Abstract
To better investigate the relationship between sequence, stability, and folding, the Streptococcus pyogenes collagenous domain CL (Gly-Xaa-Yaa)79 was divided to create three recombinant triple helix subdomains A, B, and C of almost equal size with distinctive amino acid features: an A domain high in polar residues, a B domain containing the highest concentration of Pro residues, and a very highly charged C domain. Each segment was expressed as a monomer, a linear dimer, and a linear trimer fused with the trimerization domain (V domain) in Escherichia coli. All recombinant proteins studied formed stable triple helical structures, but the stability varied depending on the amino acid sequence in the A, B, and C segments and increased as the triple helix got longer. V-AAA was found to melt at a much lower temperature (31.0 °C) than V-ABC (V-CL), whereas V-BBB melted at almost the same temperature (∼36–37 °C). When heat-denatured, the V domain enhanced refolding for all of the constructs; however, the folding rate was affected by their amino acid sequences and became reduced for longer constructs. The folding rates of all the other constructs were lower than that of the natural V-ABC protein. Amino acid substitution mutations at all Pro residues in the C fragment dramatically decreased stability but increased the folding rate. These results indicate that the thermostability of the bacterial collagen is dominated by the most stable domain in the same manner as found with eukaryotic collagens.
Keywords: Bacteria, Collagen, Protein Conformation, Protein Folding, Protein Stability, Streptococcus Pyogenes, Triple Helix
Introduction
The collagen triple helix is composed of three left-handed polyproline helices twisted into a right-handed supercoiled structure (1–4). The close packing of the three chains requires every third residue be a Gly, generating the typical (Gly-Xaa-Yaa)n repeating sequence pattern. Triple helix stability is also dependent on a high content of imino acids Pro and hydroxyproline (Hyp)3 and interchain backbone hydrogen bonds. The amino acids occupying the Xaa and Yaa positions of the (Gly-Xaa-Yaa)n sequence are biologically important, forming binding and degradation sites, as well as modulating global and local stability of the triple helix. Host-guest model peptides have been used to determine the triple helix propensity of individual residues and of pairs of residues (3). Here, the relation between amino acid sequence and triple helix stability and folding is investigated through the use of a recombinant bacterial collagen system, where it is possible to manipulate triple helix sequence and length and to make large amounts of protein for biophysical studies.
Collagen is an important structural protein in the extracellular matrix of animals, and collagen-like sequences have now been identified in prokaryotic genomes (5). Recombinant bacterial collagen-like proteins have been shown to adopt a triple helix structure, with a thermal stability similar to that seen for human collagens (6–10). Bacteria lack the prolyl hydroxylase enzyme necessary for post-translational modification of Pro to Hyp, and in the absence of Hyp, alternative strategies to stabilize the triple helix must be present, involving electrostatic interactions, polar residues, or high Pro content (5, 6). One well characterized prokaryotic collagen is Scl2 (Streptococcus pyogenes collagen-like protein 2), a cell surface protein that may help pathogenic bacteria adhere to animal cells (12, 13). The Scl2 protein contains an N-terminal globular V domain, a collagen-like CL domain, and a transmembrane domain. The Scl2.28 (Scl2 protein from serotype M28) derived protein, containing only the V and CL domains, was successfully purified from a high yield expression system in Escherichia coli (7–9). The S. pyogenes V-CL protein forms a stable triple helix with Tm = ∼36 °C, and the collagen domain alone has the same thermal stability (7–9). The N-terminal globular V domain is an α-helix containing protein that forms trimers (14), and the V domain was shown to be essential for triple helix refolding (7, 9, 14).
The S. pyogenes collagenous domain CL consists of 79 Gly-Xaa-Yaa tripeptides. The distribution of charged residues, Pro, and polar residues is not even throughout the CL domain. The CL domain can be divided into three subdomains (A, B, and C) of almost equal size with distinctive amino acid features. We hypothesize that each of these unique domains may play a critical role in the folding of the CL domain and its overall stability. The C-terminal third of CL contains the highest concentration of charges, (31 charges/78 residues, ∼38.3%), whereas the N-terminal region has the least charge, and the central region has the most Pro (supplemental Table 1). These subdomains were expressed individually in E. coli fused with the N-terminal V domain to promote trimerization and folding. In addition, each subdomain was tandemly fused to construct a homodimer (AA, BB, or CC) or a homotrimer (AAA, BBB, or CCC) again appended to the N-terminal V domain. All recombinant proteins studied formed stable triple helical structures; however, their stabilities were quite different dependent upon their amino acid compositions and lengths. The V domain promoted refolding in all segments, but the folding rate depended on sequence and length and was always less than the natural V-CL protein.
EXPERIMENTAL PROCEDURES
Construction and Expression of Recombinant Collagen Proteins
All of the constructs were obtained by inserting the PCR products into pCold II vector (15) which has an N-terminal His6 tag, using the NdeI and BamHI restriction sites. The DNA fragment of V-CΔP was synthesized by Genscript, Inc. (Piscataway, NJ) and cloned into pCold II vector using NdeI and BamHI sites.
