell P. J. (2000) Cancer Lett. 157, 169–176 [DOI] [PubMed] [Google Scholar]
- 20. Miyazaki T., Kato H., Shitara Y., Yoshikawa M., Tajima K., Masuda N., Shouji H., Tsukada K., Nakajima T, Kuwano H. (2000) Cancer 89, 955–962 [DOI] [PubMed] [Google Scholar]
- 21. Mashimo T., Watabe M., Hirota S., Hosobe S., Miura K., Tegtmeyer P. J., Rinker-Shaeffer C. W., Watabe K. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 11307–11311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mashimo T., Bandyopadhyay S., Goodarzi G., Watabe M., Pai S. K., Gross S. C., Watabe K. (2000) Biochem. Biophys. Res. Commun. 274, 370–376 [DOI] [PubMed] [Google Scholar]
- 23. Telese F., Bruni P., Donizetti A., Gianni D., D'Ambrosio C., Scaloni A, Zambrano N, Rosenfeld M. G., Russo T. (2005) EMBO Rep. 6, 77–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Baek S. H., Ohgi K. A., Rose D. W., Koo E. H., Glass C. K., Rosenfeld M. G. (2002) Cell 110, 55–67 [DOI] [PubMed] [Google Scholar]
- 25. Kim J. H., Kim B., Cai L., Choi H. J., Ohgi K. A., Tran C., Chen C., Chung C. H., Huber O., Rose D. W., Sawyers C. L., Rosenfeld M. G., Baek S. H. (2005) Nature 434, 921–926 [DOI] [PubMed] [Google Scholar]
- 26. Dong J. T., Isaacs W. B., Barrett J. C., Isaacs J. T. (1997) Genomics 41, 25–32 [DOI] [PubMed] [Google Scholar]
- 27. Marreiros A., Dudgeon K., Dao V., Grimm M. O., Czolij R., Crossley M., Jackson P. (2005) Oncogene 24, 637–649 [DOI] [PubMed] [Google Scholar]
- 28. Marreiros A., Czolij R., Yardley G., Crossley M., Jackson P. (2003) Gene 302, 155–164 [DOI] [PubMed] [Google Scholar]
- 29. van Belzen N., Dinjens W. N., Diesveld M. P., Groen N. A., van der Made A. C., Nozawa Y., Vlietstra R., Trapman J., Bosman F. T. (1997) Lab. Invest. 77, 85–92 [PubMed] [Google Scholar]
- 30. Kovacevic Z., Richardson D. R. (2006) Carcinogenesis 27, 2355–2366 [DOI] [PubMed] [Google Scholar]
- 31. Ellen T. P., Ke Q., Zhang P., Costa M. (2008) Carcinogenesis 29, 2–8 [DOI] [PubMed] [Google Scholar]
- 32. Iiizumi M., Liu W., Pai S.K., Furuta E., Watabe K. (2008) Biochim. Biophys. Acta 1786, 87–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bandyopadhyay S., Pai S. K., Gross S. C., Hirota S., Hosobe S., Miura K., Saito K., Commes T., Hayashi S., Watabe M., Watabe K. (2003) Cancer Res. 63, 1731–1736 [PubMed] [Google Scholar]
- 34. Guan R. J., Ford H. L., Fu Y., Li Y., Shaw L. M., Pardee A. B. (2000) Cancer Res. 60, 749–755 [PubMed] [Google Scholar]
- 35. Kurdistani S. K., Arizti P., Reimer C. L., Sugrue M. M., Aaronson S. A., Lee S. W. (1998) Cancer Res. 58, 4439–4444 [PubMed] [Google Scholar]
- 36. Maruyama Y., Ono M., Kawahara A., Yokoyama T., Basaki Y., Kage M., Aoyagi S., Kinoshita H., Kuwano M. (2006) Cancer Res. 66, 6233–6242 [DOI] [PubMed] [Google Scholar]
- 37. Nishio S., Ushijima K., Tsuda N., Takemoto S., Kawano K., Yamaguchi T., Nishida N., Kakuma T., Tsuda H., Kasamatsu T., Sasajima Y., Kage M., Kuwano M, Kamura T. (2008) Cancer Lett. 264, 36–43 [DOI] [PubMed] [Google Scholar]