All of the constructs were confirmed by DNA sequencing and then transformed into an E. coli BL21 strain. Cells were shaken in 1 ml of M9-casamino acid medium with 50 μg/ml ampicillin at 37 °C for 5 h. 1 mm isopropyl 1-thio-β-d-galactopyranoside was added to induce protein expression, and the cells were then shaken at room temperature. After 16 h, 1-ml cultures were centrifuged at 4 °C, and the cell pellets thus obtained were resuspended in 100 μl of lysis buffer (20 mm NaPO4 + 500 mm NaCl, pH 7.4), followed by four cycles of sonication (30 s on and 30 s off). Subsequently, the lysates were centrifuged at 13,000 rpm at 4 °C to separate soluble and insoluble proteins. The pellet fractions were resuspended in 100 μl of lysis buffer. Both the soluble and insoluble fractions were analyzed by SDS-PAGE (data not shown). If the expressed protein was present mostly in the soluble fractions, further purification of the proteins was carried out as described below. Because V-CC and V-CCC were present only in the insoluble fractions, they were not further purified.
Purification of His-tagged Proteins
A single colony of E. coli BL21 cells containing the corresponding plasmid was inoculated into 5 ml of M9-casamino acid medium with 50 μg/ml ampicillin. The culture was shaken at 37 °C for 16 h and transferred into a 250-ml M9-casamino acid medium, which was then shaken at 37 °C until A600 reached 0.8. 1 mm isopropyl 1-thio-β-d-galactopyranoside was added to induce protein expression, and the cultures were shaken at room temperature. After 16 h, the cells were collected by centrifugation at 11,500 rpm at 4 °C and resuspended in 25 ml of lysis buffer, followed by cell disruption using a French press. After ultracentrifugation at 45,000 rpm at 4 °C to remove insoluble materials, the supernatant was applied to a nickel-nitrilotriacetic acid-agarose column (25 ml) at room temperature. After washing the column with wash buffer (20 mm NaPO4, 500 mm NaCl, 20 mm imidazole, pH 7.4), elution buffers with increasing concentrations of imidazole (50, 100, 125, and 400 mm) were used for stepwise elution of proteins. The protein purity was examined by SDS-PAGE, and the concentration was determined by absorbance at 280 nm with ϵ = 9129 m−1·cm−1.
CD Spectroscopy
CD data were obtained using an AVIV Model 62DS spectropolarimeter (Aviv Associates, Inc., Lakewood, NJ). Before CD scanning, proteins were kept in 1-mm cuvettes at 4 °C for at least 24 h. CD spectra were collected from 260 nm to 195 nm at a 0.5-nm interval with an averaging time of 5 s, and each scan was repeated three times. Protein melting was examined at 220 nm with increasing temperatures, from 0 to 60 °C, in 0.33 °C steps. Proteins were equilibrated at each temperature point for 2 min, and the temperature was increased with an average rate of 0.1 °C/min. Tm was obtained by taking the peak of the first derivative of the melting curve. The thermal transition was observed at both neutral pH (PBS buffer, 20 mm NaPO4, 150 mm NaCl) and at pH 2.8 (0.1 m acetic acid).
After incubation of the samples at 65 °C for 20 min to denature the sample, protein refolding was observed at 220 nm at 0 °C for 60,000 s. The half-time was defined as the time point when the fraction folded reached 0.5. The percentage of refolding was defined as the ratio of the CD signal at 0 °C after refolding to the initial CD signal before melting.
DSC
Differential scanning calorimetry (DSC) data were obtained using a NANO DSC II Model 6100 (Calorimetry Sciences Corp., Provo, UT). Before DSC scanning, each protein was dialyzed against PBS buffer (pH 7.0) and equilibrated at 4 °C for 24 h. The dialysis buffer was used for the baseline scan. Protein scanning was carried out from 0 to 80 °C with an increasing rate of 1 °C/min.
Trypsin Digestion
20 μm purified samples in PBS buffers were incubated with 0.01 mg/ml trypsin at 15 °C for 10 min. The reaction was stopped by adding 1 mm PMSF, and then the samples were run on an SDS-PAGE. For the trypsin digestion on cell lysate, 0.005 mg/ml trypsin was added to the cell lysate, and a time course was recorded, with time equals 0, 1, 3, 7, and 24 h.
RESULTS
Subfragmentation of CL Triple Helix Domain
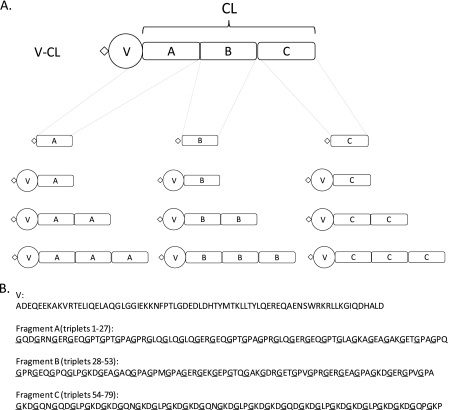
The collagenous domain (CL) of S. pyogenes Scl2 protein is highly charged (30% residues), with 12% Pro, 13% polar residues, 6% Ala, and 6% larger hydrophobic residues, but without Cys, Tyr, and Phe residues. It is an acidic protein with a predicted pI value of 5.19. As shown in Fig. 1A, this CL domain can be dissected into three unique fragments of approximately equal lengths namely on the basis of their amino acid compositions: the N-terminal 81 residues (fragment A consisting of 27 tripeptides), the central 78 residues (fragment B, 26 tripeptides), and the C-terminal 78 residues (fragment C, 26 tripeptides). Fragment A has the highest content of polar residues, fragment B the highest Pro content, and fragment C the highest charged residue content (Fig. 1B and supplemental Table 1). Each fragment contains more acidic than basic residues so that the pI value of each fragment is between 5.1 and 5.3, similar to that of the complete CL domain. Notably, fragment C contains no Arg or Glu, but only Lys and Asp with a repetitive sequence of GKDGQNGKDGLPGKD. To study the length effect of the triple helices, constructs with one, two, and three tandem repeats of each fragment were designed (Fig. 1A). For all constructs, the V trimerization domain was added at the N-terminal end.
FIGURE 1.
A, schematic diagram of the constructs including fragments of the original S. pyogenes Scl2 CL domain, showing the rod-like triple helix domain, the globular V domain, and a small diamond to represent the translation enhancing element sequence, from pCold vector (15) and His6 tag at the N terminus. Between the V and collagen domains, there is a linker with a sequence of LVPRGSP (9). B, amino acid sequences of the V domain and the individual A, B, and C fragments, with Gly residues underlined to highlight the repeating (Gly-Xaa-Yaa)n nature in the triple helix segments.
All of the proteins were expressed in E. coli at room temperature using a cold-shock vector, pCold II, as described previously (15). The V-CC and V-CCC proteins were expressed well but were not soluble. Therefore, they were not further studied. All the soluble proteins were purified on a nickel-agarose column and analyzed by SDS-PAGE (supplemental Fig. 3).
Conformation of Collagen Constructs
CD spectra of the A, B, and C constructs, recorded from 260 to 195 nm, show typical triple helical spectra with a positive peak at 220 nm (MRE220 nm ∼ +8000 deg·cm·dmol−1) and a minimum at 198 nm (MRE198 nm ∼ −60,000 deg·cm·dmol−1) (Fig. 2A). The Rpn values (ratio of the value of positive peak to that of the negative peak) of the A, B, and C collagen fragments (0.12–0.13) are comparable with those seen for mammalian collagens and model peptides, suggesting that they form complete triple helical molecules (16).
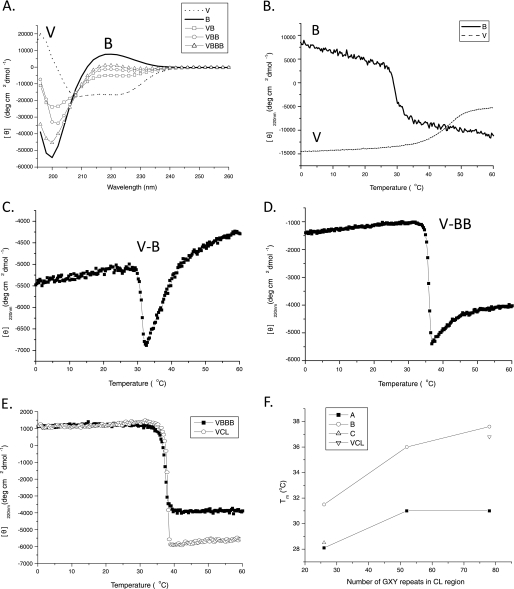
FIGURE 2.
A, circular dichroism spectra of the V domain (dashed line), the B fragment (solid line), V-B (square), V-BB (circle), and V-BBB (triangle). B, CD melting transition of the B fragment (solid line) and the transition of the isolated V domain (dashed line). C, CD melting transition of V-B. D, CD melting transition of V-BB. E, CD melting transition of V-BBB (black squares), compared with V-CL (circles). F, a plot of the melting temperature Tm of the VA, VAA, VAAA (black squares), V-B, V-BB, V-BBB (circles), and V-C (triangles) as a function of the number of (Gly-Xaa-Yaa)n units, with the value for the full length V-CL (reversed triangle).
Previous studies showed that the V domain has a typical α-helical CD spectrum, with Tm = 45 °C (14). The CD spectra of the A, B, and C constructs with the V domain include opposing contributions from the α-helix structure of the V domain (MRE222 nm = −20,100 deg·cm·dmol−1; MRE208 nm = −21,700 deg·cm·dmol−1; MRE197 nm = +24,300 deg·cm·dmol−1) and the triple helix structure (MRE220 nm = +8000 deg·cm·dmol−1; MRE198 nm = −60,000 deg·cm·dmol−1). Note that the net spectrum depends on the relative fraction of residues in each domain; there are 74 residues in the V domain, whereas there are 81 residues in A and 78 residues in B and C. For the V-B spectrum, there is a net negative MRE220 = −5000 deg·cm·dmol−1 and MRE198 = −27,000 deg·cm·dmol−1 (Fig. 2A). As the proportion of triple helical structures relative to α-helix structures increases in the longer tandem constructs, the peak at 220 nm gets higher. The MRE value of V-BBB at 220 nm is almost the same as that of the full-length V-CL molecule (Fig. 2A) (9). The CD spectra of the molecules derived from fragment A (A, V-A, V-AA, and V-AAA) or fragment C (C, V-C) showed patterns similar to those described above for fragment B (data not shown), suggesting that all three fragments are capable of forming a triple helical structure connected to the α-helical V domain and that the V and triple helix domains contribute additively to their CD spectra.