- 38. Hosoi F., Izumi H., Kawahara A., Murakami Y., Kinoshita H., Kage M., Nishio K., Kohno K., Kuwano M., Ono M. (2009) Cancer Res. 69, 4983–4991 [DOI] [PubMed] [Google Scholar]
- 39. Xu X., Sutak R., Richardson D. R. (2008) Mol. Pharmacol. 73, 833–844 [DOI] [PubMed] [Google Scholar]
- 40. Ishiguro T., Nakajima M., Naito M., Muto T., Tsuruo T. (1996) Cancer Res. 56, 875–879 [PubMed] [Google Scholar]
- 41. Ishiguro T., Nagawa H., Naito M., Tsuruo T. (2000) Jpn. J. Cancer Res. 91, 833–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hai T., Wolfgang C. D., Marsee D. K., Allen A. E., Sivaprasad U. (1999) Gene Expr. 7, 321–335 [PMC free article] [PubMed] [Google Scholar]
- 43. Lee J. H., Seo Y. W., Park S. R., Kim Y. J., Kim K. K. (2003) Cancer Res. 63, 7247–7255 [PubMed] [Google Scholar]
- 44. Luo J. L., Tan W., Ricono J. M., Korchynskyi O., Zhang M., Gonias S. L., Cheresh D. A., Karin M. (2007) Nature 446, 690–694 [DOI] [PubMed] [Google Scholar]
- 45. Konishi N., Shimada K., Nakamura M., Ishida E., Ota I., Tanaka N., Fujimoto K. (2008) Clin. Cancer Res. 14, 4408–4416 [DOI] [PubMed] [Google Scholar]
- 46. Lakshman M., Huang X., Ananthanarayanan V., Jovanovic B., Liu Y., Craft C. S., Romero D., Vary C. P., Bergan R. C. (2011) Clin. Exp. Metastasis 28, 39–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Suh J., Rabson A. B. (2004) J. Cell. Biochem. 91, 100–117 [DOI] [PubMed] [Google Scholar]
- 48. Ross J. S., Kallakury B. V., Sheehan C. E., Fisher H. A., Kaufman R. P., Jr., Kaur P., Gray K., Stringer B. (2004) Clin. Cancer Res. 10, 2466–2472 [DOI] [PubMed] [Google Scholar]
- 49. Jin R. J., Lho Y., Connelly L., Wang Y., Yu X., Saint Jean L., Case T. C., Ellwood-Yen K, Sawyers C. L., Bhowmick N. A., Blackwell T. S., Yull F. E., Matusik R. J. (2008) Cancer Res. 68, 6762–6769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhang Q., Helfand B. T., Jang T. L., Zhu L. J., Chen L., Yang X. J., Kozlowski J., Smith N., Kundu S. D., Yang G., Raji A. A., Javonovic B., Pins M., Lindholm P., Guo Y., Catalona W. J., Lee C. (2009) Clin. Cancer Res. 15, 3557–3567 [DOI] [PubMed] [Google Scholar]
- 51. El Touny L. H., Banerjee P. P. (2007) Biochem. Biophys. Res. Commun. 361, 169–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Neve R. M., Chin K., Fridlyand J., Yeh J., Baehner F. L., Fevr T., Clark L., Bayani N., Coppe J. P., Tong F., Speed T., Spellman P. T., DeVries S., Lapuk A, Wang N. J., Kuo W. L., Stilwell J. L., Pinkel D., Albertson D. G., Waldman F. M., McCormick F., Dickson R. B., Johnson M. D., Lippman M., Ethier S., Gazdar A., Gray J. W. (2006) Cancer Cell 10, 515–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Gilchrist M., Thorsson V., Li B., Rust A. G., Korb M., Roach J. C., Kennedy K., Hai T., Bolouri H., Aderem A. (2006) Nature 441, 173–178 [DOI] [PubMed] [Google Scholar]
- 54. Miyazaki K., Inoue S., Yamada K., Watanabe M., Liu Q., Watanabe T., Adachi M. T., Tanaka Y., Kitajima S. (2009) Nucleic Acids Res. 37, 1438–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.