Thermal Stability
Thermal stabilities of all of the constructs were determined by monitoring the intensity at the triple helix maximum, MRE220 nm, with increasing temperature at a rate of 0.1 °C/min. The thermal transition of fragment B occurs at Tm = 29.5 °C, a value lower than the 36.0 °C observed for the full-length CL domain, and the B transition is broader than seen for CL (Fig. 2B) (9). In addition, the CD signal of B has a sloping baseline before the major transition. For V-B, in which both V and B domains consist of similar residue numbers (74 and 78 residues, respectively), the observed melting profile shows a combination of decreasing MRE220 nm values due to triple helix unfolding and increasing MRE222 nm values due to the unfolding of the α-helix containing V domain (Fig. 2C). The first derivative of the V-B thermal transition shows a single peak at 31.5 °C, a value 2 °C higher than the Tm of B alone and lower than the value observed for the V domain alone (Tm = 45.0 °C). The Tm values increase to 36.0 °C and 37.6 °C for V-BB and V-BBB, respectively, as the length of the collagen domain increases (Fig. 2, D and E). It is noteworthy that the melting temperatures of V-BB and V-BBB reach the value reported for the full-length molecule V-CL (9). The melting of the V domain becomes less visible as the residue proportion of the α-helix in the V domain to the triple helix in the collagen domains decreases.
The stability of fragment A is Tm = 23.5 °C, showing that it is less stable than fragment B of the same length (Tm = 29.5 °C). The Tm value is higher for V-A (28.1 °C) and increases further for V-AA (Tm = 31.0 °C) and V-AAA (Tm = 31.0 °C) (supplemental Fig. 1, A–D). These tandem A constructs are less stable than the V-BB and V-BBB constructs, respectively, and the Tm values of V-AA and V-AAA never reach the V-CL Tm value. The relative proportion of the α-helix structure in the V domain to the length of the triple helix in the collagen domains again affects the shape of the transitions. In all cases, the proteins with the B domains are more stable than those with the A domains (Table 1).
TABLE 1.
Thermal stability of the constructs containing fragments of the bacterial CL domain, as determined by CD spectroscopy
| Monomer | V-Monomer | V-Dimer | V-Trimer | |
|---|---|---|---|---|
| Fragment A | 23.5 °C | 28.1 °Ca | 31.0 °C | 31.0 °C |
| Fragment B | 29.5 °C | 31.5 °C | 36.0 °C | 37.6 °C |
| Fragment C | 25.3 °C | 28.5 °C | N/Ab | N/Ab |
a A small transition at about 12.5 °C was also observed, which may reflect a small amount of misfolded, less stable species.
b Thermal stabilities of V-CC and V-CCC were not determined because they were insoluble.
The major thermal transition of fragment C is ∼ 25.3 °C, followed by a second transition near 36.5 °C (supplemental Fig. 1E); the higher transition may be due to the aggregation because visible aggregates were observed after incubation for a few days. The melting temperature of V-C is 28.5 °C, similar to that of V-A and lower than that of V-B (supplemental Fig. 1F). The melting profiles for V-CC and V-CCC were not studied due to insolubility. The melting temperatures of V-A, V-AA, V-AAA, V-B, V-BB, V-BBB, V-C, and full-length V-CL were plotted against the numbers of (Gly-Xaa-Yaa)n tripeptides (Fig. 2F), and the values were shown in Table 1.
Thermal transitions of V-A, V-B and V-C also were examined by DSC (supplemental Fig. 2). Only one major transition is seen for each sample, with Tm = 29.4, 32.4, and 29.4 °C, for V-A, V-B, and V-C, respectively. These values are slightly higher than the Tm values obtained by CD likely due to the faster DSC heating rate and the nonequilibrium conditions (17). The DSC profile of V-A also shows a small peak at 12.5 °C, consistent with the small transition observed on CD (supplemental Fig. 1B), which may reflect a small amount of a misfolded, less stable species in the protein preparations.
Because native triple helices are resistant to trypsin cleavage, the protein constructs were digested with trypsin to assess the integrity and stability of their triple helices (supplemental Fig. 3). All fragments have many potential trypsin sensitive sites (supplemental Table 1). Trypsin digestion of both purified fragments and cell lysates suggests that fragment A is the most susceptible to trypsin (supplemental Fig. 3, A and E), although it has the least number of trypsin-sensitive sites (eight sites, supplemental Table 1), suggesting this may form a looser triple helical structure. Fragments B and C are more resistant to trypsin, although there are even more trypsin-sensitive sites in the sequence, indicating a tighter triple helical structure. The proteins with two and three tandem repeating fragments (V-AA, V-AAA, V-BB, and V-BBB) were more resistant to trypsin than their corresponding smaller constructs, but all were still more susceptible to tryptic digestion than the natural V-CL protein (supplemental Fig. 3, B–D). The results indicate that the shorter constructs are more susceptible to trypsin but, intriguingly, that the V-AAA and V-BBB constructs, having the same length as V-CL, are still more trypsin susceptible than V-CL.
Stability of Proteins in Acid pH
The stability of all of the constructs was examined by CD at low pH (0.1 m acetic acid, pH 2.8) to evaluate how electrostatic interactions play roles in stabilization of the highly charged triple helix domains: V-A (21.8% charged residues), V-B (28.2%), and V-C (38.3%). It was reported previously that the V domain is denatured in acid (14) and that the triple helix CL domain shows a decreased stability with two thermal transitions (Tm at 24 and 27 °C) compared with PBS (8). The melting temperatures of all of the A, B, and C and V-A, V-B, and V-C proteins showed one thermal transition at low pH, which were lower than their respective values at neutral pH (Table 2). Fragment A, with the least amount of charges (21.8%), showed the smallest drop in Tm (ΔTm = −0.6 °C) compared with B (28.2% charged residues; ΔTm = −10.6 °C) and C (38.3% charged residues; ΔTm = −12.8 °C). The V-A protein showed a smaller decrease, ΔTm = −4.9 °C in comparison with V-B (ΔTm = −9.1 °C) and V-C (ΔTm = −14.1 °C). The decrease in thermal stability appears to correlate with the percentage of charged residues in the A, B, and C fragments, suggesting that at neutral pH, electrostatic interactions play a significant role in protein stability.
TABLE 2.
Comparison of the thermal stabilities at neutral and acidic pH, as determined by CD spectroscopy
|
Tm |
Charged residues in the collagenous domain | ||
|---|---|---|---|
| pH 7.0 | pH 2.8 | ||
| °C | % | ||
| A | 23.5 | 22.9 | 21.8 |
| V-A | 28.1 | 23.2 | 21.8 |
| B | 29.5 | 18.9 | 28.2 |
| V-B | 31.5 | 22.4 | 28.2 |
| V-CL | 35.6 | 24.2a | 29.5 |
| C | 25.3 | 12.5 | 38.3 |
| V-C | 28.5 | 14.4 | 38.3 |
a For V-CL in acidic pH, two thermal transitions were measured by DSC, 23.7 °C and 27.0 °C (8).
Refolding of Triple Helix
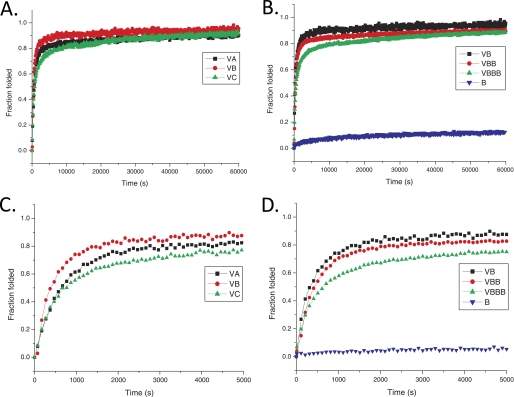
Refolding of all the fragments was examined by monitoring CD spectra at MRE220 nm after heating samples at 50 °C for 20 min followed by cooling to 0 °C. Fragments A, B, and C did not refold, consistent with previous observations that the CL domain cannot refold in the absence of the V domain. When the V domain was fused to these fragments, both V domain α-helices and the CL triple helix structure influence the CD signal at 220 nm. Studies on the isolated V domain showed that it trimerizes and is able to fold very quickly within 1 to 2 min required for cooling the sample from 50 to 0 °C.4 We assume that the V domain may be completely refolded within the first 1 min of cooling time and that refolding to the triple helical structure starts at 0 °C. Because the kinetics shown in Fig. 3 did not fit the first, second, or third order reactions for any samples, the half-time for folding (t½) was used to compare the folding rate for each sample. Protein V-B has the fastest folding rate (t½ ∼ 330 s), followed by V-A (t½ ∼ 640 s), and then V-C (t½ ∼ 770 s) (Fig. 3, A and C). After 16 h, the CD signals for V-B, V-A, and V-C reach 95.6, 90.4, and 92.6% of their individual original signals, respectively, and after 3 days of incubation at 0 °C, the CD signals of all constructs containing the V domain reached their original signal values.
FIGURE 3.
A, folding of V-A (black squares), V-B (red circles), and V-C (green triangles), showing the effect of sequence. B, folding of B (blue reversed triangles), V-B (black squares), V-BB (red circles), V-BBB (green triangles), showing the effect of length; an expanded scale is presented in C and D, which shows only the first 5000 s.
Increasing length of the triple helix led to slower folding (Table 3). For V-B, V-BB, and V-BBB, the half-time increased as the length of the collagen domain increased (330, 410, and 700 s, respectively) (Fig. 3, B and D). The final refolding efficiency after 16 h also dropped (90.0% for V-BB and 89.2% for V-BBB) as the number of collagen repeats increased, which might be due to more misfolding of proteins with increasing triple helix length. Comparison of refolding of V-BBB with that of V-CL shows that the tandem repeating construct (V-BBB) folded more slowly than V-CL, even if both have the same length of collagen domain (t½ ∼ 351 s for V-CL versus 700 s for V-BBB). Similarly, for the V-A, V-AA, and V-AAA constructs, the folding rates and the final refolding percentage decreased as the lengths increased. Although V-A folded significantly slower than V-B, the folding of V-AAA was only slightly slower than that of V-BBB.
TABLE 3.
Refolding of the bacterial collagen fragments showing the half-times of folding and the final percentages of structures recovered as determined by CD spectroscopy
| V-Monomer | V-Dimer | V-Trimer | |
|---|---|---|---|
| Fragment A | 640 s/90.4% | 670 s/85.0% | 760 s/85.0% |
| Fragment B | 330 s/95.6% | 410 s/90.0% | 700 s/89.2% |
| Fragment C | 770 s/92.6% | N/Aa | N/Aa |
a Refolding of V-CC and V-CCC was not identified because they were insoluble.
Removal of Pro Residues in V-C Construct
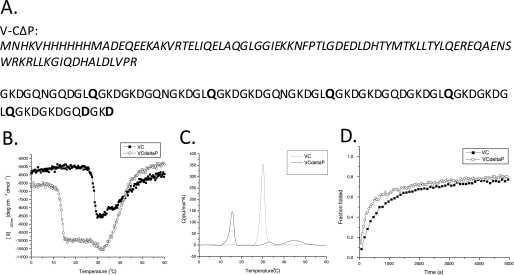
Because imino acids, Pro and Hyp are known to play an important role in stabilizing mammalian collagens, the effect of eliminating Pro residues in fragment C (containing seven Pro residues) on the thermal stability was investigated. Fragment C has the least number of Pro residues compared with fragments A and B (supplemental Table 1). Furthermore, all of these are at the Yaa positions in Gly-Xaa-Yaa triplets. All of these Pro residues are mutated to either Gln or Asp, creating V-CΔP (Fig. 4A). All GLP tripeptides were altered to GLQ, because GLQ is the second most abundant tripeptide after GLP in the Scl2 protein, and the GQPGKP sequence was mutated to GQDGKD, because Asp is the most frequently used amino acid at the Yaa position if the Xaa position is occupied by Gln or Lys in the CL triple helix. The host-guest peptide set in a peptide context predicts that substitution of a Pro residue with Gln in the Yaa position will lower the stability by ∼1.6 °C if Leu is in the Xaa position (3). On the other hand, a Pro-to-Asp substitution may decrease the stability by −3.9 °C if Lys is in the Xaa position and by ∼11.5 °C if Gln is in the Xaa position (3). By CD analysis, the V-CΔP protein showed a melting temperature at 13.9 °C, which is 14.6 °C lower than V-C (Fig. 4B). A second thermal transition with Tm of ∼ 45 °C is also observed, consistent with independent melting of the V domain in this construct.
FIGURE 4.
A, amino acid sequence of the protein constructed from the C fragment, with all Pro residues mutated to other residues, designated the V-CΔP protein. The translation enhancing element sequence, His6 tag, V domain, and the linker sequence are shown in italics. GSP from the linker sequence was removed, and the amino acid residues that replaced Pro were boldface. B, the thermal transition of the V-CΔP (circles) compared with V-C (black squares). C, DSC of V-CΔP (solid lines) compared with V-C (dotted lines). D, folding of V-CΔP (circles) compared with V-C (black squares).
DSC of V-CΔP (Fig. 4C) (1 °C/min) shows three peaks at 15.7, 32.8, and 45.0 °C. The 15.7 °C peak represents the thermal transition of the triple helix domain, which is slightly higher than the transition temperature measured by CD, as expected (17). The 45.0 °C peak is consistent with the thermal transition of V domain, which is the same value as obtained by CD for the isolated V domain. The origin of the small 32.8 °C peak is not clear, but it is not seen on a second DSC scan, suggesting that it might be due to some aggregates, which dissolve by heating and take time to reform. The 32.8 °C peak may represent soluble aggregates that can associate further with time, as visible aggregates were seen after incubation for several days at 4 °C.
Refolding kinetics of V-CΔP was monitored using MRE220 nm. Surprisingly, the Pro-free triple helix protein shows a faster folding rate than that seen for V-C (Fig. 4D). The faster folding in the absence of Pro residues suggests that formation of the triple helix structure in the C domain may have been limited by cis/trans isomerization of Pro residues.
DISCUSSION
The rod-like structure of the collagen triple helix, which precludes long range interactions, in theory enables the dissecting of the full-length molecule into fragments that maintain the original triple helix conformation. The recombinant system using a bacterial collagen protein expressed in E. coli allows manipulation of the sequence. In this study, the CL domain derived from Scl2.28 protein was dissected into three fragments of equal length, and studies were carried out on each fragment with and without the trimerization V domain or in multiple tandem repeats together with the V domain. The CD analyses of these constructs indicate that all the (Gly-Xaa-Yaa)n fragments maintain a triple helix conformation regardless of their sequences and lengths. Attachment of a V domain to the N-terminal end of the triple helix sequences does not affect either the α-helix structure of the V-domain or the triple helix conformation of the collagen domains. The maintenance of the original conformation allows comparisons to determine the effect of amino acid sequence and triple helix length on stability and folding.
Longer Triple Helix Increases Stability
Increasing triple helix length led to increased stability for A and B fragments; however, the stability reached a plateau at ∼50 triplets. Because the proteins studied here contain tandem repeats of A or B fragments, the effect of the lengths of triple helix structures on stability can be compared without any influence from the amino acid sequence. In an earlier study, it had been shown that thermal stability reached a plateau as the length of several natural and engineered S. pyogenes bacterial collagen constructs reached ∼80 triplets (18). Studies on collagen-like peptides (Pro-Hyp-Gly)n and (Pro-Pro-Gly)n also indicate the presence of a maximum for stability as the length increases to >20 tripeptides (3).
Stability Depends on Amino Acid Sequence
Sequence-dependent differences in the thermal stability of the isolated triple helix domains of the same length were observed. The most stable fragment is the central fragment B, with Tm = 29.5 °C, followed by fragment C (25.3 °C) and A (23.5 °C). As the length increases, V-BB and V-BBB reach a plateau at 36.0 °C, whereas V-AA and V-AAA plateau at a lower temperature (31.0 °C). Analysis of the amino acid sequence of fragment B reveals unique features likely contributing to the stability of the fragment; it contains more Pro residues (12) than A (10) or C (7) and also has two highly stabilizing sequences, KGD and KGE. The important contribution of imino acid residues to triple helix stability (19) was confirmed by the observed decrease in stability from 25.3 to 15.0 °C when seven Pro residues in fragment C were mutated to non-imino acid residues. In the case of V-CΔP, the significant drop of Tm was observed in the collagen domain, which shows a thermal transition widely separated from that of the more stable V domain.
Effect of V Domain on Collagen Domain Stability
The V domain was added at the N-terminal end of most of the constructs because this domain has been shown to be essential for triple helix refolding (6, 9, 14). The results here indicate the V domain does not affect the conformation of the triple helix but does increase triple helix stability by 2–5 °C for the V-A, V-B, and V-C proteins. The CD thermal transition profiles of V-A, V-B, and V-C show that the melting of the triple helix occurs first and is followed immediately by the melting of the α-helix in the V domain. The α-helix structure in the V domain unfolds at a significantly lower temperature in V-A, V-B, and V-C constructs than when in an isolated V domain (Tm = 45.0 °C). Although unfolding of both the triple helix in the collagen domain (decreasing MRE220 nm) and α-helix in the V domain (increasing MRE220 nm) can be observed in the heat denaturation experiments, the first derivative of the CD signal or the DSC profile shows only one transition. It appears that unfolding of the triple helix structure triggers subsequent unfolding of the V domain, at a temperature below the Tm of the V domain by itself. Although electron microscopy of V-CL by rotary shadowing shows a small globular domain at the terminus of a rod-like structure (7), suggesting that the V domain and the collagen domain are independent forming modular structures, it appears that as the triple helix starts to unfold, the unfolded chains may have a destabilizing effect on the V domain. In V-CΔP, the transition of the collagen domain occurs at 13.9 °C and the V domain at 45.0 °C, demonstrating that these two domains behave completely independently without apparent interactions. It is possible that interactions may only occur when the triple helix domain stability is closer to that of the V domain.
Electrostatic Interactions Are Important for Stability at Neutral pH
Comparison of thermal stability at acidic and neutral pH shows that electrostatic interactions make a significant contribution to stability in these highly charged triple helix structures. The stabilities at an acidic pH dropped most significantly for fragment C, which has an extremely high content of charged residues (38.3%), including many GKD and GKDGKD sequences, which have been shown to have highly stabilizing effects on the triple helix structures (8). In animal collagens negatively charged residues are mainly at Xaa positions and positively charged residues at Yaa positions; however, it is clear from these studies that positively charged residues in the Xaa position adjacent to negatively charged residues in the Yaa position also can have a highly stabilizing effect on collagen. All V-A, V-B, and V-C constructs show only one transition at acid pH from 14 to 23 °C, whereas the V-CL protein at an acidic pH shows two thermal transitions, at 24 and 27 °C, suggesting there may be some different domain structures.
Triple Helix Folding Is Affected by Sequence and Length
The A, B, and C domains alone are unable to refold after heat denaturation; however, when the V domain was added N-terminal to each fragment, they gained the ability to refold and attain the expected triple helix CD spectra. The requirement for the trimerization V domain to facilitate triple helix formation is reminiscent of the folding of human collagens, which frequently requires a C-terminal trimerization domain (20). Although the V domain is located originally adjacent to the N-terminal A fragment in the V-CL molecule, this V domain is also capable of promoting folding of these much shorter triple helical segments and can do this when adjacent to B and C fragments as well as A. This is consistent with the previous report that the V domain is capable of promoting folding of the heterologous Clostridium perfringens triple helix domain (6). The ability of the V domain to rapidly form trimers may bring the N-terminal ends of the (Gly-Xaa-Yaa)n chains close enough together to entropically favor the triple helix formation. It is likely that this initial triple helix formation at the N-terminal end propagates to the C-terminal end.
Interestingly, the refolding rate observed for the V-B construct is faster than seen for the V-A protein containing the original junction between the globular domain and the triple helix domain. Examination of the sequences near the N terminus of A shows a Pro-rich sequence that could act as a nucleation domain.4 However, the overall higher Pro content and stability of the B region could make this construct more favorable for rapid folding. Surprisingly, the refolding of V-CΔP was even faster than V-C, possibly because the elimination of Pro from the sequence resulted in the reduction of the time required for Pro cis-trans isomerization. Refolding studies also show that the folding rate decreases with increasing lengths of the collagen domain. This length effect could be due to the increased number of cis-trans isomerization events required for triple helix formation as the chain gets longer or to an increased potential for misfolding.
Comparison with Animal Collagen
Manipulation of animal collagen sequences has been reported to investigate the relation between sequence and stability (21, 22) where the authors expressed repetitive constructs of segments of type II collagen, using a mammalian cell expression system. In the present work, we have done similar studies using a bacterial collagen domain where the expression of bacterial collagens with different sequences and lengths is much easier than for collagens in mammalian cells. Unlike animal collagens with the more stable triple helix domains at both ends where they may serve as clamps for folding and stability (11), bacterial collagen Scl2 has its most stable triple helix region in the middle, with less stable regions at both N- and C-terminal ends.
Comparison of the repetitive V-AAA and V-BBB proteins with the original more diverse V-CL (V-ABC) protein of the same length reveals that V-BBB but not V-AAA achieves the same melting temperature as V-CL at ∼36–37 °C. It appears that bacterial collagens follow the rule proposed previously from the study on animal collagens (22), that the thermostability of collagen triple helix depends on the most thermostabile domain. Considering that almost all other bacterial collagens studied so far (6, 10, 18) have melting temperatures between 36 and 38 °C, it is reasonable to speculate that this narrow range of melting temperatures might have been evolutionarily set as bacterial collagens may be important for the interaction of these bacteria with animal hosts. Intriguingly, the fact that V-BBB refolds faster than V-AAA but still refolds more slowly than V-CL (V-ABC), suggests that the V-ABC arrangement in the CL domain might have been selected evolutionarily for the most efficient folding. Further studies of the intrinsic properties of the individual fragments and their arrangement will provide important insights into the biological roles of bacterial collagens and genetic engineering of bacterial collagens for biomaterials.
Supplementary Material
Acknowledgments
We thank Eileen Hwang and Chunying Xu for helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grant GM60048 (to B. B.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1 and Figs. 1–3.
E. Hwang, personal communication.
- Hyp
- hydroxyproline
- DSC
- differential scanning calorimetry
- CL
- collagenous domain
- deg
- degrees.
REFERENCES
- 1. Rich A., Crick F. H. (1961) J. Mol. Biol. 3, 483–506 [DOI] [PubMed] [Google Scholar]
- 2. Bella J., Eaton M., Brodsky B., Berman H. M. (1994) Science 266, 75–81 [DOI] [PubMed] [Google Scholar]
- 3. Persikov A. V., Ramshaw J. A., Brodsky B. (2005) J. Biol. Chem. 280, 19343–19349 [DOI] [PubMed] [Google Scholar]
- 4. Ramachandran G. N. (1967) in Treatise on Collagen (Ramachandran G. N. ed) Vol. 1, pp. 103–183, Academic Press, New York [Google Scholar]
- 5. Rasmussen M., Jacobsson M., Björck L. (2003) J. Biol. Chem. 278, 32313–32316 [DOI] [PubMed] [Google Scholar]
- 6. Xu C., Yu Z., Inouye M., Brodsky B., Mirochnitchenko O. Biomacromolecules 11, 348–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Xu Y., Keene D. R., Bujnicki J. M., Höök M., Lukomski S. (2002) J. Biol. Chem. 277, 27312–27318 [DOI] [PubMed] [Google Scholar]
- 8. Mohs A., Silva T., Yoshida T., Amin R., Lukomski S., Inouye M., Brodsky B. (2007) J. Biol. Chem. 282, 29757–29765 [DOI] [PubMed] [Google Scholar]
- 9. Yoshizumi A., Yu Z., Silva T., Thiagarajan G., Ramshaw J. A., Inouye M., Brodsky B. (2009) Protein Sci. 18, 1241–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Boydston J. A., Chen P., Steichen C. T., Turnbough C. L., Jr. (2005) J. Bacteriol. 187, 5310–5317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Arnold W. V., Fertala A., Sieron A. L., Hattori H., Mechling D., Bächinger H. P., Prockop D. J. (1998) J. Biol. Chem. 273, 31822–31828 [DOI] [PubMed] [Google Scholar]
- 12. Lukomski S., Nakashima K., Abdi I., Cipriano V. J., Shelvin B. J., Graviss E. A., Musser J. M. (2001) Infect Immun 69, 1729–1738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rasmussen M., Björck L. (2001) Mol. Microbiol. 40, 1427–1438 [DOI] [PubMed] [Google Scholar]
- 14. Yu Z., Mirochnitchenko O., Xu C., Yoshizumi A., Brodsky B., Inouye M. (2010) Protein Sci. 19, 775–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Qing G., Ma L. C., Khorchid A., Swapna G. V., Mal T. K., Takayama M. M., Xia B., Phadtare S., Ke H., Acton T., Montelione G. T., Ikura M., Inouye M. (2004) Nat. Biotechnol. 22, 877–882 [DOI] [PubMed] [Google Scholar]
- 16. Feng Y., Melacini G., Taulane J. P., Goodman M. (1996) Biopolymers 39, 859–872 [DOI] [PubMed] [Google Scholar]
- 17. Persikov A. V., Xu Y., Brodsky B. (2004) Protein Sci. 13, 893–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Han R., Zwiefka A., Caswell C. C., Xu Y., Keene D. R., Lukomska E., Zhao Z., Höök M., Lukomski S. (2006) Appl Microbiol. Biotechnol. 72, 109–115 [DOI] [PubMed] [Google Scholar]
- 19. Josse J., Harrington W. F. (1964) J. Mol. Biol. 9, 269–287 [DOI] [PubMed] [Google Scholar]
- 20. Khoshnoodi J., Cartailler J. P., Alvares K., Veis A., Hudson B. G. (2006) J. Biol. Chem. 281, 38117–38121 [DOI] [PubMed] [Google Scholar]
- 21. Sieron A. L., Fertala A., Ala-Kokko L., Prockop D. J. (1993) J. Biol. Chem. 268, 21232–21237 [PubMed] [Google Scholar]
- 22. Steplewski A., Majsterek I., McAdams E., Rucker E., Brittingham R. J., Ito H., Hirai K., Adachi E., Jimenez S. A., Fertala A. (2004) J. Mol. Biol. 338, 989–998 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.